Expression Profiles of Long Non-Coding RNA GAS5 and MicroRNA-222 in Younger AML Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Therapy Protocol
2.2. Growth Arrest-Specific Transcript 5 (GAS5) and MiR-222 Expression Level Analysis
2.3. Statistical Analysis and Definition of Clinical Endpoints
3. Results
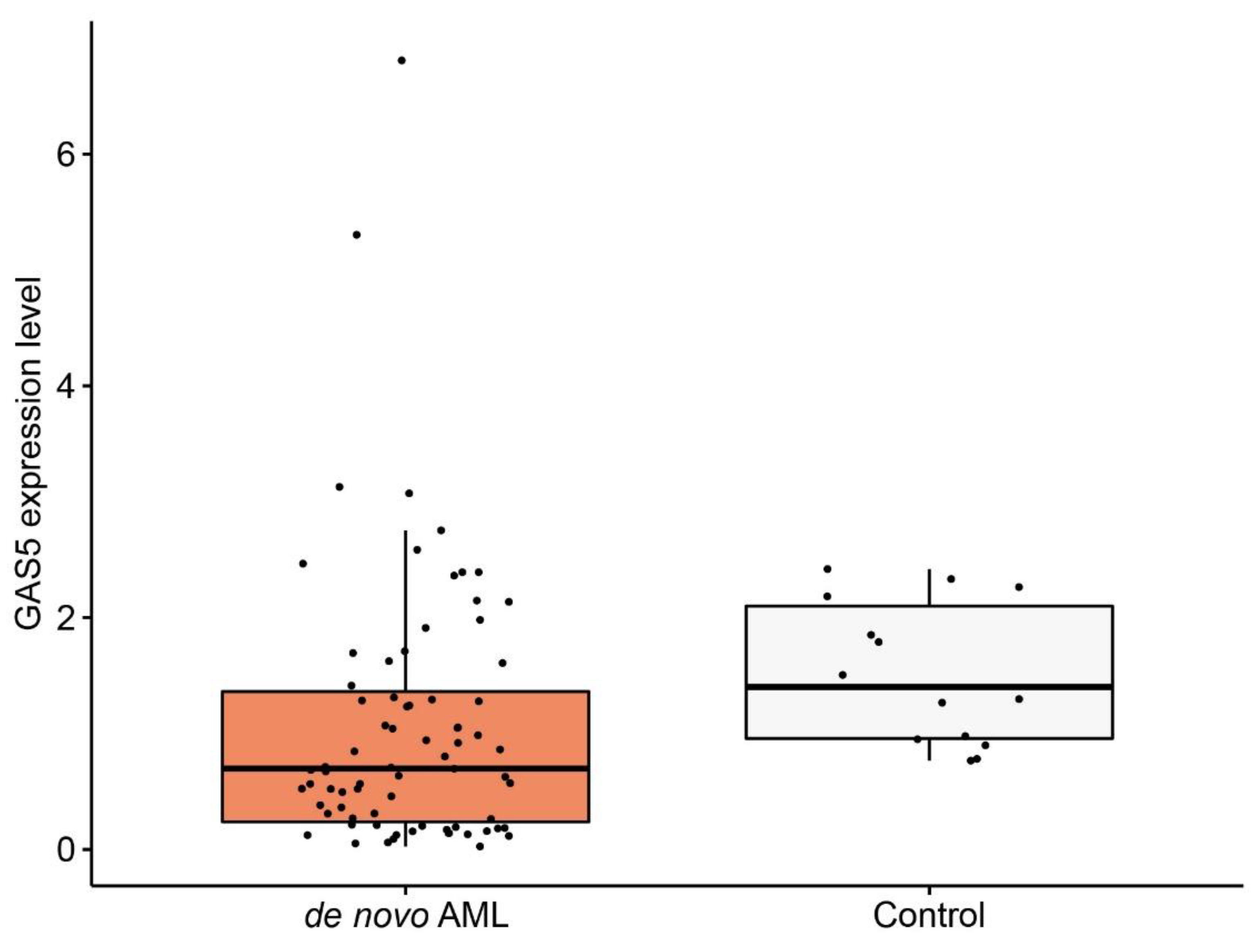
3.1. GAS5 Expression Level in De Novo Acute Myeloid Leukemia (AML) Patients
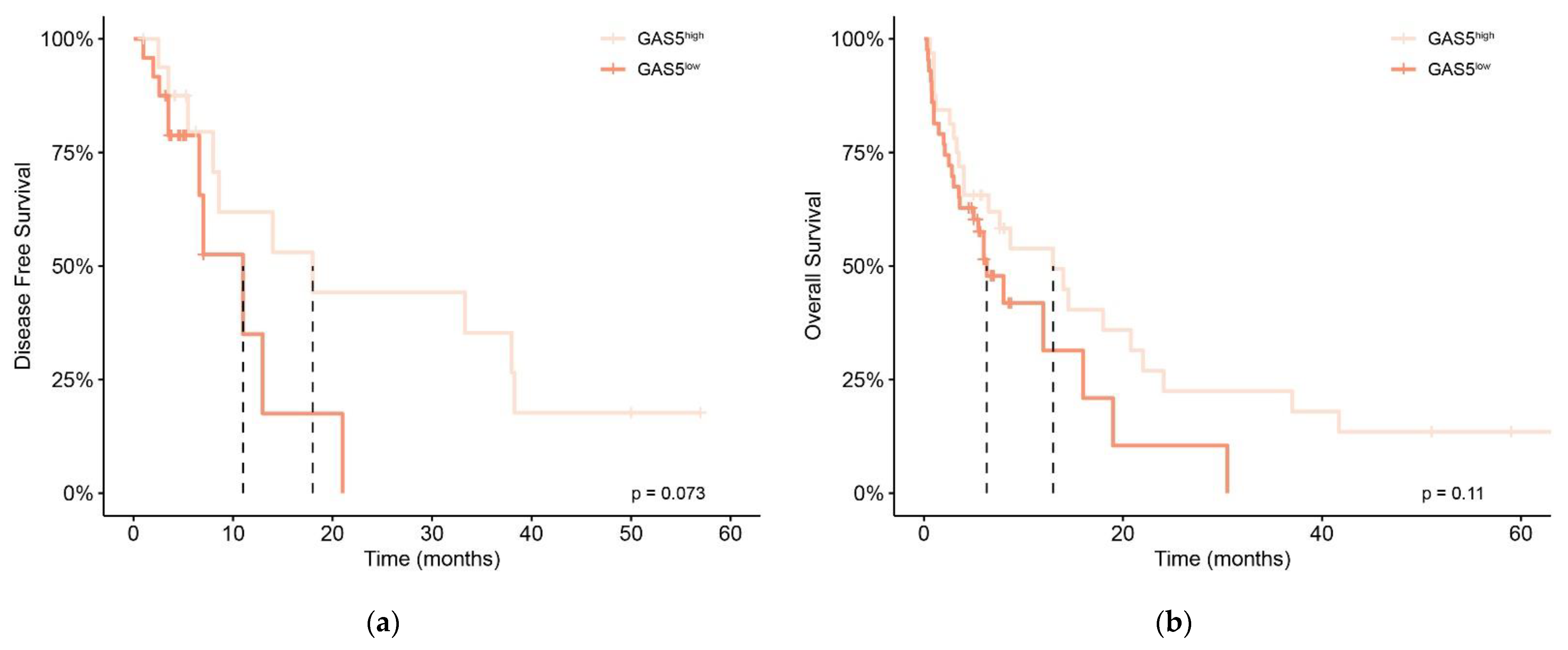
3.2. Prognostic Significance of GAS5 Expression Level in De Novo AML Patients
3.3. GAS5 Expression Level in AML-NK Patients
3.4. Prognostic Significance of MiR-222 Expression Level in AML-NK Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405, Erratum in Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Shi, X.; Zhu, Q.; Li, Q.; Liu, Y.; Yao, Y.; Song, Y. The growth arrest-specific transcript 5 (GAS5): A pivotal tumor suppressor long noncoding RNA in human cancers. Tumor Biol. 2015, 37, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.T.; Mourtada-Maarabouni, M.; Farzaneh, F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem. Soc. Trans. 2011, 39, 482–486. [Google Scholar] [CrossRef] [Green Version]
- Fleming, V.J.; Hay, M.S.; Harries, D.N.; Rees, D.W. Effects of nutrient deprivation and differentiation on the expression of growth-arrest genes (gas and gadd) in F9 embryonal carcinoma cells. Biochem. J. 1998, 330, 573–579. [Google Scholar] [CrossRef]
- Pickard, M.R.; Williams, G.T. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes 2015, 6, 484–499. [Google Scholar] [CrossRef] [Green Version]
- Mazar, J.; Rosado, A.; Shelley, J.; Marchica, J.; Westmoreland, T.J. The long non-coding RNA GAS5 differentially regulates cell cycle arrest and apoptosis through activation of BRCA1 and p53 in human neuroblastoma. Oncotarget 2017, 8, 6589–6607. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Mo, Y.-Y. Abstract 1829: Negative regulation of lncRNA GAS5 by miR-21. Mol. Cell. Biol. 2013, 20, 1558–1568. [Google Scholar] [CrossRef]
- Vesovic, N.; Tosic, N.; Djurasevic, T.K.; Andric, Z.; Zdravkovic, D.; Pavlovic, S.; Jovanovic, D. Expression pattern of circulating long non-coding RNA GAS5 as a novel biomarker in non-small cell lung cancer patients. Arch. Med. Sci. 2020, 16, 161. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, S.; Yang, X.; Li, X.; Chen, R. Association between polymorphism in the promoter region of lncRNA GAS5 and the risk of colorectal cancer. Biosci. Rep. 2019, 39, 20190091. [Google Scholar] [CrossRef] [Green Version]
