A Host-Directed Approach to the Detection of Infection in Hard-to-Heal Wounds
Abstract
:1. Introduction
2. Wound Infection
2.1. Distinguishing Colonization from Infection
2.2. The Role of Biofilm in Chronic Infection
2.3. Current Approaches in the Diagnosis of Wound Infection Status
2.3.1. Clinical Observation
2.3.2. Microbiological Investigation
2.3.3. PCR and Sequencing-Based Technologies
2.3.4. Existing Biomarkers and Uses
2.3.5. Electronic Noses and Imaging
2.4. Wound Healing: An Overview
2.4.1. The Role of Inflammation in Wound Healing and Chronic Wounds
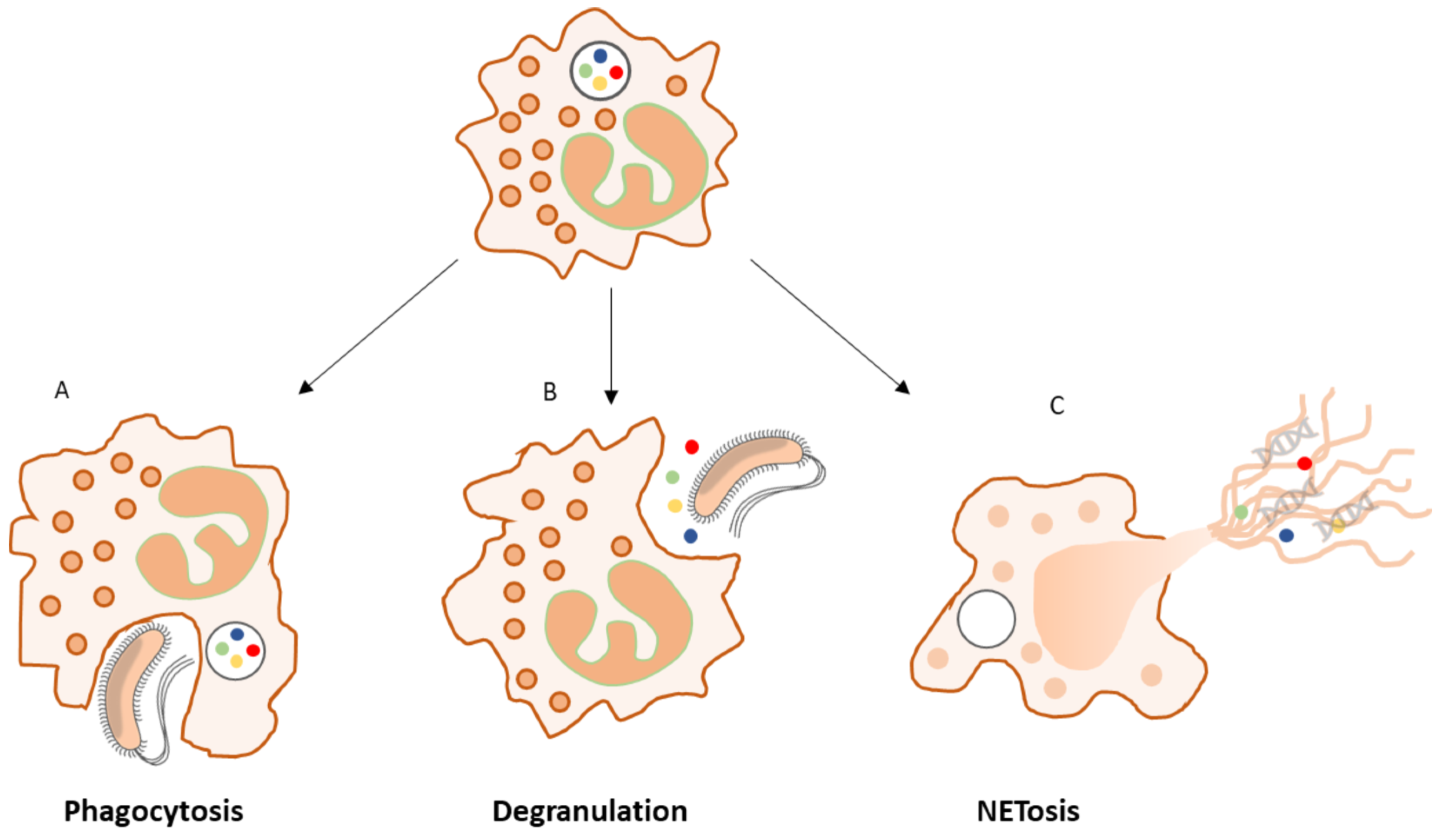
2.4.2. The Role of Neutrophils in the Inflammatory Phase
2.4.3. Neutrophil Granules: A Rich Source of Proteases and Peroxidases
2.5. Scenarios of Wound Healing
2.5.1. Scenario 1: The Healing Wound
2.5.2. Scenario 2: Acute (Early Onset) Infection
2.5.3. Scenario 3: Chronic (Prolonged, Local) Infection
3. Towards a New Approach to Wound Infection Detection
3.1. Neutrophil Enzymes as Markers to Detect Wound Infection
3.2. A Window of Opportunity
3.3. The Four Pillars of Wound Infection Detection
- I.
