Development of a Novel Evaluation Method for Endoscopic Ultrasound-Guided Fine-Needle Biopsy in Pancreatic Diseases Using Artificial Intelligence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Patients
2.3. EUS-FNB Procedure
2.4. Specimen Processing for EUS-FNB
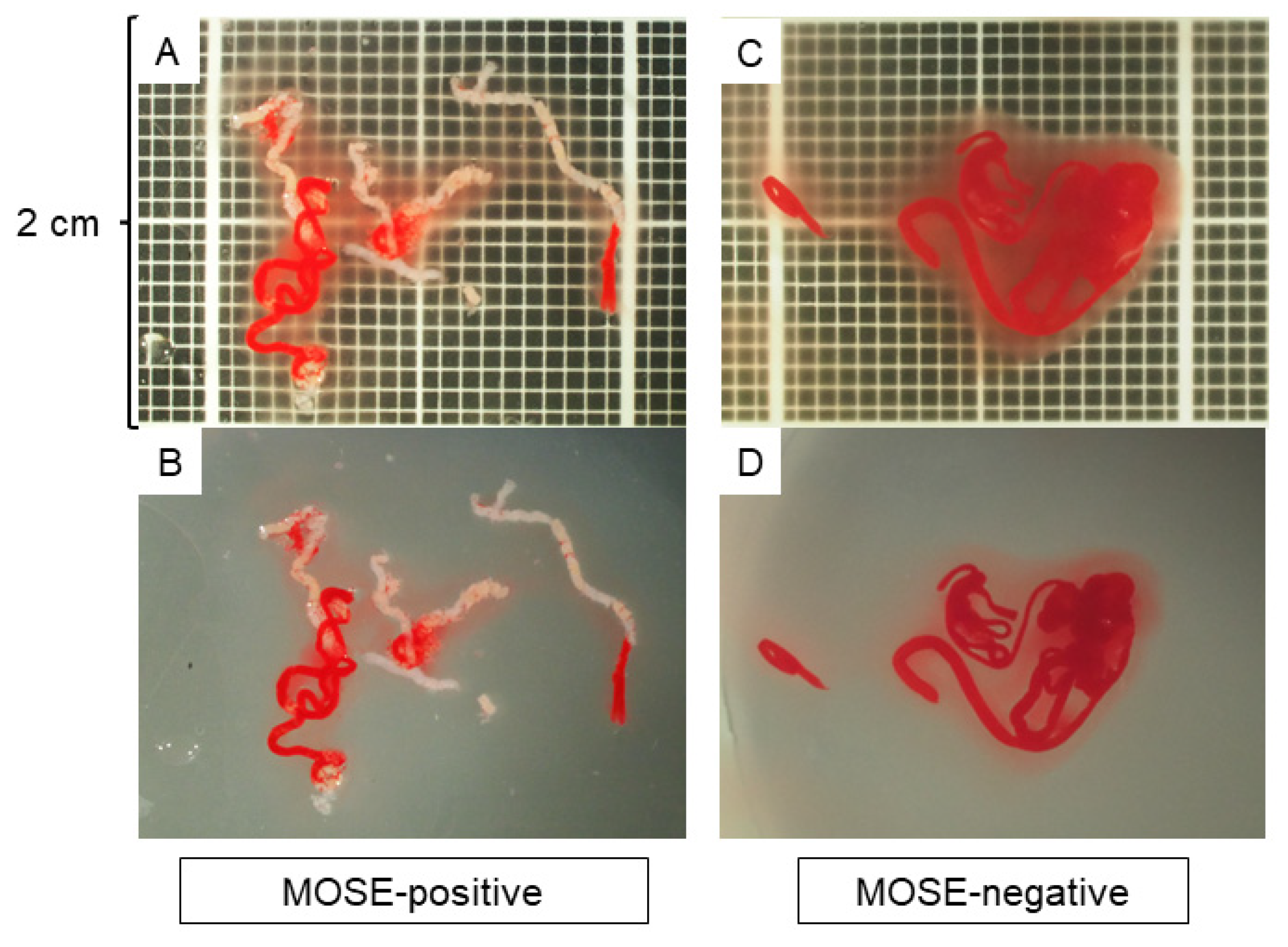
2.5. MOSE and Imaging EUS-FNB Specimens
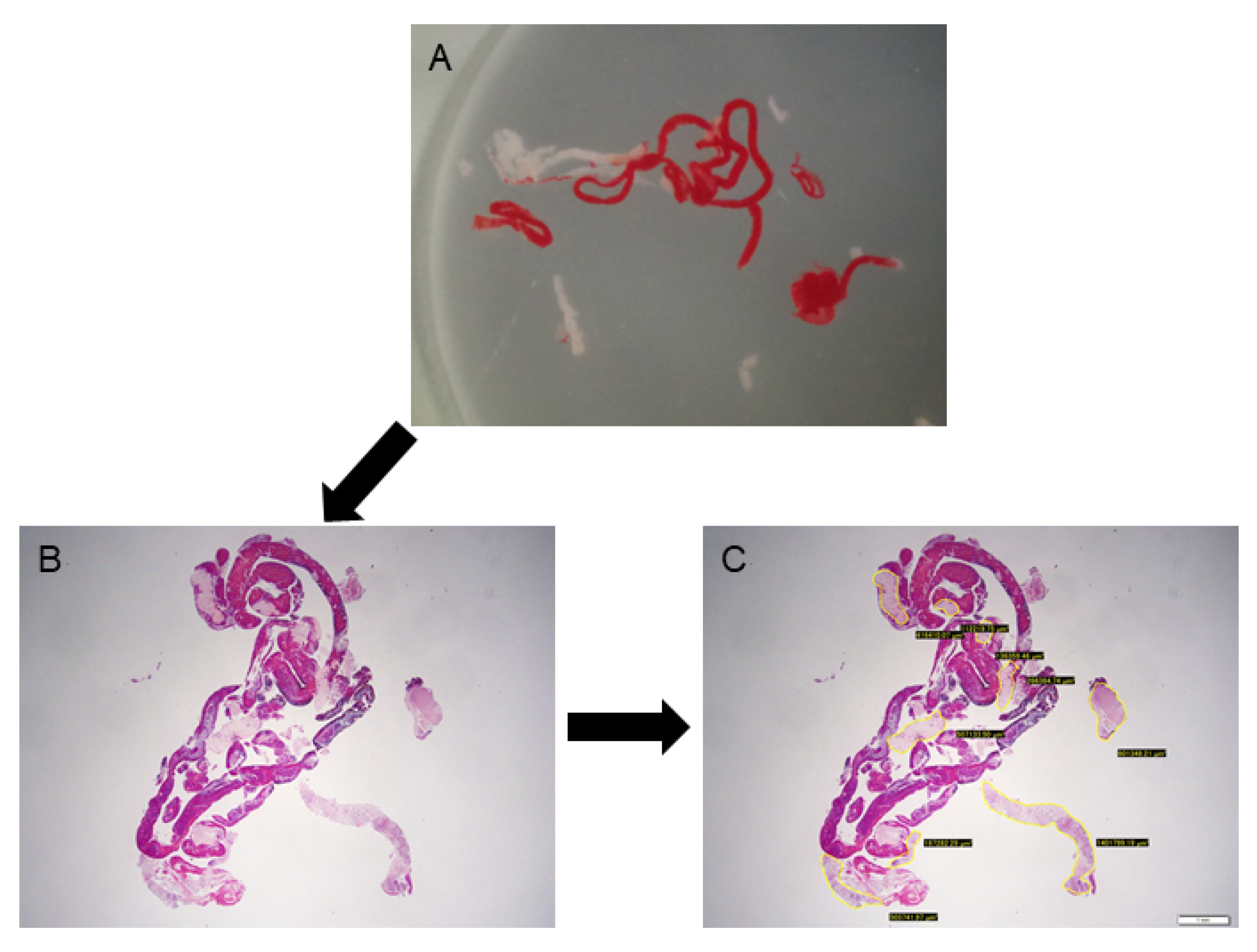
2.6. Histology Evaluation
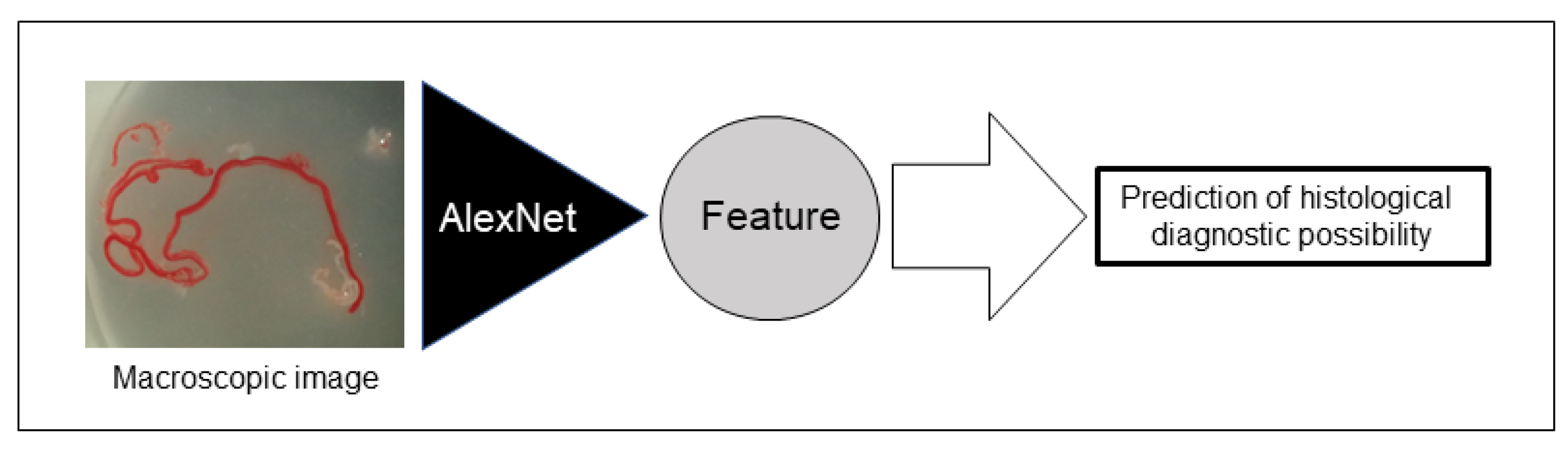
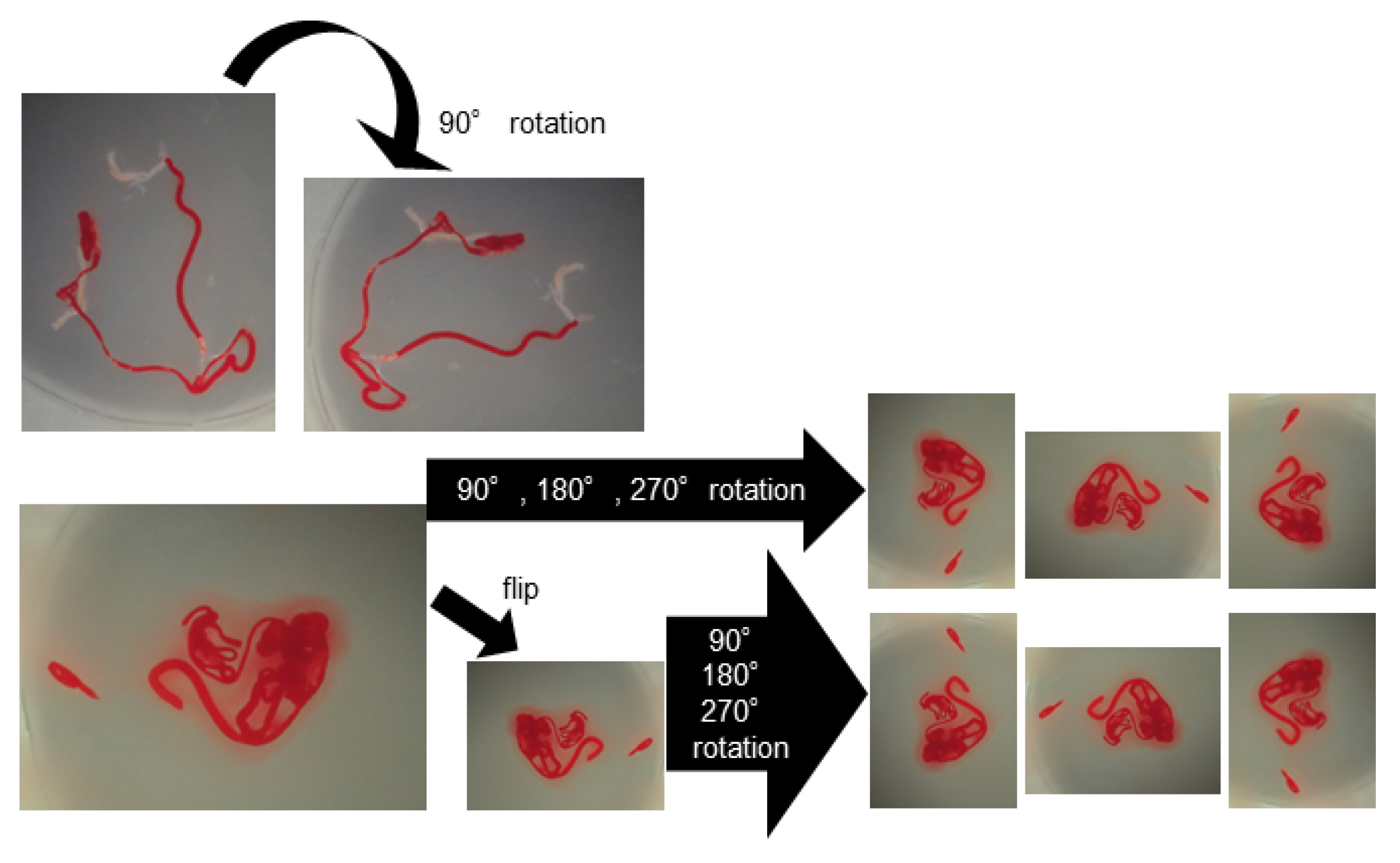
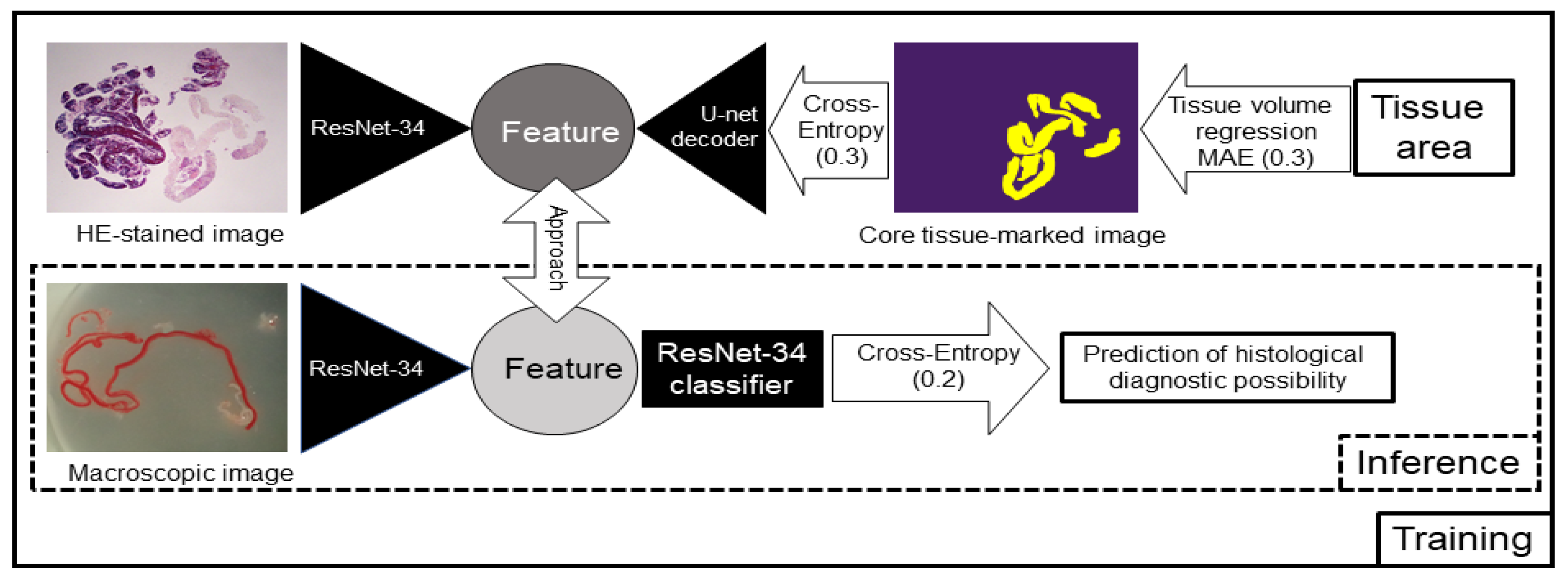
2.7. AI-Based Evaluation Using Deep Learning
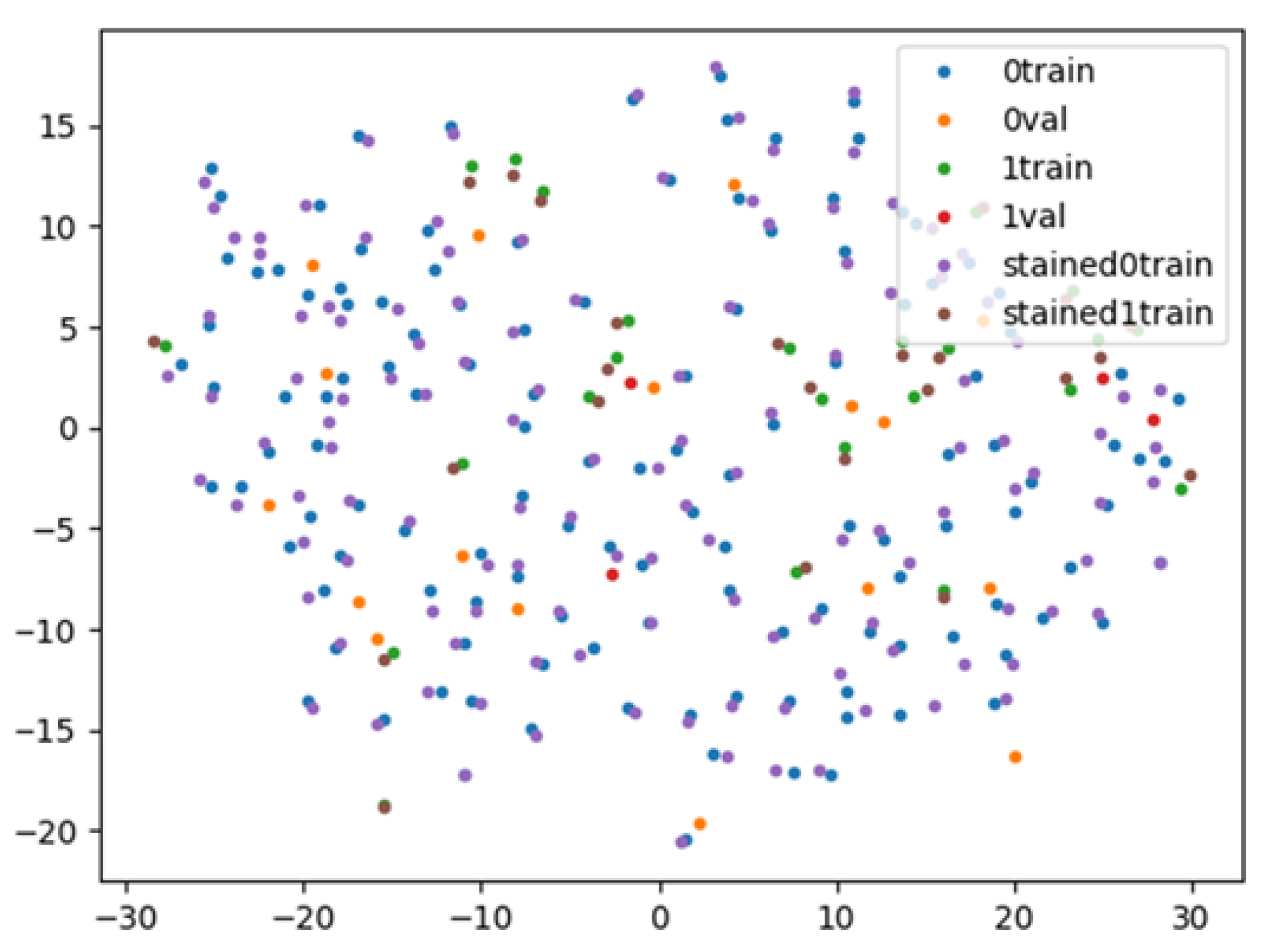
2.8. AI-Based Evaluation Using Contrastive Learning
2.9. Evaluation Items
2.10. Statistical Analysis
3. Results
3.1. AI-Based Evaluation Using Deep Learning
3.2. AI-Based Evaluation Using Contrastive Learning
3.3. Interobserver Agreement among the Endosonographers Performing MOSE
3.4. Association between Lesion Features and MOSE Positivity
3.5. Evaluation of Tissue Sample Area and Diagnostic Accuracy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vilmann, P.; Jacobsen, G.K.; Henriksen, F.W.; Hancke, S. Endoscopic ultrasonography with guided fine needle aspiration biopsy in pancreatic disease. Gastrointest Endosc. 1992, 38, 172–173. [Google Scholar] [CrossRef]
- Ishikawa, T.; Kawashima, H.; Ohno, E.; Tanaka, H.; Sakai, D.; Iida, T.; Nishio, R.; Yamamura, T.; Furukawa, K.; Nakamura, M.; et al. Clinical Impact of EUS-Guided Fine Needle Biopsy Using a Novel Franseen Needle for Histological Assessment of Pancreatic Diseases. Can J. Gastroenterol. Hepatol. 