- Avgeris, M.; Tsilimantou, A.; Levis, P.K.; Tokas, T.; Sideris, D.C.; Stravodimos, K.; Ardavanis, A.; Scorilas, A. Loss of GAS5 tumour suppressor lncRNA: An independent molecular cancer biomarker for short-term relapse and progression in bladder cancer patients. Br. J. Cancer 2018, 119, 1477–1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimta, A.-A.; Tomuleasa, C.; Sahnoune, I.; Calin, G.; Berindan-Neagoe, I. Long Non-coding RNAs in Myeloid Malignancies. Front. Oncol. 2019, 9, 1048. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Zhang, D.-Y.; Li, X.; Yuan, X.-Q.; Yang, Y.-L.; Zhu, K.-W.; Zeng, H.; Li, X.-L.; Cao, S.; Zhou, H.-H.; et al. Long non-coding RNA GAS5 polymorphism predicts a poor prognosis of acute myeloid leukemia in Chinese patients via affecting hematopoietic reconstitution. Leuk. Lymphoma 2017, 58, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Ketab, F.N.G.; Gharesouran, J.; Ghafouri-Fard, S.; Dastar, S.; Mazraeh, S.A.; Hosseinzadeh, H.; Moradi, M.; Javadlar, M.; Hiradfar, A.; Rezamand, A.; et al. Dual biomarkers long non-coding RNA GAS5 and its target, NR3C1, contribute to acute myeloid leukemia. Exp. Mol. Pathol. 2020, 114, 104399. [Google Scholar] [CrossRef]
- Rodriguez, P.; Paculova, H.; Kogut, S.; Heath, J.; Schjerven, H.; Frietze, S. Non-Coding RNA Signatures of B-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2021, 22, 2683. [Google Scholar] [CrossRef]
- Gasic, V.; Stankovic, B.; Zukic, B.; Janic, D.; Dokmanovic, L.; Krstovski, N.; Lazic, J.; Milosevic, G.; Lucafò, M.; Stocco, G.; et al. Expression pattern of long non-coding RNA growth arrest-specific 5 in the remission induction therapy in childhood acute lymphoblastic leukemia. J. Med. Biochem. 2019, 38, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Ye, Y.; Zhao, S.-J. LncRNA Gas5 acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p in papillary thyroid carcinoma. Oncotarget 2017, 9, 3519–3530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isken, F.; Steffen, B.; Merk, S.; Dugas, M.; Markus, B.; Tidow, N.; Zühlsdorf, M.; Illmer, T.; Thiede, C.; Berdel, W.E.; et al. Identification of acute myeloid leukaemia associated microRNA expression patterns. Br. J. Haematol. 