p53 Immunohistochemistry and Mutation Types Mismatching in High-Grade Serous Ovarian Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Targeted Next-Generation Sequencing
2.2. p53 Immunohistochemistry
2.3. Dataset Visualization and Statistical Analysis
3. Results
3.1. p53 Immunohistochemistry Results
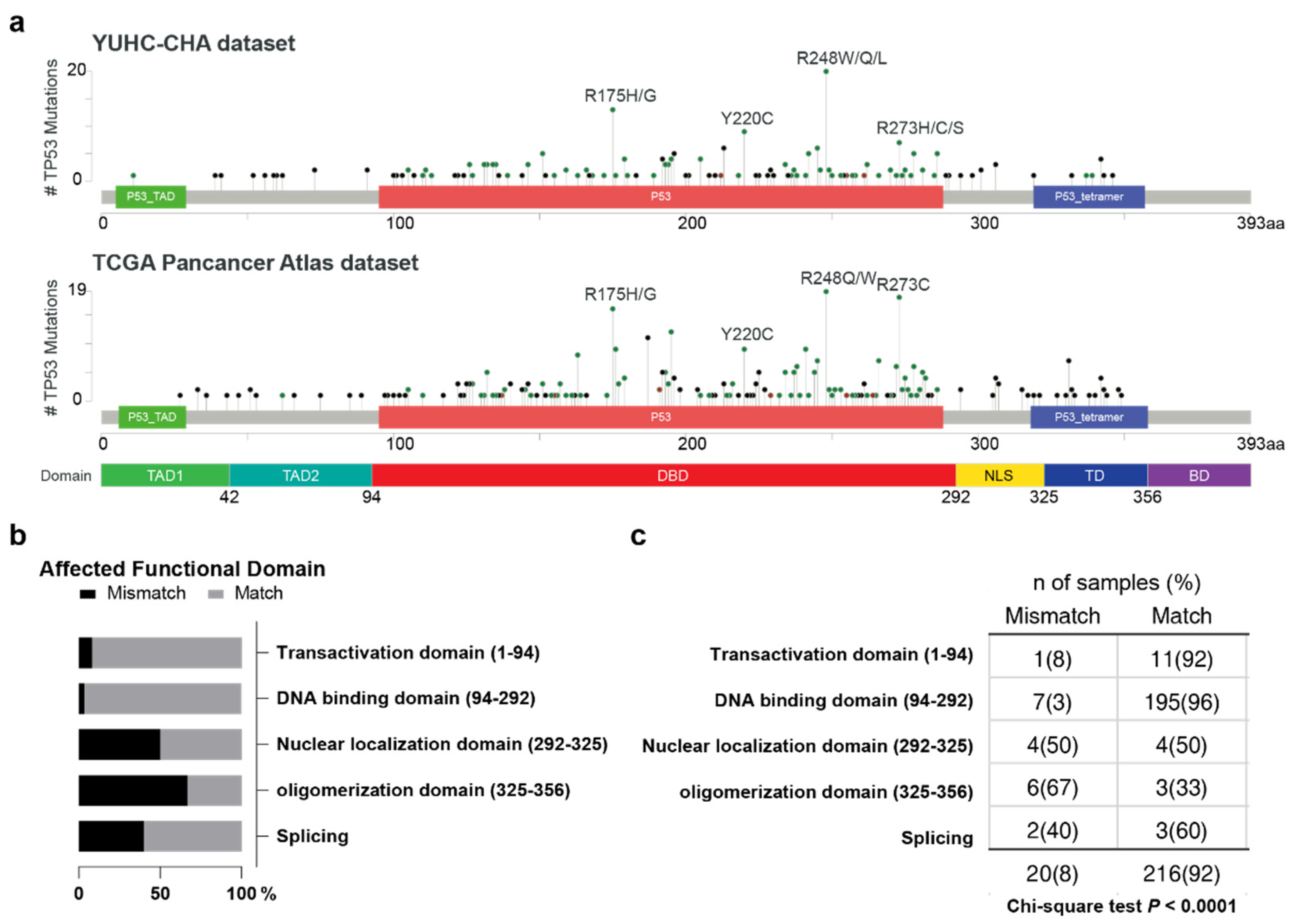
3.2. IHC and NGS Data Correlation, Stratified by p53 Functional Domain
3.2.1. Transactivation Domain (Amino Acid 1–94)
3.2.2. DNA-Binding Domain (AA 94–292)
- A clear cell carcinoma of a TP53 G105R missense mutation (variant allele frequency (VAF) 49.9%) showed a WT-like staining pattern. This was in contrast with another HGSCa of a TP53 G105V missense mutation, but concordant “all” patterned staining. The clear cell carcinoma case presented with additional PIK3CA missense mutation.
