Reliability as a Precondition for Trust—Segmentation Reliability Analysis of Radiomic Features Improves Survival Prediction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Data
2.3. Training of the Segmentation Network
2.4. Plausibility of Segmentation Outlines
2.5. Feature Computation
2.6. Feature Selection
- “high ICC”, containing all features with an ICC value >0.99;
- “low ICC”, containing all features with an ICC value <0.75;
- “all ICC”, containing all features irrespective of the ICC values.
2.7. Survival Prediction
2.8. Statistical Analysis
3. Results
3.1. Lesion Segmentation by the Neural Network
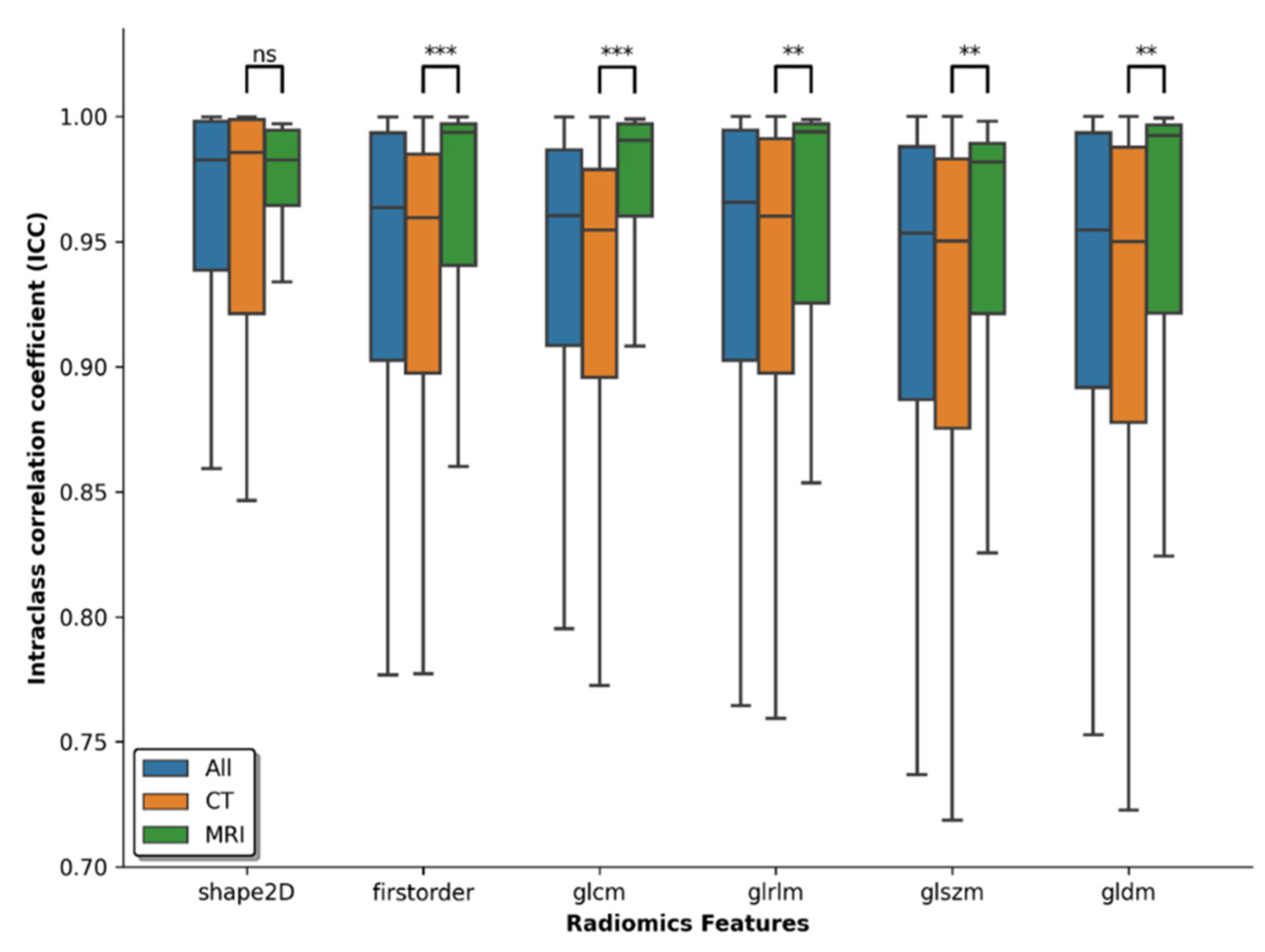
3.2. Reliability of Features
3.3. Survival Analysis Employing Feature Reliability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

References
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.K.; Truhn, D. The Long Route to Standardized Radiomics: Unraveling the Knot from the End. Radiology 2020, 295, 339–341. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, R.; Pastor-Juan, M.d.R.; Canales-Vázquez, J.; Castro-García, M.; Villas, M.V.; Mansilla Legorburo, F.; Sabater, S. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology 2018, 288, 407–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-Based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The Facts and the Challenges of Image Analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Owens, C.A.; Peterson, C.B.; Tang, C.; Koay, E.J.; Yu, W.; Mackin, D.S.; Li, J.; Salehpour, M.R.; Fuentes, D.T.; Court, L.E.; et al. Lung Tumor Segmentation Methods: Impact on the Uncertainty of Radiomics Features for Non-Small Cell Lung Cancer. PLoS ONE 2018, 13, e0205003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Simpson, G.; Young, L.; Ford, J.; Dogan, N.; Wang, L. Impact of Contouring Variability on Oncological PET Radiomics Features in the Lung. Sci. Rep. 2020, 10, 369. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, C.F.; Tezcan, K.C.; Chaitanya, K.; Hötker, A.M.; Muehlematter, U.J.; Schawkat, K.; Becker, A.S.; Donati, O.; Konukoglu, E. PHiSeg: Capturing Uncertainty in Medical Image Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; Volume 11765, pp. 119–127. ISBN 978-3-030-32244-1. [Google Scholar]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas Glioma MRI Collections with Expert Segmentation Labels and Radiomic Features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [Green Version]
- Aerts, H.J.W.L.; Wee, L.; Rios Velazquez, E.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; et al. Data From NSCLC-Radiomics 2019. Available online: https://wiki.cancerimagingarchive.net/display/Public/NSCLC-Radiomics (accessed on 5 December 2021).
