An Antibody-Aptamer-Hybrid Lateral Flow Assay for Detection of CXCL9 in Antibody-Mediated Rejection after Kidney Transplantation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Samples, Data and Ethic Vote
2.2. Determination of Biomarkers
2.3. Statistics
2.4. Generation and Selection of Monoclonal Antibody against CXCL9
2.5. Aptamer Sequence
2.6. Further Components of the LFA
2.7. Preparation of Gold Nanoparticle-Aptamer Conjugates (AuNP-G123)
2.8. Preparation of Test Strips for the LFA
2.9. LFA Evaluation with Technical and Spiked Samples
3. Results
3.1. Patients
3.1.1. Characteristics of the Two Independent Patient Cohorts
3.1.2. Immunosuppression and Graft Function
3.1.3. Panel-Reactive Antibodies (PRA) and DSAs
3.2. Biomarkers
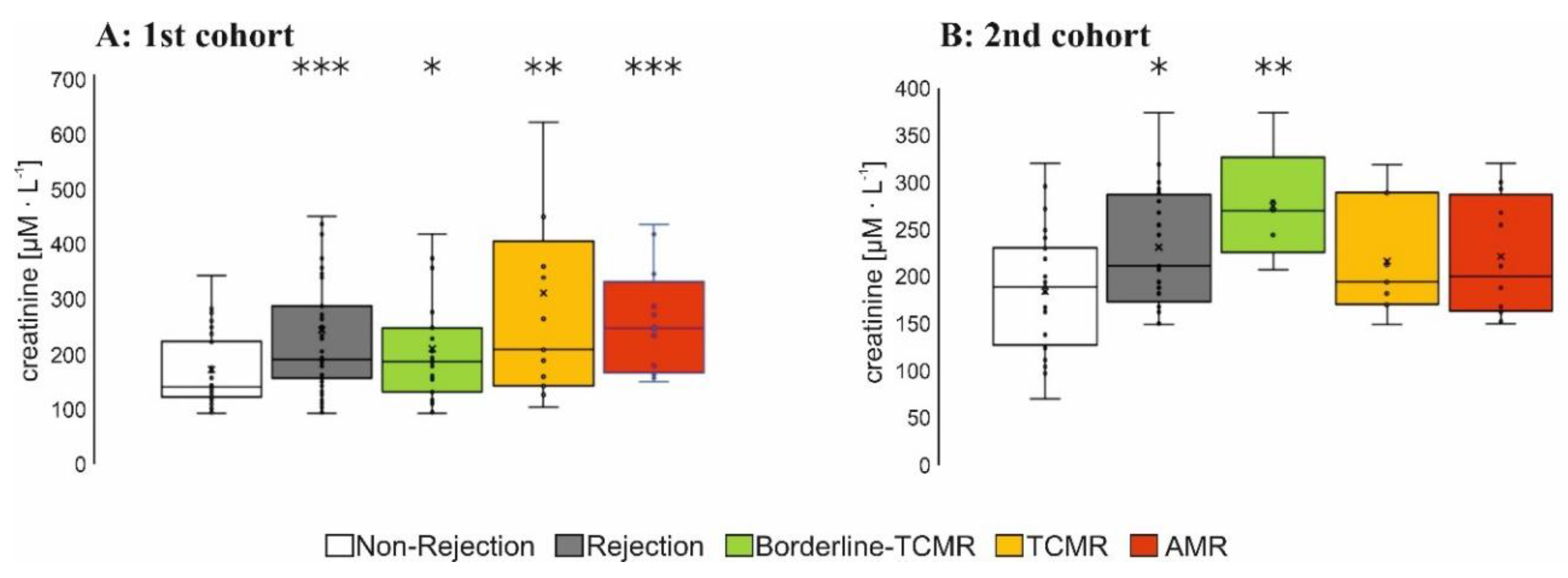
3.2.1. Plasma Level of Relevant Biomarkers in Patients with AMR
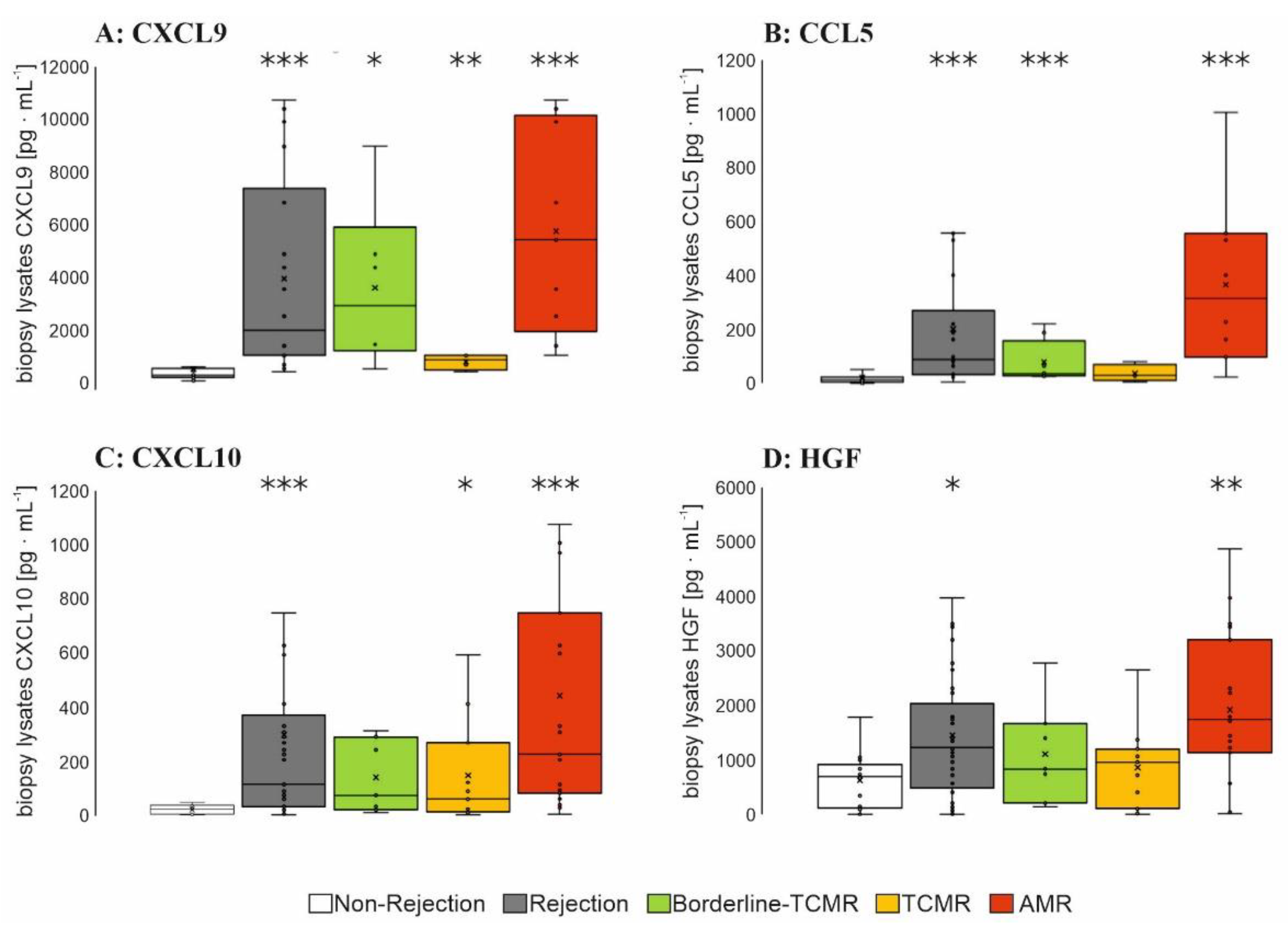
3.2.2. Biopsy Lysate Screening
3.2.3. Urine Screening
3.3. CXCL9-Specific LFA
3.3.1. Selection of a High-Affinity Monoclonal Antibody for the CXCL9-Specific LFA
3.3.2. Coupling of a CXCL9-Binding Aptamer (AuNP-G123) to AuNPs
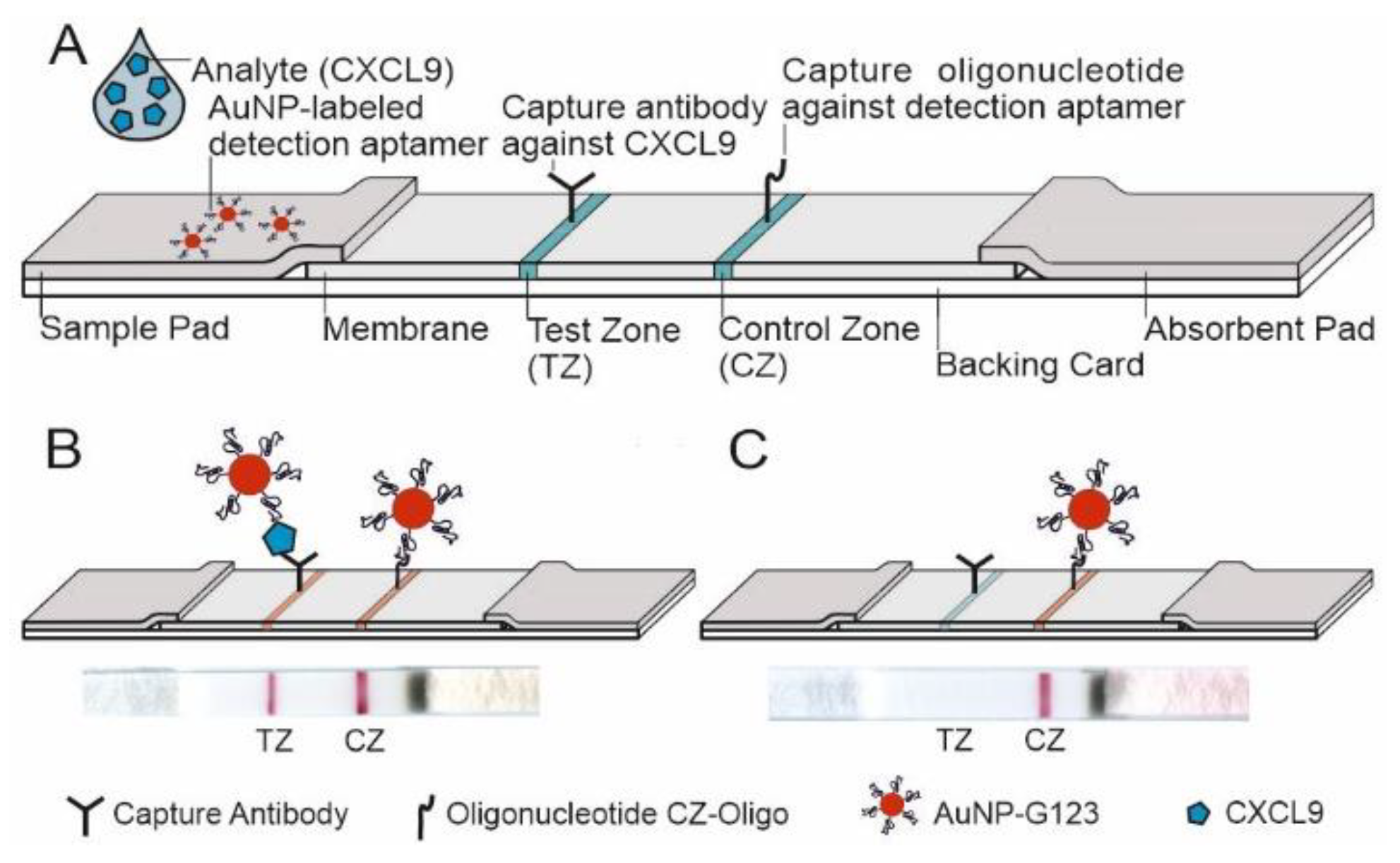
3.3.3. Design of the LFA and LOD
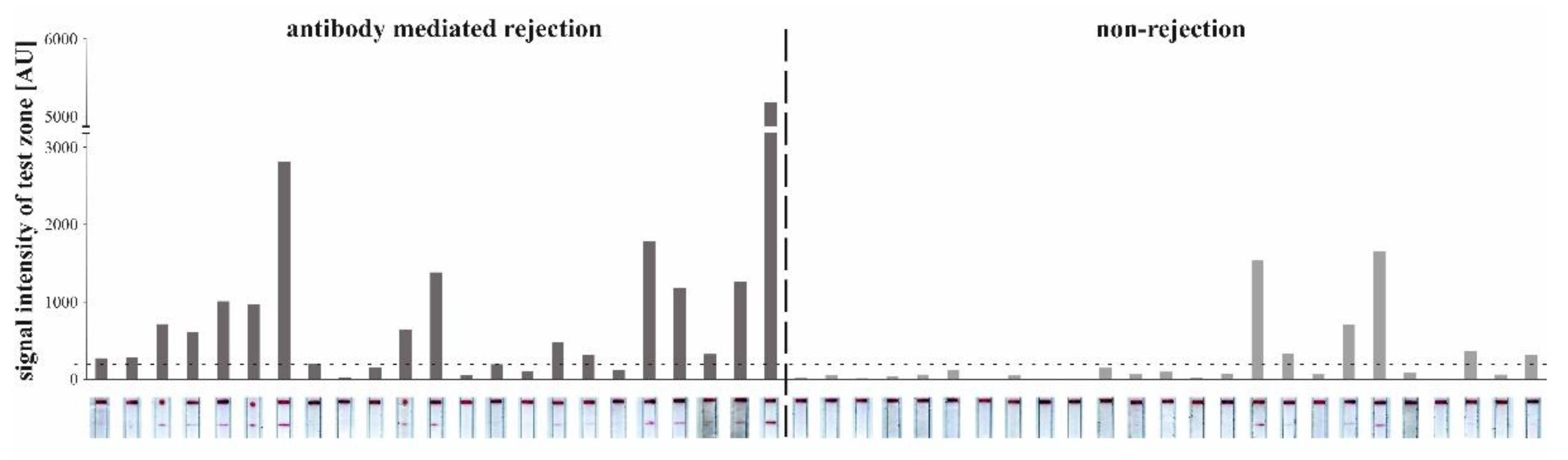
3.3.4. Detection of CXCL9 by LFA in Patients with AMR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loupy, A.; Lefaucheur, C. Antibody-Mediated Rejection of Solid-Organ Allografts. N. Engl. J. Med. 2018, 379, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R. Anti-Interleukin 6 Therapeutics for Chronic Antibody-Mediated Rejection In Kidney Transplant Recipients. Exp. Clin. Transplant. 2022. [Google Scholar] [CrossRef] [PubMed]
- Halloran, P.F.; de Freitas, D.G.; Einecke, G.; Famulski, K.S.; Hidalgo, L.G.; MengeL, M.; Reeve, J.; Sellares, J.; Sis, B. An integrated view of molecular changes, histopathology and outcomes in kidney transplants. Am. J. Transplant. 2010, 10, 2223–2230. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.J.; Goto, R. Mechanisms of rejection: Current perspectives. Transplantation 2012, 93, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Haas, M.; Loupy, A.; Lefaucheur, C.; Roufosse, C.; Glotz, D.; Seron, D.; Nankivell, B.J.; Halloran, P.F.; Colvin, R.B.; Akalin, E.; et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am. J. Transplant. 2018, 18, 293–307. [Google Scholar] [CrossRef] [Green Version]
- Schinstock, C.A.; Cosio, F.; Cheungpasitporn, W.; Dadhania, D.M.; Everly, M.J.; Samaniego-Picota, M.D.; Cornell, L.; Stegall, M.D. The Value of Protocol Biopsies to Identify Patients With De Novo Donor-Specific Antibody at High Risk for Allograft Loss. Am. J. Transplant. 2017, 17, 1574–1584. [Google Scholar] [CrossRef] [Green Version]