- A HGSCa of probable peritoneal origin was found to have a nonsense mutation of TP53 (S183*) (VAF 42.7%), but the IHC showed “all” patterned staining. Additional variants were found in RET, RB1 and ARID1A, all of unknown significance.
- A HGSCa presented with a TP53 in-frame deletion (F212_H214del), but the staining showed diffuse and strong nuclear immunoreactivity. This case has been described in our earlier report [16].
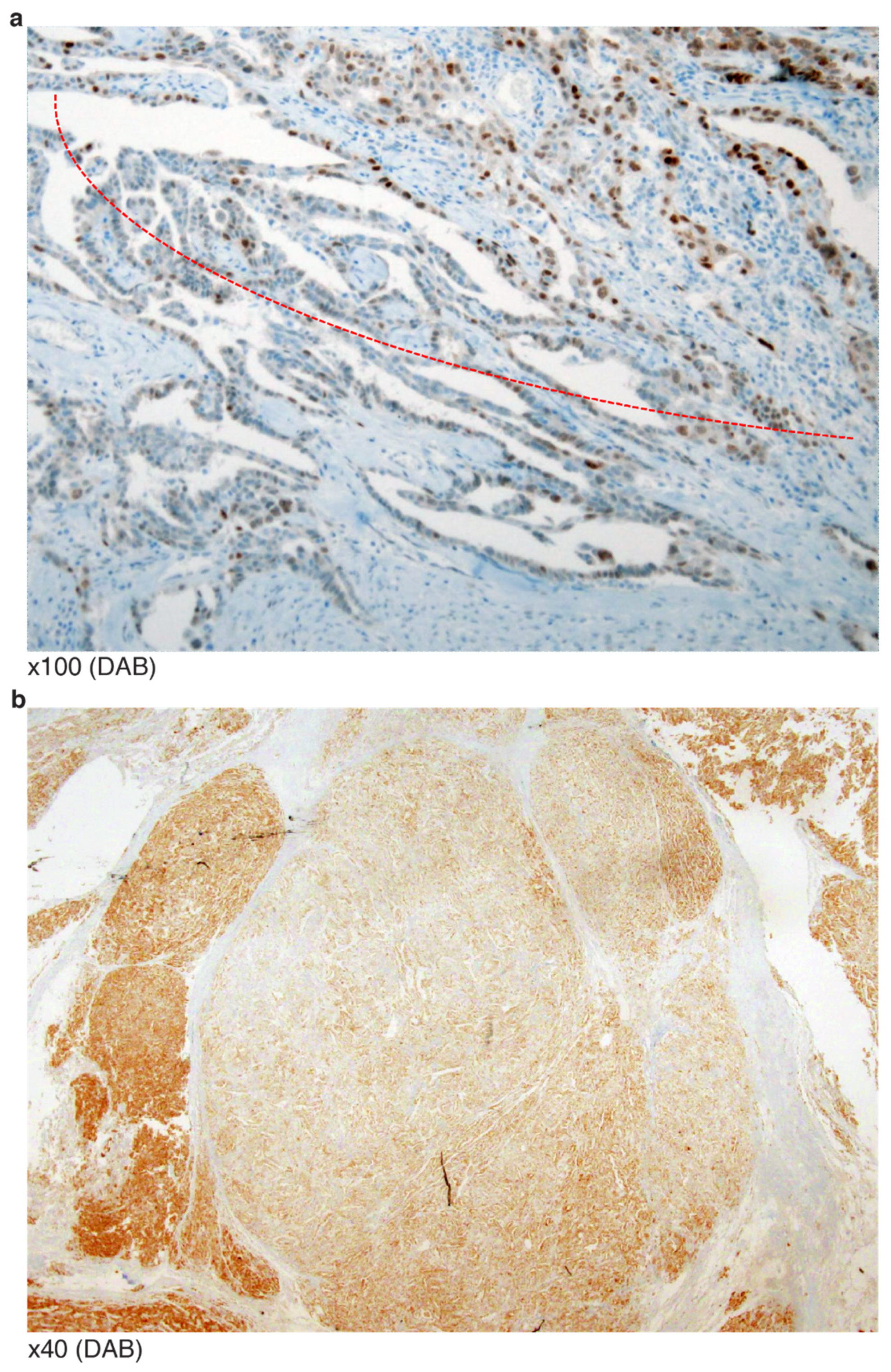
- An omental HGSCa with extrapelvic peritoneal metastasis revealed a TP53 G262 in-frame deletion at the end of DBD (VAF 32.9%). IHC showed a mosaic staining pattern (Figure 4a). There was no additional mutation/copy number alteration directly associated with p53 protein degradation.
- Similar to the above case (G262 in-frame deletion), an endometrioid carcinoma was reported as having a TP53 S262_G262dup in-frame insertion (VAF 16.7%). However, the staining revealed zonal heterogeneity with some WT-like staining patterns (Figure 4b). In contrast, two HGSCa cases of I254S fs*91 frameshift deletion (VAF 30.3%) and I255del in-frame deletion (VAF 42.1%) presented as having negative or “none” patterned staining. This implies that some non-NMs occurring beyond amino acid 261 of DBD may generate p53 proteins that do not degrade like those with mutations or the earlier amino acid 255 of DBD.
- A HGSCa presented with a missense mutation of R273H but negative IHC staining, potentially explainable by its low VAF (0.75%).