2019, 2019, 8581743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishikawa, T.; Mohamed, R.; Heitman, S.J.; Turbide, C.; Kumar, P.R.; Goto, H.; Hirooka, Y.; Belletrutti, P.J. Diagnostic yield of small histological cores obtained with a new EUS-guided fine needle biopsy system. Surg. Endosc. 2017, 31, 5143–5149. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Garcia, J.; Dominguez-Munoz, J.E.; Abdulkader, I.; Larino-Noia, J.; Eugenyeva, E.; Lozano-Leon, A.; Forteza-Vila, J. Influence of on-site cytopathology evaluation on the diagnostic accuracy of endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) of solid pancreatic masses. Am. J. Gastroenterol. 2011, 106, 1705–1710. [Google Scholar] [CrossRef] [PubMed]
- Crino, S.F.; Larghi, A.; Bernardoni, L.; Parisi, A.; Frulloni, L.; Gabbrielli, A.; Parcesepe, P.; Scarpa, A.; Manfrin, E. Touch imprint cytology on endoscopic ultrasound fine-needle biopsy provides comparable sample quality and diagnostic yield to standard endoscopic ultrasound fine-needle aspiration specimens in the evaluation of solid pancreatic lesions. Cytopathology 2019, 30, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Aadam, A.A.; Wani, S.; Amick, A.; Shah, J.N.; Bhat, Y.M.; Hamerski, C.M.; Klapman, J.B.; Muthusamy, V.R.; Watson, R.R.; Rademaker, A.W.; et al. A randomized controlled cross-over trial and cost analysis comparing endoscopic ultrasound fine needle aspiration and fine needle biopsy. Endosc. Int. Open. 2016, 4, E497–E505. [Google Scholar] [CrossRef] [Green Version]