- Bartel, G.; Regele, H.; Wahrmann, M.; Huttary, N.; Exner, M.; Hörl, W.H.; Böhmig, G.A. Posttransplant HLA alloreactivity in stable kidney transplant recipients-incidences and impact on long-term allograft outcomes. Am. J. Transplant. 2008, 8, 2652–2660. [Google Scholar] [CrossRef]
- Gebel, H.M.; Bray, R.A. HLA antibody detection with solid phase assays: Great expectations or expectations too great? Am. J. Transplant. 2014, 14, 1964–1975. [Google Scholar] [CrossRef]
- Eskandary, F.; Bond, G.; Kozakowski, N.; Regele, H.; Marinova, L.; Wahrmann, M.; Kikić, Ž.; Haslacher, H.; Rasoul-Rockenschaub, S.; Kaltenecker, C.C.; et al. Diagnostic Contribution of Donor-Specific Antibody Characteristics to Uncover Late Silent Antibody-Mediated Rejection-Results of a Cross-Sectional Screening Study. Transplantation 2017, 101, 631–641. [Google Scholar] [CrossRef]
- Tait, B.D.; Süsal, C.; Gebel, H.M.; Nickerson, P.W.; Zachary, A.A.; Claas, F.H.J.; Reed, E.F.; Bray, R.A.; Campbell, P.; Chapman, J.R.; et al. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation 2013, 95, 19–47. [Google Scholar] [CrossRef] [Green Version]
- Schinstock, C.A.; Gandhi, M.J.; Stegall, M.D. Interpreting Anti-HLA Antibody Testing Data: A Practical Guide for Physicians. Transplantation 2016, 100, 1619–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, M. The significance of C4d staining with minimal histologic abnormalities. Curr. Opin. Organ Transplant. 2010, 15, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Halloran, P.F.; Venner, J.M.; Madill-Thomsen, K.S.; Einecke, G.; Parkes, M.D.; Hidalgo, L.G.; Famulski, K.S. Review: The transcripts associated with organ allograft rejection. Am. J. Transplant. 2018, 18, 785–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnamoorthy, S.; Kyeso, Y. Challenges of Diagnosing Antibody-Mediated Rejection: The Role of Invasive and Non-Invasive Biomarkers. Medicina (Kaunas) 2021, 57, 439. [Google Scholar] [CrossRef] [PubMed]
- Quaglia, M.; Merlotti, G.; Guglielmetti, G.; Castellano, G.; Cantaluppi, V. Recent Advances on Biomarkers of Early and Late Kidney Graft Dysfunction. Int. J. Mol. Sci. 2020, 21, 5404. [Google Scholar] [CrossRef] [PubMed]
- Suviolahti, E.; Ge, S.; Nast, C.C.; Mirocha, J.; Karasyov, A.; White, M.; Jordan, S.C.; Toyoda, M. Genes associated with antibody-dependent cell activation are overexpressed in renal biopsies from patients with antibody-mediated rejection. Transpl. Immunol. 2015, 32, 9–17. [Google Scholar] [CrossRef]