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [Green Version]
- Armato Samuel, G., III; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. Data From LIDC-IDRI 2015. Available online: https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI (accessed on 5 December 2021).
- Armato, S.G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans: The LIDC/IDRI Thoracic CT Database of Lung Nodules. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef]
- Bilic, P.; Christ, P.F.; Vorontsov, E.; Chlebus, G.; Chen, H.; Dou, Q.; Fu, C.-W.; Han, X.; Heng, P.-A.; Hesser, J.; et al. The Liver Tumor Segmentation Benchmark (LiTS). arXiv 2019, arXiv:1901.04056. [Google Scholar]
- Heller, N.; Sathianathen, N.; Kalapara, A.; Walczak, E.; Moore, K.; Kaluzniak, H.; Rosenberg, J.; Blake, P.; Rengel, Z.; Oestreich, M.; et al. The KiTS19 Challenge Data: 300 Kidney Tumor Cases with Clinical Context, CT Semantic Segmentations, and Surgical Outcomes. arXiv 2020, arXiv:1904.00445. [Google Scholar]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv 2019, arXiv:1811.02629. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Haarburger, C.; Müller-Franzes, G.; Weninger, L.; Kuhl, C.; Truhn, D.; Merhof, D. Radiomics Feature Reproducibility under Inter-Rater Variability in Segmentations of CT Images. Sci. Rep. 2020, 10, 12688. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Liljequist, D.; Elfving, B.; Skavberg Roaldsen, K. Intraclass Correlation—A Discussion and Demonstration of Basic Features. PLoS ONE 2019, 14, e0219854. [Google Scholar] [CrossRef] [Green Version]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef] [Green Version]
- Carrasco, J.L.; Jover, L. Estimating the Generalized Concordance Correlation Coefficient through Variance Components. Biometrics 2003, 59, 849–858. [Google Scholar] [CrossRef]
- Cox, D.R. Regression Models and Life-Tables. In Breakthroughs in Statistics; Kotz, S., Johnson, N.L., Eds.; Springer Series in Statistics; Springer: New York, NY, USA, 1992; pp. 527–541. ISBN 978-0-387-94039-7. [Google Scholar]
- Pölsterl, S. Scikit-Survival: A Library for Time-to-Event Analysis Built on Top of Scikit-Learn. J. Mach. Learn. Res. 2020, 21, 1–6. [Google Scholar]
- van Velden, F.H.P.; Kramer, G.M.; Frings, V.; Nissen, I.A.; Mulder, E.R.; de Langen, A.J.; Hoekstra, O.S.; Smit, E.F.; Boellaard, R. Repeatability of Radiomic Features in Non-Small-Cell Lung Cancer [18F]FDG-PET/CT Studies: Impact of Reconstruction and Delineation. Mol. Imaging Biol. 2016, 18, 788–795. [Google Scholar] [CrossRef] [Green Version]
- Suter, Y.; Knecht, U.; Alão, M.; Valenzuela, W.; Hewer, E.; Schucht, P.; Wiest, R.; Reyes, M. Radiomics for Glioblastoma Survival Analysis in Pre-Operative MRI: Exploring Feature Robustness, Class Boundaries, and Machine Learning Techniques. Cancer Imaging 2020, 20, 55. [Google Scholar] [CrossRef]
- Kadoya, N.; Tanaka, S.; Kajikawa, T.; Tanabe, S.; Abe, K.; Nakajima, Y.; Yamamoto, T.; Takahashi, N.; Takeda, K.; Dobashi, S.; et al. Homology-based Radiomic Features for Prediction of the Prognosis of Lung Cancer Based on CT-based Radiomics. Med. Phys. 2020, 47, 2197–2205. [Google Scholar] [CrossRef]
- Fu, J.; Singhrao, K.; Zhong, X.; Gao, Y.; Qi, S.X.; Yang, Y.; Ruan, D.; Lewis, J.H. An Automatic Deep Learning–Based Workflow for Glioblastoma Survival Prediction Using Preoperative Multimodal MR Images: A Feasibility Study. Adv. Radiat. Oncol. 2021, 6, 100746. [Google Scholar] [CrossRef]
- Truhn, D.; Schrading, S.; Haarburger, C.; Schneider, H.; Merhof, D.; Kuhl, C. Radiomic versus Convolutional Neural Networks Analysis for Classification of Contrast-Enhancing Lesions at Multiparametric Breast MRI. Radiology 2019, 290, 290–297. [Google Scholar] [CrossRef]