2007, 140, 153–161. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, P.; Liu, J.; Zheng, J.; Liu, Y.; Chen, J.; Xue, Y. Gas5 Exerts Tumor-suppressive Functions in Human Glioma Cells by Targeting miR-222. Mol. Ther. 2015, 23, 1899–1911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Gu, J.; Lu, H. The GAS5/miR-222 Axis Regulates Proliferation of Gastric Cancer Cells Through the PTEN/Akt/mTOR Pathway. Dig. Dis. Sci. 2017, 62, 3426–3437. [Google Scholar] [CrossRef]
- Jing, Z.; Gao, L.; Wang, H.; Chen, J.; Ben Nie, B.; Hong, Q. Long non-coding RNA GAS5 regulates human B lymphocytic leukaemia tumourigenesis and metastasis by sponging miR-222. Cancer Biomark. 2019, 26, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanovic, M.; Tosic, N.; Colovic, N.; Karan-Djurasevic, T.; Spasovski, V.; Radmilovic, M.; Nikcevic, G.; Suvajdzic-Vukovic, N.; Tomin, D.; Vidovic, A.; et al. Prognostic Impact ofNPM1Mutations in Serbian Adult Patients with Acute Myeloid Leukemia. Acta Haematol. 2012, 128, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Gyorffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff Finder: A Comprehensive and Straightforward Web Application Enabling Rapid Biomarker Cutoff Optimization. PLoS ONE 2012, 7, e51862. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Gao, X.; Tian, X. High expression of long intergenic non-coding RNA LINC00662 contributes to malignant growth of acute myeloid leukemia cells by upregulating ROCK1 via sponging microRNA-340-5p. Eur. J. Pharmacol. 2019, 859, 172535. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, P.; Zhao, Y.; Liu, B. The role of long non-coding RNAs and downstream signaling pathways in leukemia progression. Hematol. Oncol. 2021, 39, 27–40. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Bai, J.; He, A. MicroRNA: An important regulator in acute myeloid leukemia. Cell Biol. Int. 2017, 41, 936–945. [Google Scholar] [CrossRef]
- Diederichs, S.; Haber, D.A. Sequence Variations of MicroRNAs in Human Cancer: Alterations in Predicted Secondary Structure Do Not Affect Processing. Cancer Res. 2006, 66, 6097–6104. [Google Scholar] [CrossRef] [Green Version]