- Parkes, M.D.; Halloran, P.F.; Hidalgo, L.G. Evidence for CD16a-Mediated NK Cell Stimulation in Antibody-Mediated Kidney Transplant Rejection. Transplantation 2017, 101, e102–e111. [Google Scholar] [CrossRef]
- Ho, J.; Wiebe, C.; Gibson, I.W.; Rush, D.N.; Nickerson, P.W. Immune monitoring of kidney allografts. Am. J. Kidney Dis. 2012, 60, 629–640. [Google Scholar] [CrossRef]
- Raju, R.; Malloy, A.; Shah, T.; Smith, R.; Oaks, M.; Hosenpud, J.D. Alloimmune induction of endothelial cell-derived interferon-gamma-inducible chemokines. Transplantation 2003, 75, 1072–1074. [Google Scholar] [CrossRef]
- Luster, A.D.; Unkeless, J.C.; Ravetch, J.V. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 1985, 315, 672–676. [Google Scholar] [CrossRef]
- Demmers, M.W.H.J.; Baan, C.C.; van Beelen, E.; Ijzermans, J.N.M.; Weimar, W.; Rowshani, A.T. Differential effects of activated human renal epithelial cells on T-cell migration. PLoS ONE 2013, 8, e64916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, L.; Wiehler, F.; Bräsen, J.H.; Gwinner, W.; Greite, R.; Kreimann, K.; Thorenz, A.; Derlin, K.; Teng, B.; Rong, S.; et al. Chemokine CXCL13 as a New Systemic Biomarker for B-Cell Involvement in Acute T Cell-Mediated Kidney Allograft Rejection. Int. J. Mol. Sci. 2019, 20, 2552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krupickova, L.; Fialova, M.; Novotny, M.; Svachova, V.; Mezerova, K.; Cecrdlova, E.; Viklicky, O.; Striz, I. Chemokine Profiles Are Affected in Serum of Patients with Acute Rejection of Kidney Allograft. Mediat. Inflamm. 2021, 2021, 5513690. [Google Scholar] [CrossRef] [PubMed]
- Piotti, G.; Palmisano, A.; Maggiore, U.; Buzio, C. Vascular endothelium as a target of immune response in renal transplant rejection. Front. Immunol. 2014, 5, 505. [Google Scholar] [CrossRef] [Green Version]
- Cross, A.R.; Glotz, D.; Mooney, N. The Role of the Endothelium during Antibody-Mediated Rejection: From Victim to Accomplice. Front. Immunol. 2018, 9, 106. [Google Scholar] [CrossRef] [Green Version]
- Madill-Thomsen, K.S.; Böhmig, G.A.; Bromberg, J.; Einecke, G.; Eskandary, F.; Gupta, G.; Hidalgo, L.G.; Myslak, M.; Viklicky, O.; Perkowska-Ptasinska, A.; et al. Donor-Specific Antibody Is Associated with Increased Expression of Rejection Transcripts in Renal Transplant Biopsies Classified as No Rejection. J. Am. Soc. Nephrol. 2021, 32, 2743–2758. [Google Scholar] [CrossRef]
- Guzzi, F.; Cirillo, L.; Buti, E.; Becherucci, F.; Errichiello, C.; Roperto, R.M.; Hunter, J.P.; Romagnani, P. Urinary Biomarkers for Diagnosis and Prediction of Acute Kidney Allograft Rejection: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 6889. [Google Scholar] [CrossRef]