- The first can be considered as “screening”. In this setting, inexpensive reagents are used regularly to ascertain infection status. If, over time, higher levels are detected, an incipient infection may be suspected, and appropriate action taken. Such an inflection point in biomarker levels is also a reasonable point to initiate any other measures such as additional wound hygiene [112], antisepsis and antimicrobial dressings. Monitoring via screening is particularly relevant to fast-changing settings such as surgical wounds, where early intervention could happen on the scale of hours or days. In the post-surgical setting, regular testing of fluids either from drains or sutures may prove prudent as a means to initiate therapy whilst bacterial numbers are low, if a wound does not immediately progress to healing.
- II.
- The second opportunity is more relevant to longer-term or hard-to-heal wounds and can be considered “providing more certainty” or “disambiguation” of an unclear clinical picture. This applies more to situations where wound healing is delayed, but classical signs of infection are not apparent. Under these conditions, sub-clinical levels of infection may be interfering with the resolution and healing processes. However, because these are not visible, they may go untreated. This may be particularly the case for biofilm which may be underestimated by microbiological analysis. In such cases, the use of biomarkers as measures of infection may detect the underlying cause of wound stasis and provide a new impetus or therapy direction [2,113]. Indeed, the fact that biofilm induces a “frustrated” hyper-inflammatory state, detection of elevated enzymes in chronic wounds could indirectly confirm the presence of biofilm.
- III.
- The third opportunity is “monitoring following diagnosis”. In this setting, the impact or success of the measures taken should be assessed in real time if possible. Thus, ineffective wound hygiene measures can be recognized by a resurgence in biomarkers before a return to suppuration is observed. In various studies, the application of antimicrobial agents has been associated with a reduction in biomarkers, suggesting that the use of these substances reduced lysis of immune cells. Alternatively, no change or a further increase in biomarkers would signal a failure of therapy and possibly resistance to the agent(s) in use.
- IV.
- The fourth opportunity relates to the common problem of persister cells, biofilm, and the re-emergence of infection after cessation of therapy. Thus, “monitoring of resolved infections”, especially in unstable patients is a means to validate remission or to detect progression or reversion to infection before it is severe. This is particularly relevant to those with immune suppression or multiple wounds.
4. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolcott, R.D.; Rhoads, D.D.; Dowd, S.E. Biofilms and chronic wound inflammation. J. Wound Care 2008, 17, 333–341. [Google Scholar] [CrossRef]
- Bowler, P.G.; Duerden, B.I.; Armstrong, D.G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001, 14, 244–269. [Google Scholar] [CrossRef] [Green Version]
- Thomson, P.D.; Smith, D.J. What is infection? Am. J. Surg. 1994, 167, S7–S11. [Google Scholar] [CrossRef]
- Morton, L.M.; Phillips, T.J. Wound healing and treating wounds: Differential diagnosis and evaluation of chronic wounds. J. Am. Acad. Dermatol. 2016, 74, 589–605. [Google Scholar] [CrossRef]
- Hurlow, J.; Bowler, P.G. Acute and chronic wound infections: Microbiological, immunological, clinical, and therapeutic distinctions. J. Wound Care 2022, 31, 436–445. [Google Scholar] [CrossRef]
- Haalboom, M.; Blokhuis-Arkes, M.H.E.; Beuk, R.J.; Meerwaldt, R.; Klont, R.; Schijffelen, M.J.; Bowler, P.G.; Burnet, M.; Sigl, E.; van der Palen, J.A.M. Culture results from wound biopsy versus wound swab: Does it matter for the assessment of wound infection? Clin. Microbiol. Infect. 2019, 25, 629.e7–629.e12. [Google Scholar] [CrossRef]