- Huang, L.; Chen, J.; Hu, W.; Xu, X.; Liu, D.; Wen, J.; Lu, J.; Cao, J.; Zhang, J.; Gu, Y.; et al. Assessment of a Radiomic Signature Developed in a General NSCLC Cohort for Predicting Overall Survival of ALK-Positive Patients With Different Treatment Types. Clin. Lung Cancer 2019, 20, e638–e651. [Google Scholar] [CrossRef]
- Shi, Z.; Zhovannik, I.; Traverso, A.; Dankers, F.J.W.M.; Deist, T.M.; Kalendralis, P.; Monshouwer, R.; Bussink, J.; Fijten, R.; Aerts, H.J.W.L.; et al. Distributed Radiomics as a Signature Validation Study Using the Personal Health Train Infrastructure. Sci. Data 2019, 6, 218. [Google Scholar] [CrossRef] [Green Version]
- Haarburger, C.; Weitz, P.; Rippel, O.; Merhof, D. Image-Based Survival Prediction for Lung Cancer Patients Using CNNS. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1197–1201. [Google Scholar]
- Meyer, M.; Ronald, J.; Vernuccio, F.; Nelson, R.C.; Ramirez-Giraldo, J.C.; Solomon, J.; Patel, B.N.; Samei, E.; Marin, D. Reproducibility of CT Radiomic Features within the Same Patient: Influence of Radiation Dose and CT Reconstruction Settings. Radiology 2019, 293, 583–591. [Google Scholar] [CrossRef]
- Zhao, B.; Tan, Y.; Tsai, W.-Y.; Qi, J.; Xie, C.; Lu, L.; Schwartz, L.H. Reproducibility of Radiomics for Deciphering Tumor Phenotype with Imaging. Sci. Rep. 2016, 6, 23428. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The Bridge between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and Limitations of Radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Dataset | Tumor Entity | Patients [n] | Lesions [n] | Age * [Years] | Sex ** | Cropping [Pixels] | Modality | Reference |
|---|---|---|---|---|---|---|---|---|
| NSCLC | Non-small cell lung carcinoma | 421 | 487 | 68 ± 10 | 290M, 131W | 128 × 128 | CT | [1,10,11] |
| LIDC | Lung cancer | 875 | 1175 | NA | NA | 128 × 128 | CT | [11,12,13] |
| LiTS | Liver tumor | 131 | 908 | NA | NA | 192 × 192 | CT | [14] |
| KiTS | Kidney tumor | 210 | 253 | 58 ± 14 | 123M, 87W | 192 × 192 | CT | [15] |
| BraTS | Glioblastoma and Lower Grade Glioma | 335 | 335 | 61 ± 13 UNK: 19 | NA | 192 × 128 | MRI | [9,16,17] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller-Franzes, G.; Nebelung, S.; Schock, J.; Haarburger, C.; Khader, F.; Pedersoli, F.; Schulze-Hagen, M.; Kuhl, C.; Truhn, D. Reliability as a Precondition for Trust—Segmentation Reliability Analysis of Radiomic Features Improves Survival Prediction. Diagnostics 2022, 12, 247. https://doi.org/10.3390/diagnostics12020247
Müller-Franzes G, Nebelung S, Schock J, Haarburger C, Khader F, Pedersoli F, Schulze-Hagen M, Kuhl C, Truhn D. Reliability as a Precondition for Trust—Segmentation Reliability Analysis of Radiomic Features Improves Survival Prediction. Diagnostics. 2022; 12(2):247. https://doi.org/10.3390/diagnostics12020247
Chicago/Turabian StyleMüller-Franzes, Gustav, Sven Nebelung, Justus Schock, Christoph Haarburger, Firas Khader, Federico Pedersoli, Maximilian Schulze-Hagen, Christiane Kuhl, and Daniel Truhn. 2022. "Reliability as a Precondition for Trust—Segmentation Reliability Analysis of Radiomic Features Improves Survival Prediction" Diagnostics 12, no. 2: 247. https://doi.org/10.3390/diagnostics12020247
APA StyleMüller-Franzes, G., Nebelung, S., Schock, J., Haarburger, C., Khader, F., Pedersoli, F., Schulze-Hagen, M., Kuhl, C., & Truhn, D. (2022). Reliability as a Precondition for Trust—Segmentation Reliability Analysis of Radiomic Features Improves Survival Prediction. Diagnostics, 12(2), 247. https://doi.org/10.3390/diagnostics12020247