- Adachi, T.; Nakamura, Y. Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application. Molecules 2019, 24, 4229. [Google Scholar] [CrossRef] [Green Version]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Chen, L.; Chikkanna, A.; Chen, S.; Brusius, I.; Sbuh, N.; Veedu, R.N. Development of nucleic acid aptamer-based lateral flow assays: A robust platform for cost-effective point-of-care diagnosis. Theranostics 2021, 11, 5174–5196. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Mao, X.; Phillips, J.A.; Xu, H.; Tan, W.; Zeng, L. Aptamer-nanoparticle strip biosensor for sensitive detection of cancer cells. Anal. Chem. 2009, 81, 10013–10018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad Raston, N.H.; Nguyen, V.-T.; Gu, M.B. A new lateral flow strip assay (LFSA) using a pair of aptamers for the detection of Vaspin. Biosens. Bioelectron. 2017, 93, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.-R.; Sekhon, S.S.; Rhee, S.-K.; Ko, J.H.; Ahn, J.-Y.; Min, J.; Kim, Y.-H. Aptamer-Based Paper Strip Sensor for Detecting Vibrio fischeri. ACS Comb. Sci. 2018, 20, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Schüling, T.; Eilers, A.; Scheper, T.; Walter, J. Aptamer-based lateral flow assays. AIMS Bioeng. 2018, 5, 78–102. [Google Scholar] [CrossRef]
- Wu, W.; Li, J.; Pan, D.; Li, J.; Song, S.; Rong, M.; Li, Z.; Gao, J.; Lu, J. Gold nanoparticle-based enzyme-linked antibody-aptamer sandwich assay for detection of Salmonella Typhimurium. ACS Appl. Mater. Interfaces 2014, 6, 16974–16981. [Google Scholar] [CrossRef]
- Mahmoud, M.; Ruppert, C.; Rentschler, S.; Laufer, S.; Deigner, H.-P. Combining aptamers and antibodies: Lateral flow quantification for thrombin and interleukin-6 with smartphone readout. Sens. Actuators B Chem. 2021, 333, 129246. [Google Scholar] [CrossRef]
- Kang, J.; Kim, M.-G. Advancements in DNA-assisted Immunosensors. BioChip J. 2020, 14, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Immenschuh, S.; Zilian, E.; Dämmrich, M.E.; Schwarz, A.; Gwinner, W.; Becker, J.U.; Blume, C.A. Indicators of treatment responsiveness to rituximab and plasmapheresis in antibody-mediated rejection after kidney transplantation. Transplantation 2015, 99, 56–62. [Google Scholar] [CrossRef]
- Loupy, A.; Haas, M.; Solez, K.; Racusen, L.; Glotz, D.; Seron, D.; Nankivell, B.J.; Colvin, R.B.; Afrouzian, M.; Akalin, E.; et al. The Banff 2015 Kidney Meeting Report: Current Challenges in Rejection Classification and Prospects for Adopting Molecular Pathology. Am. J. Transplant. 2017, 17, 28–41. [Google Scholar] [CrossRef]