- Bowler, P.G. The 10(5) bacterial growth guideline: Reassessing its clinical relevance in wound healing. Ostomy Wound Manag. 2003, 49, 44–53. [Google Scholar]
- Cutting, K.F.; White, R. Defined and refined: Criteria for identifying wound infection revisited. Br. J. Community Nurs. 2004, 9, S6–S15. [Google Scholar] [CrossRef] [PubMed]
- Blokhuis-Arkes, M.H.E.; Haalboom, M.; van der Palen, J.; Heinzle, A.; Sigl, E.; Guebitz, G.; Beuk, R. Rapid enzyme analysis as a diagnostic tool for wound infection: Comparison between clinical judgment, microbiological analysis, and enzyme analysis. Wound Repair Regen. 2015, 23, 345–352. [Google Scholar] [CrossRef]
- Schiffer, D.; Verient, V.; Luschnig, D.; Blokhuis-Arkes, M.H.E.; van der Palen, J.; Gamerith, C.; Burnet, M.; Sigl, E.; Heinzle, A.; Guebitz, G.M. Lysozyme-responsive polymer systems for detection of infection. Eng. Life Sci. 2015, 15, 368–375. [Google Scholar] [CrossRef]
- Schiffer, D.; Tegl, G.; Heinzle, A.; Sigl, E.; Metcalf, D.; Bowler, P.; Burnet, M.; Guebitz, G.M. Enzyme-responsive polymers for microbial infection detection. Expert Rev. Mol. Diagn. 2015, 15, 1125–1131. [Google Scholar] [CrossRef]
- Tegl, G.; Schiffer, D.; Sigl, E.; Heinzle, A.; Guebitz, G.M. Biomarkers for infection: Enzymes, microbes, and metabolites. Appl. Microbiol. Biotechnol. 2015, 99, 4595–4614. [Google Scholar] [CrossRef]
- Schiffer, D.; Tegl, G.; Vielnascher, R.; Weber, H.; Herrero-Rollett, A.; Sigl, E.; Heinzle, A.; Guebitz, G.M. Myeloperoxidase-responsive materials for infection detection based on immobilized aminomethoxyphenol. Biotechnol. Bioeng. 2016, 113, 2553–2560. [Google Scholar] [CrossRef]
- Metcalf, D.G.; Haalboom, M.; Bowler, P.G.; Gamerith, C.; Sigl, E.; Heinzle, A.; Burnet, M.W.M. Elevated wound fluid pH correlates with increased risk of wound infection. Wound Med. 2019, 26, 100166. [Google Scholar] [CrossRef]
- Webb, R. A chronic case of confusion. J. Wound Care 2017, 26, 421. [Google Scholar] [CrossRef] [Green Version]
- Edwards, R.; Harding, K.G. Bacteria and wound healing. Curr. Opin. Infect. Dis. 2004, 17, 91–96. [Google Scholar] [CrossRef]
- White, R.J.; Cutting, K.F. Critical colonization--the concept under scrutiny. Ostomy Wound Manag. 2006, 52, 50–56. [Google Scholar]
- Bowler, P.G.; Davies, B.J. The microbiology of infected and noninfected leg ulcers. Int. J. Dermatol. 1999, 38, 573–578. [Google Scholar] [CrossRef]
- White, R.J.; Cutting, K.F. Critical colonisation of chronic wounds: Microbial mechanisms. Wounds UK 2008, 4, 70–78. [Google Scholar]
- Malone, M.; Bjarnsholt, T.; McBain, A.J.; James, G.A.; Stoodley, P.; Leaper, D.; Tachi, M.; Schultz, G.; Swanson, T.; Wolcott, R.D. The prevalence of biofilms in chronic wounds: A systematic review and meta-analysis of published data. J. Wound Care 2017, 26, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Wolcott, R.D. Biofilms cause chronic infections. J. Wound Care 2017, 26, 423–425. [Google Scholar] [CrossRef] [Green Version]
- Bowler, P.G. Antibiotic resistance and biofilm tolerance: A combined threat in the treatment of chronic infections. J. Wound Care 2018, 27, 273–277. [Google Scholar] [CrossRef]
- Chen, L.; Wen, Y.M. The role of bacterial biofilm in persistent infections and control strategies. Int. J. Oral Sci. 2011, 3, 66–73. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Bowler, P.; Murphy, C.; Wolcott, R. Biofilm exacerbates antibiotic resistance: Is this a current oversight in antimicrobial stewardship? Antimicrob. Resist. Infect. Control 2020, 9, 162. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fischer, E.R.; DeLeo, F.R.; Otto, M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar] [CrossRef] [Green Version]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [Green Version]
- Percival, S.L.; Hill, K.E.; Malic, S.; Thomas, D.W.; Williams, D.W. Antimicrobial tolerance and the significance of persister cells in recalcitrant chronic wound biofilms. Wound Repair Regen. 2011, 19, 1–9. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef] [Green Version]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS 2013, 121, 1–58. [Google Scholar] [CrossRef]