- Bang, J.Y.; Hawes, R.; Varadarajulu, S. A meta-analysis comparing ProCore and standard fine-needle aspiration needles for endoscopic ultrasound-guided tissue acquisition. Endoscopy 2016, 48, 339–349. [Google Scholar] [CrossRef]
- Mohamadnejad, M.; Mullady, D.; Early, D.S.; Collins, B.; Marshall, C.; Sams, S.; Yen, R.; Rizeq, M.; Romanas, M.; Nawaz, S.; et al. Increasing Number of Passes Beyond 4 Does Not Increase Sensitivity of Detection of Pancreatic Malignancy by Endoscopic Ultrasound-Guided Fine-Needle Aspiration. Clin. Gastroenterol. Hepatol. 2017, 15, 1071–1078. [Google Scholar] [CrossRef]
- Iwashita, T.; Yasuda, I.; Mukai, T.; Doi, S.; Nakashima, M.; Uemura, S.; Mabuchi, M.; Shimizu, M.; Hatano, Y.; Hara, A.; et al. Macroscopic on-site quality evaluation of biopsy specimens to improve the diagnostic accuracy during EUS-guided FNA using a 19-gauge needle for solid lesions: A single-center prospective pilot study (MOSE study). Gastrointest. Endosc. 2015, 81, 177–185. [Google Scholar] [CrossRef]
- Chong, C.C.N.; Lakhtakia, S.; Nguyen, N.; Hara, K.; Chan, W.K.; Puri, R.; Almadi, M.A.; Ang, T.L.; Kwek, A.; Yasuda, I.; et al. Endoscopic ultrasound-guided tissue acquisition with or without macroscopic on-site evaluation: Randomized controlled trial. Endoscopy 2020, 52, 856–863. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, T.; Kubota, K.; Mori, M.; Kikuchi, Y.; Katsuta, L.; Kasahara, M.; Oda, G.; Ishiba, T.; Nakagawa, T.; Tateishi, U. Distinction between benign and malignant breast masses at breast ultrasound using deep learning method with convolutional neural network. Jpn. J. Radiol. 2019, 37, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.W.; Cheung, C.Y.; Lim, G.; Tan, G.S.W.; Quang, N.D.; Gan, A.; Hamzah, H.; Garcia-Franco, R.; San Yeo, I.Y.; Lee, S.Y.; et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA 2017, 318, 2211–2223. [Google Scholar] [CrossRef] [PubMed]