- A frameshift insertion at the c-terminal end of DBD (L289F fs*57) presented as complex staining pattens of cytoplasmic retained signals together with heavy nuclear staining of p53 (Figure 5a). It is possible that the mutation may have disrupted the nuclear localization site.
3.2.3. Nuclear Localizing Sites (AA 292–325)
3.2.4. Oligomerization Domain (AA 325–356)
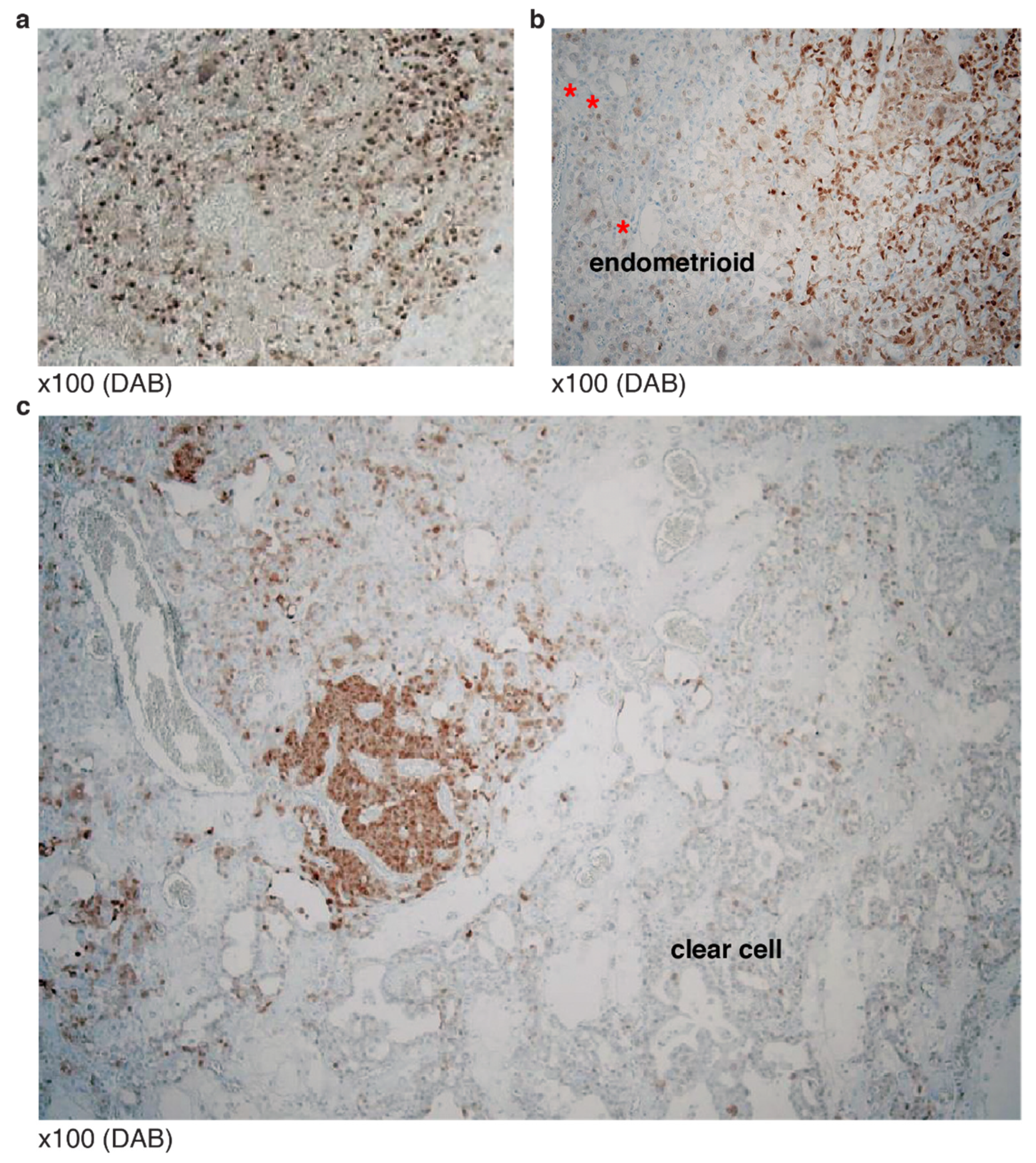
3.3. Typical Mosaic Patterns of p53 IHC in Mixed Carcinoma
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leroy, B.; Ballinger, M.L.; Baran-Marszak, F.; Bond, G.L.; Braithwaite, A.; Concin, N.; Donehower, L.A.; El-Deiry, W.S.; Fenaux, P.; Gaidano, G.; et al. Recommended guidelines for validation, quality control, and reporting of tp53 variants in clinical practice. Cancer Res. 2017, 77, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Vang, R.; Levine, D.A.; Soslow, R.A.; Zaloudek, C.; Shih Ie, M.; Kurman, R.J. Molecular alterations of tp53 are a defining feature of ovarian high-grade serous carcinoma: A rereview of cases lacking tp53 mutations in the cancer genome atlas ovarian study. Int. J. Gynecol. Pathol. 2016, 35, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Bouaoun, L.; Sonkin, D.; Ardin, M.; Hollstein, M.; Byrnes, G.; Zavadil, J.; Olivier, M. Tp53 variations in human cancers: New lessons from the iarc tp53 database and genomics data. Hum. Mutat. 2016, 37, 865–876. [Google Scholar] [CrossRef]
- Hainaut, P.; Olivier, M.; Wiman, K.G. P53 in the Clinics; Springer Science & Business Media LLC: New York, NY, USA, 2012. [Google Scholar]