- Percival, S.L.; Bowler, P.G. Biofilms and Their Potential Role in Wound Healing. Wounds 2004, 7, 234–240. [Google Scholar]
- Bowler, P. The role of bacterial communities in wound healing. J. Tissue Viability 2005, 15, 19. [Google Scholar] [CrossRef]
- Metcalf, D.; Bowler, P.; Parsons, D. Wound Biofilm and Therapeutic Strategies. In Microbial Biofilms, 1st ed.; Dhanasekaran, D., Thajuddi, N., Eds.; InTechOpen: London, UK, 2016. [Google Scholar]
- Attinger, C.; Wolcott, R. Clinically Addressing Biofilm in Chronic Wounds. Adv. Wound Care 2012, 1, 127–132. [Google Scholar] [CrossRef] [Green Version]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Geisinger, E.; Isberg, R.R. Interplay Between Antibiotic Resistance and Virulence During Disease Promoted by Multidrug-Resistant Bacteria. J. Infect. Dis. 2017, 215, S9–S17. [Google Scholar] [CrossRef] [Green Version]
- Glaudemans, A.W.; Uçkay, I.; Lipsky, B.A. Challenges in diagnosing infection in the diabetic foot. Diabet. Med. 2015, 32, 748–759. [Google Scholar] [CrossRef]
- Li, S.; Renick, P.; Senkowsky, J.; Nair, A.; Tang, L. Diagnostics for Wound Infections. Adv. Wound Care 2021, 10, 317–327. [Google Scholar] [CrossRef]
- Bertesteanu, S.; Triaridis, S.; Stankovic, M.; Lazar, V.; Chifiriuc, M.C.; Vlad, M.; Grigore, R. Polymicrobial wound infections: Pathophysiology and current therapeutic approaches. Int. J. Pharm. 2014, 463, 119–126. [Google Scholar] [CrossRef]
- Wu, Y.-K.; Cheng, N.-C.; Cheng, C.-M. Biofilms in Chronic Wounds: Pathogenesis and Diagnosis. Trends Biotech. 2019, 37, 505–517. [Google Scholar] [CrossRef]
- Bamberg, R.; Sullivan, P.; Conner-Kerr, T. Diagnosis of wound infections: Current culturing practices of U.S. Wound care professionals. Wounds 2002, 14, 314–328. [Google Scholar]
- Rondas, A.A.; Halfens, R.J.; Schols, J.M.; Thiesen, K.P.; Trienekens, T.A.; Stobberingh, E.E. Is a wound swab for microbiological analysis supportive in the clinical assessment of infection of a chronic wound? Future Microbiol. 2015, 10, 1815–1824. [Google Scholar] [CrossRef] [PubMed]
- Fazli, M.; Bjarnsholt, T.; Kirketerp-Møller, K.; Jørgensen, B.; Andersen, A.S.; Krogfelt, K.A.; Givskov, M.; Tolker-Nielsen, T. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 2009, 47, 4084–4089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLeod, A.S.; Mansbridge, J.N. The Innate Immune System in Acute and Chronic Wounds. Adv. Wound Care 2016, 5, 65–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enoch, S.; Grey, J.E.; Harding, K.G. ABC of wound healing. Non-surgical and drug treatments. Br. Med. J. Clin. Res. Ed. 2006, 332, 900–903. [Google Scholar] [CrossRef] [Green Version]
- Bowler, P.; Davies, B.J. The Microbiology of Acute and Chronic Wounds. Wounds 1999, 11, 72–78. [Google Scholar]
- Malone, M.; Johani, K.; Jensen, S.O.; Gosbell, I.B.; Dickson, H.G.; Hu, H.; Vickery, K. Next Generation DNA Sequencing of Tissues from Infected Diabetic Foot Ulcers. EBioMedicine 2017, 21, 142–149. [Google Scholar] [CrossRef]
- Maugeri, G.; Lychko, I.; Sobral, R.; Roque, A.C.A. Identification and Antibiotic-Susceptibility Profiling of Infectious Bacterial Agents: A Review of Current and Future Trends. Biotech. J. 2019, 14, e1700750. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [Green Version]
- Gupta, D.; Agarwal, R.; Aggarwal, A.N.; Singh, N.; Mishra, N.; Khilnani, G.C.; Samaria, J.K.; Gaur, S.N.; Jindal, S.K.; Pneumonia Guidelines Working Group. Guidelines for diagnosis and management of community- and hospital-acquired pneumonia in adults: Joint ICS/NCCP(I) recommendations. Lung India 2012, 29, S27–S62. [Google Scholar] [CrossRef]
- Allaband, C.; McDonald, D.; Vázquez-Baeza, Y.; Minich, J.J.; Tripathi, A.; Brenner, D.A.; Loomba, R.; Smarr, L.; Sandborn, W.J.; Schnabl, B.; et al. Microbiome 101: Studying, Analyzing, and Interpreting Gut Microbiome Data for Clinicians. Clin. Gastroenterol. Hepatol. 2019, 17, 218–230. [Google Scholar] [CrossRef]