- Sis, B.; MengeL, M.; Haas, M.; Colvin, R.B.; Halloran, P.F.; Racusen, L.C.; Solez, K.; Baldwin, W.M.; Bracamonte, E.R.; Broecker, V.; et al. Banff ‘09 meeting report: Antibody mediated graft deterioration and implementation of Banff working groups. Am. J. Transplant. 2010, 10, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Egelkamp, J.; Chichelnitskiy, E.; Kühne, J.F.; Wandrer, F.; Daemen, K.; Keil, J.; Bräsen, J.H.; Schmitz, J.; Bellmàs-Sanz, R.; Iordanidis, S.; et al. Back signaling of HLA class I molecules and T/NK cell receptor ligands in epithelial cells reflects the rejection-specific microenvironment in renal allograft biopsies. Am. J. Transplant. 2019, 19, 2692–2704. [Google Scholar] [CrossRef] [PubMed]
- Luft, T.; Dietrich, S.; Falk, C.; Conzelmann, M.; Hess, M.; Benner, A.; Neumann, F.; Isermann, B.; Hegenbart, U.; Ho, A.D.; et al. Steroid-refractory GVHD: T-cell attack within a vulnerable endothelial system. Blood 2011, 118, 1685–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef]
- Sudan, K.; Vijayan, V.; Madyaningrana, K.; Gueler, F.; Igarashi, K.; Foresti, R.; Motterlini, R.; Immenschuh, S. TLR4 activation alters labile heme levels to regulate BACH1 and heme oxygenase-1 expression in macrophages. Free Radic. Biol. Med. 2019, 137, 131–142. [Google Scholar] [CrossRef]
- Phung, N.L.; Walter, J.G.; Jonczyk, R.; Seiler, L.K.; Scheper, T.; Blume, C. Development of an Aptamer-Based Lateral Flow Assay for the Detection of C-Reactive Protein Using Microarray Technology as a Prescreening Platform. ACS Comb. Sci. 2020, 22, 617–629. [Google Scholar] [CrossRef]
- Seiler, L.K.; Jonczyk, R.; Lindner, P.; Phung, N.L.; Falk, C.S.; Kaufeld, J.; Gwinner, W.; Scheffner, I.; Immenschuh, S.; Blume, C. A new lateral flow assay to detect sIL-2R during T-cell mediated rejection after kidney transplantation. Analyst 2021. [Google Scholar] [CrossRef]
- Piñeiro, G.J.; Montagud-Marrahi, E.; Ríos, J.; Ventura-Aguiar, P.; Cucchiari, D.; Revuelta, I.; Lozano, M.; Cid, J.; Cofan, F.; Esforzado, N.; et al. Influence of Persistent Inflammation in Follow-Up Biopsies After Antibody-Mediated Rejection in Kidney Transplantation. Front. Med. 2021, 8, 761919. [Google Scholar] [CrossRef]
- Nickerson, P.W. What have we learned about how to prevent and treat antibody-mediated rejection in kidney transplantation? Am. J. Transplant. 2020, 20 (Suppl. 4), 12–22. [Google Scholar] [CrossRef]
- Einecke, G.; Reeve, J.; Gupta, G.; Böhmig, G.A.; Eskandary, F.; Bromberg, J.S.; Budde, K.; Halloran, P.F. Factors associated with kidney graft survival in pure antibody-mediated rejection at the time of indication biopsy: Importance of parenchymal injury but not disease activity. Am. J. Transplant. 2021, 21, 1391–1401. [Google Scholar] [CrossRef]