- Rommer, A.; Steinleitner, K.; Hackl, H.; Schneckenleithner, C.; Engelmann, M.; Scheideler, M.; Vlatkovic, I.; Kralovics, R.; Cerny-Reiterer, S.; Valent, P.; et al. Overexpression of primary microRNA 221/222 in acute myeloid leukemia. BMC Cancer 2013, 13, 364. [Google Scholar] [CrossRef] [Green Version]
- Goustin, A.S.; Thepsuwan, P.; Kosir, M.A.; Lipovich, L. The Growth-Arrest-Specific (GAS)-5 Long Non-Coding RNA: A Fascinating lncRNA Widely Expressed in Cancers. Non-Coding RNA 2019, 5, 46. [Google Scholar] [CrossRef] [Green Version]
- Mourtada-Maarabouni, M.; Hasan, A.M.; Farzaneh, F.; Williams, G.T. Inhibition of Human T-Cell Proliferation by Mammalian Target of Rapamycin (mTOR) Antagonists Requires Noncoding RNA Growth-Arrest-Specific Transcript 5 (GAS5). Mol. Pharmacol. 2010, 78, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, H.; Li, Y.; Li, L.; Hou, W.; You, Z. Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol. Rep. 2016, 36, 3241–3250. [Google Scholar] [CrossRef] [Green Version]
- Bian, D.; Shi, W.; Shao, Y.; Li, P.; Song, G. Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in melanoma. Am. J. Transl. Res. 2017, 9, 1509–1520. [Google Scholar] [PubMed]
- Ravegnini, G.; Cargnin, S.; Sammarini, G.; Zanotti, F.; Bermejo, J.L.; Hrelia, P.; Terrazzino, S.; Angelini, S. Prognostic Role of miR-221 and miR-222 Expression in Cancer Patients: A Systematic Review and Meta-Analysis. Cancers 2019, 11, 970. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Sang, Y.; Sun, T.; Kong, P.; Zhang, L.; Dai, Y.; Cao, Y.; Tao, Z.; Liu, W. Emerging roles and mechanisms of microRNA-222-3p in human cancer (Review). Int. J. Oncol. 2021, 58, 20. [Google Scholar] [CrossRef]
- Morotti, A.; Panuzzo, C.; Crivellaro, S.; Carrá, G.; Torti, D.; Guerrasio, A.; Saglio, G. The role of PTEN in myeloid malignancies. Hematol. Rep. 2015, 7, 5844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seval, G.C.; Ozcan, M. Treatment of Acute Myeloid Leukemia in Adolescent and Young Adult Patients. J. Clin. Med. 2015, 4, 441–459. [Google Scholar] [CrossRef] [Green Version]