- Horie, Y.; Yoshio, T.; Aoyama, K.; Yoshimizu, S.; Horiuchi, Y.; Ishiyama, A.; Hirasawa, T.; Tsuchida, T.; Ozawa, T.; Ishihara, S.; et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastroint. Endosc. 2019, 89, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, T.; Aoyama, K.; Tanimoto, T.; Ishihara, S.; Shichijo, S.; Ozawa, T.; Ohnishi, T.; Fujishiro, M.; Matsuo, K.; Fujisaki, J.; et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018, 21, 653–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Linares Perez, M.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019, 68, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Kuwahara, T.; Hara, K.; Mizuno, N.; Okuno, N.; Matsumoto, S.; Obata, M.; Kurita, Y.; Koda, H.; Toriyama, K.; Onishi, S.; et al. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin. Transl. Gastroenterol. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- Naito, Y.; Tsuneki, M.; Fukushima, N.; Koga, Y.; Higashi, M.; Notohara, K.; Aishima, S.; Ohike, N.; Tajiri, T.; Yamaguchi, H.; et al. A deep learning model to detect pancreatic ductal adenocarcinoma on endoscopic ultrasound-guided fine-needle biopsy. Sci. Rep. 2021, 11, 8454. [Google Scholar] [CrossRef]
- Matsuzaki, I.; Miyahara, R.; Hirooka, Y.; Funasaka, K.; Ohno, E.; Nakamura, M.; Kawashima, H.; Nukaga, A.; Shimoyama, Y.; Goto, H. Forward-viewing versus oblique-viewing echoendoscopes in the diagnosis of upper GI subepithelial lesions with EUS-guided FNA: A prospective, randomized, crossover study. Gastroint. Endosc. 2015, 82, 287–295. [Google Scholar] [CrossRef]