- De Oliveira, F.P.; Pires, B.M.F.B.; de Cássia Ferreira de Almeida Silva, K.; de Carvalho, B.T.F.; Teixeira, L.A.; de Paula, G.R.; de Oliveira, B.G.R.B. Prevalence, Antimicrobial Susceptibility, and Clonal Diversity of Pseudomonas aeruginosa in Chronic Wounds. J. Wound Ostomy Cont. Nurs. 2017, 44, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.D.; Dowd, S.E. A rapid molecular method for characterising bacterial bioburden in chronic wounds. J. Wound Care 2008, 17, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Verbanic, S.; Shen, Y.; Lee, J.; Deacon, J.M.; Chen, I.A. Microbial predictors of healing and short-term effect of debridement on the microbiome of chronic wounds. npj Biofilms Microbiomes 2020, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Jacombs, A.; Vickery, K.; Merten, S.L.; Pennington, D.G.; Deva, A.K. Chronic Biofilm Infection in Breast Implants Is Associated with an Increased T-Cell Lymphocytic Infiltrate: Implications for Breast Implant–Associated Lymphoma. Plast. Reconstr. Surg. 2015, 135, 1057e–1059e. [Google Scholar] [CrossRef]
- Kommedal, Ø.; Lekang, K.; Langeland, N.; Wiker, H.G. Characterisation of polymicrobial clinical samples using a set of group-specific briad-range primers targeting the 16rRNA gene followed by DNA sequencing and RipSeq analysis. J Med Micro. 2011, 60, 927–936. [Google Scholar] [CrossRef]
- Clarridge, J.E., 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef]
- Percival, S.L.; Hill, K.E.; Williams, D.W.; Hooper, S.J.; Thomas, D.W.; Costerton, J.W. A review of the scientific evidence for biofilms in wounds. Wound Repair Regen. 2012, 20, 647–657. [Google Scholar] [CrossRef]
- Yang, S.; Rothman, R.E. PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Petralia, S.; Conoci, S. PCR Technologies for Point of Care Testing: Progress and Perspectives. ACS Sens. 2017, 2, 876–891. [Google Scholar] [CrossRef]
- Sproston, N.R.; Ashworth, J.J. Role of C-Reactive Protein at Sites of Inflammation and Infection. Front. Immunol. 2018, 9, 754. [Google Scholar] [CrossRef] [Green Version]
- Faix, J.D. Biomarkers of sepsis. Crit. Rev. Clin. Lab. Sci. 2013, 50, 23–36. [Google Scholar] [CrossRef] [Green Version]
- Biron, B.M.; Ayala, A.; Lomas-Neira, J.L. Biomarkers for Sepsis: What Is and What Might Be? Biomark. Insights 2015, 10 (Suppl. 4), 7–17. [Google Scholar] [CrossRef] [Green Version]
- Venge, P. Human neutrophil lipocalin (HNL) as a biomarker of acute infections. Upsala J. Med. Sci. 2018, 123, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Slade, E.A.; Thorn, R.M.; Young, A.E.; Reynolds, D.M. Real-time detection of volatile metabolites enabling species-level discrimination of bacterial biofilms associated with wound infection. J. Appl. Microbiol. 2022, 132, 1558–1572. [Google Scholar] [CrossRef] [PubMed]
- Saviauk, T.; Kiiski, J.P.; Nieminen, M.K.; Tamminen, N.N.; Roine, A.N.; Kumpulainen, P.S.; Hokkinen, L.J.; Karjalainen, M.T.; Vuento, R.E.; Aittoniemi, J.J.; et al. Electronic nose in the detection of wound infection bacteria from bacterial cultures: A proof-of-principle study. Eur. Surg. Res. 2018, 59, 1–11. [Google Scholar] [CrossRef]
- Janowska, A.; Davini, G.; Romanelli, M.; Oranges, T.; Iannone, M.; Dini, V. The association between pH and fluorescence as non-invasive diagnostic tools in chronic wounds. Int. J. Lower Extrem. Wounds 2021. [Google Scholar] [CrossRef] [PubMed]
- Ellis, S.; Lin, E.J.; Tartar, D. Immunology of Wound Healing. Curr. Dermatol. Rep. 2018, 7, 350–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultz, G.S.; Chin, G.A.; Moldawer, L.; Diegelmann, R.F. Principles of Wound Healing. In Mechanisms of Vascular Disease: A Reference Book for Vascular Specialists; Fitridge, R., Thompson, M., Eds.; University of Adelaide Press: Adelaide, Australia, 2011; p. 23. [Google Scholar]