- Fang, P.; Xiang, L.; Chen, W.; Li, S.; Huang, S.; Li, J.; Zhuge, L.; Jin, L.; Feng, W.; Chen, Y.; et al. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3. Innate Immun. 2019, 25, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Varier, K.M.; Dhandapani, H.; Liu, W.; Song, J.; Wang, C.; Hu, A.; Ben-David, Y.; Shen, X.; Li, Y.; Gajendran, B. An immunotherapeutic approach to decipher the role of long non-coding RNAs in cancer progression, resistance and epigenetic regulation of immune cells. J. Exp. Clin. Cancer Res. 2021, 40, 242. [Google Scholar] [CrossRef]
- Zhai, S.; Xu, Z.; Xie, J.; Zhang, J.; Wang, X.; Peng, C.; Li, H.; Chen, H.; Shen, B.; Deng, X. Epigenetic silencing of LncRNA LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene 2021, 40, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lu, Y.; Wang, H.; Han, X.; Mao, J.; Li, J.; Yu, L.; Wang, B.; Fan, S.; Yu, X.; et al. miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed. Pharmacother. 2016, 79, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Panneerselvam, J.; Srivastava, A.; Muralidharan, R.; Wang, Q.; Zheng, W.; Zhao, L.; Chen, A.; Zhao, Y.D.; Munshi, A.; Ramesh, R. IL-24 modulates the high mobility group (HMG) A1/miR222 /AKT signaling in lung cancer cells. Oncotarget 2016, 7, 70247–70263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Parameter | Total (n = 75) | GAS5 Expression | p | |
|---|---|---|---|---|
| GAS5high (n = 32) | GAS5low (n = 43) | |||
| Sex | 0.78 | |||
| Male (%) | 42 (56) | 19 (45) | 23 (55) | |
| Female (%) | 33 (44) | 13 (39) | 20 (61) | |
| Age, years, median (range) | 50 (18–62) | 51 (24–62) | 48 (18–62) | 0.44 |
| WBC (White Blood Cells) count, ×109/L, median (range) | 17.1 (1–348.8) | 9.8 (1–183.7) | 22.3 (1.2–348.8) | 0.09 |
| Hemoglobin (g/L), median (range) | 97 (2–166) | 98.5 (2–153) | 97 (24–166) | 0.62 |
| Platelets (×109/L), median (range) | 54 (1–422) | 48.5 (12–216) | 54 (1–422) | 0.59 |
| LDH (U/L), median (range) | 287.5 (153–4169) | 175 (153–4196) | 462.5 (2–2904) | 0.04 |
| Peripheral blood blast (%) median (range) | 16 (0–98) | 13.5 (0–92) | 21 (0–98) | 0.43 |
| Bone marrow blasts (%) median (range) | 63 (21–97) | 60.5 (21–97) | 67(21–97) | 0.81 |
| CD34 (%) | 0.31 | |||
| present | 46 (61) | 20 (43) | 26 (57) | |
| absent | 22 (39) | 6 (27) | 16 (73) | |
| FAB (%) | 0.15 | |||
| M0 | 6 (8) | 3 (50) | 3 (50) | |
| M1 | 10 (13) | 5 (50) | 5 (50) | |
| M2 | 17 (23) | 10 (59) | 7 (41) | |
| M4 | 28 (37) | 12 (43) | 16 (57) | |