- Ishikawa, T.; Kawashima, H.; Ohno, E.; Suhara, H.; Hayashi, D.; Hiramatsu, T.; Matsubara, H.; Suzuki, T.; Kuwahara, T.; Ishikawa, E.; et al. Usefulness of endoscopic ultrasound-guided fine-needle biopsy for the diagnosis of autoimmune pancreatitis using a 22-gauge Franseen needle: A prospective multicenter study. Endoscopy 2020, 52, 978–985. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data-Ger. 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A Simple Framework for Contrastive Learning of Visual Representations. Pr. Mach Learn Res. 2020, 119, 1597–1607. [Google Scholar]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef] [Green Version]
- Chang, K.J.; Albers, C.G.; Erickson, R.A.; Butler, J.A.; Wuerker, R.B.; Lin, F. Endoscopic ultrasound-guided fine needle aspiration of pancreatic carcinoma. Am. J. Gastroenterol. 1994, 89, 263–266. [Google Scholar] [PubMed]
- Cleveland, P.; Gill, K.R.; Coe, S.G.; Woodward, T.A.; Raimondo, M.; Jamil, L.; Gross, S.A.; Heckman, M.G.; Crook, J.E.; Wallace, M.B. An evaluation of risk factors for inadequate cytology in EUS-guided FNA of pancreatic tumors and lymph nodes. Gastroint. Endosc. 2010, 71, 1194–1199. [Google Scholar] [CrossRef]
- Klapman, J.B.; Logrono, R.; Dye, C.E.; Waxman, I. Clinical impact of on-site cytopathology interpretation on endoscopic ultrasound-guided fine needle aspiration. Am. J. Gastroenterol. 2003, 98, 1289–1294. [Google Scholar] [CrossRef]