- Mühlbacher, J.; Doberer, K.; Kozakowski, N.; Regele, H.; Camovic, S.; Haindl, S.; Bond, G.; Haslacher, H.; Eskandary, F.; Reeve, J.; et al. Non-invasive Chemokine Detection: Improved Prediction of Antibody-Mediated Rejection in Donor-Specific Antibody-Positive Renal Allograft Recipients. Front. Med. 2020, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Aizenstein, B.D.; Puchalski, A.; Burmania, J.A.; Hamawy, M.M.; Knechtle, S.J. Elevation of CXCR3-binding chemokines in urine indicates acute renal-allograft dysfunction. Am. J. Transplant. 2004, 4, 432–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, I.A.; Spiegler, S.; Kiss, E.; Gauer, S.; Sichler, O.; Scheuermann, E.H.; Ackermann, H.; Pfeilschifter, J.M.; Geiger, H.; Gröne, H.-J.; et al. Prediction of acute renal allograft rejection by urinary monokine induced by IFN-gamma (MIG). J. Am. Soc. Nephrol. 2005, 16, 1849–1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanssen, O.; Weekers, L.; Lovinfosse, P.; Jadoul, A.; Bonvoisin, C.; Bouquegneau, A.; Grosch, S.; Huynen, A.; Anglicheau, D.; Hustinx, R.; et al. Diagnostic yield of 18 F-FDG PET/CT imaging and urinary CXCL9/creatinine levels in kidney allograft subclinical rejection. Am. J. Transplant. 2020, 20, 1402–1409. [Google Scholar] [CrossRef]
- Blydt-Hansen, T.D.; Sharma, A.; Gibson, I.W.; Wiebe, C.; Sharma, A.P.; Langlois, V.; Teoh, C.W.; Rush, D.; Nickerson, P.; Wishart, D.; et al. Validity and utility of urinary CXCL10/Cr immune monitoring in pediatric kidney transplant recipients. Am. J. Transplant. 2021, 21, 1545–1555. [Google Scholar] [CrossRef]
- Hricik, D.E.; Nickerson, P.; Formica, R.N.; Poggio, E.D.; Rush, D.; Newell, K.A.; Goebel, J.; Gibson, I.W.; Fairchild, R.L.; Riggs, M.; et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am. J. Transplant. 2013, 13, 2634–2644. [Google Scholar] [CrossRef] [Green Version]
- Tinel, C.; Devresse, A.; Vermorel, A.; Marx, D.; Caillard, S.; Legendre, C.; Rabant, M.; Anglicheau, D. An Optimized Integrative Model Using Urinary Chemokines for Noninvasive Diagnosis of Acute Allograft Rejection. Am. J. Transplant. 2020, 20, 313–314. [Google Scholar] [CrossRef]
- Tinel, C.; Vermorel, A.; Picciotto, D.; Morin, L.; Devresse, A.; Sauvaget, V.; Lebreton, X.; Aouni, L.; Prié, D.; Brabant, S.; et al. Deciphering the Prognostic and Predictive Value of Urinary CXCL10 in Kidney Recipients with BK Virus Reactivation. Front. Immunol. 2020, 11, 604353. [Google Scholar] [CrossRef]