| M5 | 14 (19) | 2 (14) | 12 (86) | |
| Prognostic group—ELN (%) | 0.04 | |||
| Favorable | 12 (16) | 4 (33) | 8 (67) | |
| Intermediate | 44 (59) | 24 (55) | 20 (45) | |
| Adverse | 19 (25) | 4 (21) | 15 (79) | |
| Complete remission (%) | 1.0 | |||
| success | 41 (55) | 17 (41) | 24 (59) | |
| failure | 34 (45) | 15 (44) | 19 (56) | |
| Relapse (%) | 0.38 | |||
| yes | 22 (51) | 11 (50) | 11 (50) | |
| no | 19 (49) | 6 (32) | 13 (68) | |
| Parameter | Total (n = 39) | GAS5 Expression | p | |
|---|---|---|---|---|
| GAS5high (n = 16) | GAS5low (n = 23) | |||
| FLT3-ITD mutations | 0.32 | |||
| present (%) | 12 (31) | 3 (25) | 9 (75) | |
| absent (%) | 27 (69) | 13 (48) | 14 (52) | |
| NPM1mutations | 0.21 | |||
| present (%) | 13 (33) | 3 (25) | 10 (75) | |
| absent (%) | 26 (67) | 13 (50) | 13 (50) | |
| FLT3-ITD/NPM1 status(risk group) | 0.05 | |||
| FLT3-ITD−/NPM1− (intermediate) | 20 (51) | 12 (60) | 8 (40) | |
| FLT3-ITD+ (adverse) | 12 (31) | 3 (25) | 9 (75) | |
| NPM1+ (favorable) | 7 (18) | 1 (14) | 6 (86) | |
| miR-222 expression status | 0.40 | |||
| high (%) | 19 (49) | 6 (32) | 13 (68) | |
| low (%) | 20 (51) | 10 (50) | 10 (50) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlovic, D.; Tosic, N.; Zukic, B.; Pravdic, Z.; Vukovic, N.S.; Pavlovic, S.; Gasic, V. Expression Profiles of Long Non-Coding RNA GAS5 and MicroRNA-222 in Younger AML Patients. Diagnostics 2022, 12, 86. https://doi.org/10.3390/diagnostics12010086
Pavlovic D, Tosic N, Zukic B, Pravdic Z, Vukovic NS, Pavlovic S, Gasic V. Expression Profiles of Long Non-Coding RNA GAS5 and MicroRNA-222 in Younger AML Patients. Diagnostics. 2022; 12(1):86. https://doi.org/10.3390/diagnostics12010086
Chicago/Turabian StylePavlovic, Djordje, Natasa Tosic, Branka Zukic, Zlatko Pravdic, Nada Suvajdzic Vukovic, Sonja Pavlovic, and Vladimir Gasic. 2022. "Expression Profiles of Long Non-Coding RNA GAS5 and MicroRNA-222 in Younger AML Patients" Diagnostics 12, no. 1: 86. https://doi.org/10.3390/diagnostics12010086
APA StylePavlovic, D., Tosic, N., Zukic, B., Pravdic, Z., Vukovic, N. S., Pavlovic, S., & Gasic, V. (2022). Expression Profiles of Long Non-Coding RNA GAS5 and MicroRNA-222 in Younger AML Patients. Diagnostics, 12(1), 86. https://doi.org/10.3390/diagnostics12010086