- Kaneko, J.; Ishiwatari, H.; Sasaki, K.; Satoh, T.; Sato, J.; Matsubayashi, H.; Yabuuchi, Y.; Kishida, Y.; Yoshida, M.; Ito, S.; et al. Macroscopic on-site evaluation of biopsy specimens for accurate pathological diagnosis during EUS-guided fine needle biopsy using 22-G Franseen needle. Endosc. Ultrasound. 2020, 9, 385–391. [Google Scholar] [CrossRef]
- Ki, E.L.L.; Lemaistre, A.I.; Fumex, F.; Gincul, R.; Lefort, C.; Lepilliez, V.; Pujol, B.; Napoleon, B. Macroscopic onsite evaluation using endoscopic ultrasound fine needle biopsy as an alternative to rapid onsite evaluation. Endosc. Int. Open. 2019, 7, E189–E194. [Google Scholar] [CrossRef] [Green Version]
- Oh, D.; Seo, D.W.; Hong, S.M.; Song, T.J.; Park, D.H.; Lee, S.S.; Lee, S.K.; Kim, M.H. The impact of macroscopic on-site evaluation using filter paper in EUS-guided fine-needle biopsy. Endosc. Ultrasound 2019, 8, 342–347. [Google Scholar] [CrossRef]
- So, H.; Seo, D.W.; Hwang, J.S.; Ko, S.W.; Oh, D.; Song, T.J.; Park, D.H.; Lee, S.K.; Kim, M.H. Macroscopic on-site evaluation after EUS-guided fine needle biopsy may replace rapid on-site evaluation. Endosc. Ultrasound 2021, 10, 111–115. [Google Scholar] [CrossRef]
- Okuwaki, K.; Masutani, H.; Kida, M.; Yamauchi, H.; Iwai, T.; Miyata, E.; Hasegawa, R.; Kaneko, T.; Imaizumi, H.; Watanabe, M.; et al. Diagnostic efficacy of white core cutoff lengths obtained by EUS-guided fine-needle biopsy using a novel 22G franseen biopsy needle and sample isolation processing by stereomicroscopy for subepithelial lesions. Endosc. Ultrasound 2020, 9, 187–192. [Google Scholar] [CrossRef] [PubMed]