- Tokunaga, R.; Zhang, W.; Naseem, M.; Puccini, A.; Berger, M.D.; Soni, S.; McSkane, M.; Baba, H.; Lenz, H.-J. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat. Rev. 2018, 63, 40–47. [Google Scholar] [CrossRef]
- Ellis, S.L.; Gysbers, V.; Manders, P.M.; Li, W.; Hofer, M.J.; Müller, M.; Campbell, I.L. The cell-specific induction of CXC chemokine ligand 9 mediated by IFN-gamma in microglia of the central nervous system is determined by the myeloid transcription factor PU.1. J. Immunol. 2010, 185, 1864–1877. [Google Scholar] [CrossRef] [Green Version]
- Liao, F.; Rabin, R.L.; Yannelli, J.R.; Koniaris, L.G.; Vanduri, P.; Farber, J.M. Human Mig Chemokine: Biochemical and Functional Characterization. J. Exp. Med. 1995, 182, 1301–1314. [Google Scholar] [CrossRef]
- Farber, J.M. A macrophage mRNA selectively induced by gamma-interferon encodes a member of the platelet factor 4 family of cytokines. Proc. Natl. Acad. Sci. USA 1990, 87, 5238–5242. [Google Scholar] [CrossRef] [Green Version]
- Farber, J.M. HuMig: A new human member of the chemokine family of cytokines. Biochem. Biophys. Res. Commun. 1993, 192, 223–230. [Google Scholar] [CrossRef]
- Yan, Q.; Jiang, H.; Wang, B.; Sui, W.; Zhou, H.; Zou, G. Expression and Significance of RANTES and MCP-1 in Renal Tissue With Chronic Renal Allograft Dysfunction. Transplant. Proc. 2016, 48, 2034–2039. [Google Scholar] [CrossRef] [Green Version]
- Pavlova, Y.; Viklicky, O.; Slatinska, J.; Bürgelova, M.; Süsal, C.; Skibova, J.; Honsová, E.; Striz, I.; Kolesar, L.; Slavcev, A. Soluble CD30 and Hepatocyte growth factor as predictive markers of antibody-mediated rejection of the kidney allograft. Transpl. Immunol. 2011, 25, 72–76. [Google Scholar] [CrossRef]
- Rabant, M.; Amrouche, L.; Morin, L.; Bonifay, R.; Lebreton, X.; Aouni, L.; Benon, A.; Sauvaget, V.; Le Vaillant, L.; Aulagnon, F.; et al. Early Low Urinary CXCL9 and CXCL10 Might Predict Immunological Quiescence in Clinically and Histologically Stable Kidney Recipients. Am. J. Transplant. 2016, 16, 1868–1881. [Google Scholar] [CrossRef]
- Dragun, D.; Catar, R.; Philippe, A. Non-HLA antibodies in solid organ transplantation: Recent concepts and clinical relevance. Curr. Opin. Organ Transplant. 2013, 18, 430–435. [Google Scholar] [CrossRef] [Green Version]
- Cardinal, H.; Dieudé, M.; Hébert, M.-J. The Emerging Importance of Non-HLA Autoantibodies in Kidney Transplant Complications. J. Am. Soc. Nephrol. 2017, 28, 400–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oellerich, M.; Shipkova, M.; Asendorf, T.; Walson, P.D.; Schauerte, V.; Mettenmeyer, N.; Kabakchiev, M.; Hasche, G.; Gröne, H.-J.; Friede, T.; et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am. J. Transplant. 2019, 19, 3087–3099. [Google Scholar] [CrossRef] [Green Version]