- Rodrigues, M.; Kosaric, N.; Bonham, C.A.; Gurtner, G.C. Wound Healing: A Cellular Perspective. Physiol. Rev. 2018, 99, 665–706. [Google Scholar] [CrossRef]
- Eming, S.A.; Krieg, T.; Davidson, J.M. Inflammation in Wound Repair: Molecular and Cellular Mechanisms. J. Investig. Dermatol. 2007, 127, 514–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landén, N.X.; Li, D.; Ståhle, M. Transition from inflammation to proliferation: A critical step during wound healing. Cell. Mol. Life Sci. 2016, 73, 3861–3885. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Richmond, A. Chemokine Regulation of Neutrophil Infiltration of Skin Wounds. Adv. Wound Care 2015, 4, 631–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larouche, J.; Sheoran, S.; Maruyama, K.; Martino, M.M. Immune Regulation of Skin Wound Healing: Mechanisms and Novel Therapeutic Targets. Adv. Wound Care 2018, 7, 209–231. [Google Scholar] [CrossRef]
- Bratton, D.L.; Henson, P.M. Neutrophil clearance: When the party is over, clean-up begins. Trends Immunol. 2011, 32, 350–357. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, S.; Rosowski, E.E.; Huttenlocher, A. Neutrophil migration in infection and wound repair: Going forward in reverse. Nature Rev. Immunol. 2016, 16, 378–391. [Google Scholar] [CrossRef] [Green Version]
- Rosales, C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front. Physiol. 2018, 9, 113. [Google Scholar] [CrossRef]
- Sheshachalam, A.; Srivastava, N.; Mitchell, T.; Lacy, P.; Eitzen, G. Granule Protein Processing and Regulated Secretion in Neutrophils. Front. Immunol. 2014, 5, 448. [Google Scholar] [CrossRef]
- Cassatella, M.A.; Östberg, N.K.; Tamassia, N.; Soehnlein, O. Biological Roles of Neutrophil-Derived Granule Proteins and Cytokines. Trends Immunol. 2019, 40, 648–664. [Google Scholar] [CrossRef]
- Cramer, E.M.; Breton-Gorius, J. Ultrastructural localization of lysozyme in human neutrophils by immunogold. J. Leukoc. Biol. 1987, 41, 242–247. [Google Scholar] [CrossRef]
- Majewski, P.; Majchrzak-Gorecka, M.; Grygier, B.; Skrzeczynska-Moncznik, J.; Osiecka, O.; Cichy, J. Inhibitors of Serine Proteases in Regulating the Production and Function of Neutrophil Extracellular Traps. Front. Immunol. 2016, 7, 261. [Google Scholar] [CrossRef] [Green Version]
- McCarty, S.M.; Percival, S.L. Proteases and Delayed Wound Healing. Adv. Wound Care 2013, 2, 438–447. [Google Scholar] [CrossRef]
- Buchstein, N.; Hoffmann, D.; Smola, H.; Lang, S.; Paulsson, M.; Niemann, C.; Krieg, T.; Eming, S.A. Alternative proteolytic processing of hepatocyte growth factor during wound repair. Am. J. Pathol. 2009, 174, 2116–2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uribe-Querol, E.; Rosales, C. Control of Phagocytosis by Microbial Pathogens. Front. Immunol. 2017, 8, 1368. [Google Scholar] [CrossRef] [Green Version]
- Segal, A.W. How neutrophils kill microbes. Ann. Rev. Immunol. 2005, 23, 197–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukacs, G.L.; Rotstein, O.D.; Grinstein, S. Phagosomal acidification is mediated by a vacuolar-type H(+)-ATPase in murine macrophages. J. Biol. Chem. 1990, 265, 21099–21107. [Google Scholar] [CrossRef]
- Nordenfelt, P.; Tapper, H. Phagosome dynamics during phagocytosis by neutrophils. J. Leukoc. Biol. 2011, 90, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.T.; Green, E.R.; Mecsas, J. Neutrophils to the ROScue: Mechanisms of NADPH Oxidase Activation and Bacterial Resistance. Front. Cell. Infect. Microbiol. 2017, 7, 373. [Google Scholar] [CrossRef]
- Vyas, J.M.; Van der Veen, A.G.; Ploegh, H.L. The known unknowns of antigen processing and presentation. Nature Rev. Immunol. 2008, 8, 607–618. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.D.; Malachowa, N.; DeLeo, F.R. Influence of Microbes on Neutrophil Life and Death. Front. Cell. Infect. Microbiol. 2017, 7, 159. [Google Scholar] [CrossRef] [Green Version]
- Wolcott, R.; Costerton, J.W.; Raoult, D.; Cutler, S.J. The polymicrobial nature of biofilm infection. Clin. Microbiol. Infect. 2013, 19, 107–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowler, P.G. Wound pathophysiology, infection and therapeutic options. Ann. Med. 2002, 34, 419–427. [Google Scholar] [CrossRef]