| Deep Learning Group | Contrastive Learning Group | |
|---|---|---|
| N = 63 | N = 96 | |
| Age, median (IQR) | 65 (58–72) | 68 (60–74.75) |
| Sex, Male, N (%) | 42 (66.7) | 59 (61.5) |
| Lesion size, median (IQR), mm | 24 (20–35.5) | 25 (20–35) |
| Final diagnosis, N (%) | ||
| Pancreatic ductal adenocarcinoma | 41 (65.1) | 66 (68.8) |
| Mass-forming pancreatitis | 11 (17.5) | 13 (13.5) |
| Autoimmune pancreatitis | 8 (12.7) | 11 (11.5) |
| Pancreatic neuroendocrine tumour | 1 (1.6) | 3 (3.1) |
| Pancreatic metastasis | 1 (1.6) | 2 (2.1) |
| Intraductal papillary mucinous carcinoma | 1 (1.6) | 1 (1.0) |
| Histology | Histology | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Diagnosable | Undiagnosable | Total | Diagnosable | Undiagnosable | Total | ||||
| AI | Diagnosable | 139 | 23 | 162 | MOSE | Diagnosable | 72 | 9 | 81 |
| Undiagnosable | 61 | 75 | 136 | Undiagnosable | 9 | 8 | 17 | ||
| Total | 200 | 98 | 298 | Total | 81 | 17 | 98 | ||
| Sensitivity | 85.8% | Sensitivity | 88.9% | ||||||
| Specificity | 55.2% | Specificity | 47.1% | ||||||
| Accuracy | 71.8% | Accuracy | 81.6% | ||||||
| PPV | 69.5% | PPV | 88.9% | ||||||
| NPV | 76.5% | NPV | 47.1% | ||||||
| Histology | Histology | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Diagnosable | Undiagnosable | Total | Diagnosable | Undiagnosable | Total | ||||
| AI | Diagnosable | 131 | 13 | 144 | MOSE | Diagnosable | 129 | 13 | 142 |
| Undiagnosable | 14 | 15 | 29 | Undiagnosable | 16 | 15 | 31 | ||
| Total | 145 | 28 | 173 | Total | 145 | 28 | 173 | ||
| Sensitivity | 90.3% | Sensitivity | 88.9% | ||||||