- Seiler, L.K. Lateral Flow Test zur Abstoßungsdiagnostik mittels Antikörper. Ph.D. Thesis, Leibniz University Hannover, Hannover, Germany, 2021. [Google Scholar]
- Siegl, J.; Nikolin, C.; Phung, N.L.; Thoms, S.; Blume, C.; Mayer, G. Split–Combine Click-SELEX Reveals Ligands Recognizing the Transplant Rejection Biomarker CXCL9. ACS Chem. Biol. 2022. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. Chemokines. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef] [Green Version]




| Name | Sequence | Modification | Target |
|---|---|---|---|
| G123 | CACGACGCAAGGGACCACAGGGAGGGAGGGTGGGCAAAGGGCCCTAAGTCCGTAACAAAAACACAGCACGACACCGCAGAGGCA | C6-SS-HEG-5′-3′ | CXCL9 |
| CZ-Oligonucleotide | TGCCTCTGCGGTGT CGTGCT | biotin-HEG-HEG-5′-3′ | G123 |
| Control-Oligonucleotide | CACGACGCAAGGGACCACAGGAGGAGAGTAGGCGATACACGACGTAGCGCAGATAGGACCAAGCAGCACGACACCGCAGAGGCA | C6-SS-HEG-5′-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seiler, L.K.; Phung, N.L.; Nikolin, C.; Immenschuh, S.; Erck, C.; Kaufeld, J.; Haller, H.; Falk, C.S.; Jonczyk, R.; Lindner, P.; et al. An Antibody-Aptamer-Hybrid Lateral Flow Assay for Detection of CXCL9 in Antibody-Mediated Rejection after Kidney Transplantation. Diagnostics 2022, 12, 308. https://doi.org/10.3390/diagnostics12020308
Seiler LK, Phung NL, Nikolin C, Immenschuh S, Erck C, Kaufeld J, Haller H, Falk CS, Jonczyk R, Lindner P, et al. An Antibody-Aptamer-Hybrid Lateral Flow Assay for Detection of CXCL9 in Antibody-Mediated Rejection after Kidney Transplantation. Diagnostics. 2022; 12(2):308. https://doi.org/10.3390/diagnostics12020308
Chicago/Turabian StyleSeiler, Lisa K., Ngoc Linh Phung, Christoph Nikolin, Stephan Immenschuh, Christian Erck, Jessica Kaufeld, Hermann Haller, Christine S. Falk, Rebecca Jonczyk, Patrick Lindner, and et al. 2022. "An Antibody-Aptamer-Hybrid Lateral Flow Assay for Detection of CXCL9 in Antibody-Mediated Rejection after Kidney Transplantation" Diagnostics 12, no. 2: 308. https://doi.org/10.3390/diagnostics12020308
APA StyleSeiler, L. K., Phung, N. L., Nikolin, C., Immenschuh, S., Erck, C., Kaufeld, J., Haller, H., Falk, C. S., Jonczyk, R., Lindner, P., Thoms, S., Siegl, J., Mayer, G., Feederle, R., & Blume, C. A. (2022). An Antibody-Aptamer-Hybrid Lateral Flow Assay for Detection of CXCL9 in Antibody-Mediated Rejection after Kidney Transplantation. Diagnostics, 12(2), 308. https://doi.org/10.3390/diagnostics12020308