- Wolcott, R.; Sanford, N.; Gabrilska, R.; Oates, J.L.; Wilkinson, J.E.; Rumbaugh, K.P. Microbiota is a primary cause of pathogenesis of chronic wounds. J. Wound Care 2016, 25, S33–S43. [Google Scholar] [CrossRef]
- Hornef, M.W.; Wick, M.J.; Rhen, M.; Normark, S. Bacterial strategies for overcoming host innate and adaptive immune responses. Nat. Immunol. 2002, 3, 1033–1040. [Google Scholar] [CrossRef]
- Wilgus, T.A.; Roy, S.; McDaniel, J.C. Neutrophils and Wound Repair: Positive Actions and Negative Reactions. Adv. Wound Care 2013, 2, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014, 15, 1017–1025. [Google Scholar] [CrossRef] [Green Version]
- Yipp, B.G.; Petri, B.; Salina, D.; Jenne, C.N.; Scott, B.N.V.; Zbytnuik, L.D.; Pittman, K.; Asaduzzaman, M.; Wu, K.; Meijndert, H.C.; et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat. Med. 2012, 18, 1386–1393. [Google Scholar] [CrossRef] [Green Version]
- Hahn, S.; Giaglis, S.; Chowdury, C.S.; Hösli, I.; Hasler, P. Modulation of neutrophil NETosis: Interplay between infectious agents and underlying host physiology. Sem. Immunopathol. 2013, 35, 439–453. [Google Scholar] [CrossRef]
- Wong, S.L.; Demers, M.; Martinod, K.; Gallant, M.; Wang, Y.; Goldfine, A.B.; Kahn, C.R.; Wagner, D.D. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat. Med. 2015, 21, 815–819. [Google Scholar] [CrossRef] [Green Version]
- Manfredi, A.A.; Ramirez, G.A.; Rovere-Querini, P.; Maugeri, N. The Neutrophil’s Choice: Phagocytose vs Make Neutrophil Extracellular Traps. Front. Immunol. 2018, 9, 288. [Google Scholar] [CrossRef] [Green Version]
- Van Avondt, K.; Hartl, D. Mechanisms and disease relevance of neutrophil extracellular trap formation. Eur. J. Clin. Investig. 2018, 48, e12919. [Google Scholar] [CrossRef] [Green Version]
- Radic, M. Clearance of Apoptotic Bodies, NETs, and Biofilm DNA: Implications for Autoimmunity. Front. Immunol. 2014, 5, 365. [Google Scholar] [CrossRef] [Green Version]
- Parsons, D.; Metcalf, D.G. Understanding local barriers to wound healing. In Next-Generation Antimicrobial Dressings: AQUACEL™ Ag+ Extra™ and Ribbon; Wounds International: London, UK, 2014; Available online: www.woundsinternational.com.
- De Bont, C.M.; Boelens, W.C.; Pruijn, G.J.M. NETosis, complement, and coagulation: A triangular relationship. Cell. Mol. Immunol. 2019, 16, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Neutrophils in tissue injury and repair. Cell Tissue Res. 2018, 371, 531–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Németh, T.; Sperandio, M.; Mócsai, A. Neutrophils as emerging therapeutic targets. Nat. Rev. Drug Discov. 2020, 19, 253–275. [Google Scholar] [CrossRef]
- Hasmann, A.; Wehrschuetz-Sigl, E.; Marold, A.; Wiesbauer, H.; Schoeftner, R.; Gewessler, U.; Kandelbauer, A.; Schiffer, D.; Schneider, K.P.; Binder, B.; et al. Analysis of myeloperoxidase activity in wound fluids as a marker of infection. Ann. Clin. Biochem. 2013, 50, 245–254. [Google Scholar] [CrossRef]
- Hasmann, A.; Gewessler, U.; Hulla, E.; Schneider, K.P.; Binder, B.; Francesko, A.; Tzanov, T.; Schintler, M.; van der Palen, J.; Guebitz, G.M.; et al. Sensor materials for the detection of human neutrophil elastase and cathepsin G activity in wound fluid. Exp. Dermatol. 2011, 20, 508–513. [Google Scholar] [CrossRef]
- Hasmann, A.; Wehrschuetz-Sigl, E.; Kanzler, G.; Gewessler, U.; Hulla, E.; Schneider, K.P.; Binder, B.; Schintler, M.; Guebitz, G.M. Novel peptidoglycan-based diagnostic devices for detection of wound infection. Diagn. Microbiol. Infect. Dis. 2011, 71, 12–23. [Google Scholar] [CrossRef]
- Murphy, C.; Atkin, L.; Swanson, T.; Tachi, M.; Tan, Y.K.; Vega de Ceniga, M.; Weir, D.; Wolcott, R. International consensus document. Defying hard-to-heal wounds with an early antibiofilm intervention strategy: Wound hygiene. J. Wound Care 2020, 29 (Suppl. 3b), S1–S28. [Google Scholar] [CrossRef] [Green Version]