| Specificity | 53.5% | Specificity | 53.5% | ||||||
| Accuracy | 84.4% | Accuracy | 83.2% | ||||||
| PPV | 90.9% | PPV | 90.8% | ||||||
| NPV | 51.7% | NPV | 48.4% | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishikawa, T.; Hayakawa, M.; Suzuki, H.; Ohno, E.; Mizutani, Y.; Iida, T.; Fujishiro, M.; Kawashima, H.; Hotta, K. Development of a Novel Evaluation Method for Endoscopic Ultrasound-Guided Fine-Needle Biopsy in Pancreatic Diseases Using Artificial Intelligence. Diagnostics 2022, 12, 434. https://doi.org/10.3390/diagnostics12020434
Ishikawa T, Hayakawa M, Suzuki H, Ohno E, Mizutani Y, Iida T, Fujishiro M, Kawashima H, Hotta K. Development of a Novel Evaluation Method for Endoscopic Ultrasound-Guided Fine-Needle Biopsy in Pancreatic Diseases Using Artificial Intelligence. Diagnostics. 2022; 12(2):434. https://doi.org/10.3390/diagnostics12020434
Chicago/Turabian StyleIshikawa, Takuya, Masato Hayakawa, Hirotaka Suzuki, Eizaburo Ohno, Yasuyuki Mizutani, Tadashi Iida, Mitsuhiro Fujishiro, Hiroki Kawashima, and Kazuhiro Hotta. 2022. "Development of a Novel Evaluation Method for Endoscopic Ultrasound-Guided Fine-Needle Biopsy in Pancreatic Diseases Using Artificial Intelligence" Diagnostics 12, no. 2: 434. https://doi.org/10.3390/diagnostics12020434
APA StyleIshikawa, T., Hayakawa, M., Suzuki, H., Ohno, E., Mizutani, Y., Iida, T., Fujishiro, M., Kawashima, H., & Hotta, K. (2022). Development of a Novel Evaluation Method for Endoscopic Ultrasound-Guided Fine-Needle Biopsy in Pancreatic Diseases Using Artificial Intelligence. Diagnostics, 12(2), 434. https://doi.org/10.3390/diagnostics12020434