- Metcalf, D.G.; Bowler, P.G. Clinician perceptions of wound biofilm. Int. Wound J. 2016, 13, 717–725. [Google Scholar] [CrossRef]
- Witko-Sarsat, V.; Rieu, P.; Descamps-Latscha, B.; Lesavre, P.; Halbwachs-Mecarelli, L. Neutrophils: Molecules, Functions and Pathophysiological Aspects. Lab. Investig. 2000, 80, 617–653. [Google Scholar] [CrossRef] [Green Version]
- Thakur, A.; Mikkelsen, H.; Jungersen, G. Intracellular Pathogens: Host Immunity and Microbial Persistence Strategies. J. Immunol. Res. 2019, 2019, 1356540. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.D.; DeLeo, F.R. Neutrophil apoptosis and the resolution of infection. Immunol. Res. 2009, 43, 25–61. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, F.; Marwitz, S.; Holz, O.; Kirsten, A.; Bahmer, T.; Waschki, B.; Magnussen, H.; Rabe, K.F.; Goldmann, T.; Uddin, M.; et al. Neutrophil extracellular trap formation and extracellular DNA in sputum of stable COPD patients. Respir. Med. 2015, 109, 1360–1362. [Google Scholar] [PubMed]






| Protein Name | Protein Function |
|---|---|
| Primary granules–Main marker: Myeloperoxidase (MPO) | |
| Myeloperoxidase (MPO) | Hypochlorite deactivation of microbial and granular proteins |
| Human neutrophil elastase (HNE) | Serine protease, immune cell activation, C5a- reactions |
| Cathepsin G (Cat G) | Antibacterial Serine protease, complement C3 cleavage |
| Azurocidin | Antibacterial activity, chemoattractant |
| Neutrophil defensins | Antimicrobial activities |
| Myeloblastin | Serine protease supporting neutrophil migration |
| Lysozyme | Lysis of bacterial cell walls, also detected in primary granules of progenitor cells [79] |
| CD63 antigen | Surface receptor of TIMP1 |
| Cap57 (BPI) | Bactericidal protein |
| Secondary (specific) granules–Main marker: Lactoferrin | |
| Lactoferrin | Iron binding and transport |
| Lipocalin 2 | Iron-trafficking, involvement in innate immunity and apoptosis |
| Lysozyme | Lysis of bacterial cell walls |
| Chitinase-3-like protein 1 | Carbohydrate(chitin)-binding lectin |
| Cytochrome B558 | Membrane component of the phagocyte O2-producing NADPH oxidase |
| Collagenase | Cellular migration |
| CD11b/CD18 | Adhesion complex (Integrin), endocytosis of R-G-D-C3b bound particles |
| fMLP-R | Formyl peptide receptor 1 |
| Tertiary granules–Main marker: Gelatinase | |
| Matrix metalloproteinase (MMP)-9 | Supports migration by cleaving collagen/gelatin |
| MMP-8 | Cleavage of collagens |
| Lysozyme | Lysis of bacterial peptidoglycan |
| Cathelicidin | Antibacterial pro-peptide |
| Ficolin-1 | PAMP receptor |
| Quaternary granules/Secretory Vesicles | |
| fMLP-R | Formyl peptide receptor 1 |
| CD11b/CD18 | Adhesion complex (Integrin), endocytosis of R-G-D-C3b bound particles |
| Cytochrome B558 | Membrane component of the phagocyte O2-producing NADPH oxidase |
| Alkaline phosphatase | Detoxification of lipopolysaccharide (LPS), anti-inflammatory |
| CR1 | Complement receptor type 1 (CR1), mediates binding to particles that activated complement |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burnet, M.; Metcalf, D.G.; Milo, S.; Gamerith, C.; Heinzle, A.; Sigl, E.; Eitel, K.; Haalboom, M.; Bowler, P.G. A Host-Directed Approach to the Detection of Infection in Hard-to-Heal Wounds. Diagnostics 2022, 12, 2408. https://doi.org/10.3390/diagnostics12102408
Burnet M, Metcalf DG, Milo S, Gamerith C, Heinzle A, Sigl E, Eitel K, Haalboom M, Bowler PG. A Host-Directed Approach to the Detection of Infection in Hard-to-Heal Wounds. Diagnostics. 2022; 12(10):2408. https://doi.org/10.3390/diagnostics12102408
Chicago/Turabian StyleBurnet, Michael, Daniel G. Metcalf, Scarlet Milo, Clemens Gamerith, Andrea Heinzle, Eva Sigl, Kornelia Eitel, Marieke Haalboom, and Philip G. Bowler. 2022. "A Host-Directed Approach to the Detection of Infection in Hard-to-Heal Wounds" Diagnostics 12, no. 10: 2408. https://doi.org/10.3390/diagnostics12102408