- Elliott, K.; McQuaid, S.; Salto-Tellez, M.; Maxwell, P. Immunohistochemistry should undergo robust validation equivalent to that of molecular diagnostics. J. Clin. Pathol. 2015, 68, 766–770. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. Tp53 mutations in human cancers: Origins, consequences, and clinical use. Csh Perspect Biol. 2010, 2, a001008. [Google Scholar] [CrossRef]
- McAlpine, J.N.; Porter, H.; Kobel, M.; Nelson, B.H.; Prentice, L.M.; Kalloger, S.E.; Senz, J.; Milne, K.; Ding, J.; Shah, S.P.; et al. Brca1 and brca2 mutations correlate with tp53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod. Pathol. 2012, 25, 740–750. [Google Scholar] [CrossRef]
- Kim, S.I.; Lee, M.; Kim, H.S.; Chung, H.H.; Kim, J.W.; Park, N.H.; Song, Y.S. Effect of brca mutational status on survival outcome in advanced-stage high-grade serous ovarian cancer. J. Ovarian Res. 2019, 12, 40. [Google Scholar] [CrossRef]
- Eoh, K.J.; Kim, H.M.; Lee, J.Y.; Kim, S.; Kim, S.W.; Kim, Y.T.; Nam, E.J. Mutation landscape of germline and somatic brca1/2 in patients with high-grade serous ovarian cancer. BMC Cancer 2020, 20, 204. [Google Scholar] [CrossRef]
- Nougaret, S.; Lakhman, Y.; Gonen, M.; Goldman, D.A.; Micco, M.; D’Anastasi, M.; Johnson, S.A.; Juluru, K.; Arnold, A.G.; Sosa, R.E.; et al. High-grade serous ovarian cancer: Associations between brca mutation status, ct imaging phenotypes, and clinical outcomes. Radiology 2017, 285, 472–481. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the association for molecular pathology, american society of clinical oncology, and college of american pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Piskorz, A.M.; Bosse, T.; Jimenez-Linan, M.; Rous, B.; Brenton, J.D.; Gilks, C.B.; Kobel, M. P53 immunohistochemistry is an accurate surrogate for tp53 mutational analysis in endometrial carcinoma biopsies. J. Pathol. 2020, 250, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.Y.; Woo, H.Y.; Kim, H.S. Targeted genomic sequencing reveals novel tp53 in-frame deletion mutations leading to p53 overexpression in high-grade serous tubo-ovarian carcinoma. Anticancer. Res. 2019, 39, 2883–2889. [Google Scholar] [CrossRef]
- Bouchalova, P.; Nenutil, R.; Muller, P.; Hrstka, R.; Appleyard, M.V.; Murray, K.; Jordan, L.B.; Purdie, C.A.; Quinlan, P.; Thompson, A.M.; et al. Mutant p53 accumulation in human breast cancer is not an intrinsic property or dependent on structural or functional disruption but is regulated by exogenous stress and receptor status. J. Pathol. 2014, 233, 238–246. [Google Scholar] [CrossRef]
- Kobel, M.; Piskorz, A.M.; Lee, S.; Lui, S.; LePage, C.; Marass, F.; Rosenfeld, N.; Mes Masson, A.M.; Brenton, J.D. Optimized p53 immunohistochemistry is an accurate predictor of tp53 mutation in ovarian carcinoma. J. Pathol. Clin. Res. 2016, 2, 247–258. [Google Scholar] [CrossRef]
- Fernandez-Pol, S.; Ma, L.; Ohgami, R.S.; Arber, D.A. Immunohistochemistry for p53 is a useful tool to identify cases of acute myeloid leukemia with myelodysplasia-related changes that are tp53 mutated, have complex karyotype, and have poor prognosis. Modern Pathol. 2017, 30, 382–392. [Google Scholar] [CrossRef]
- Guedes, L.B.; Almutairi, F.; Haffner, M.C.; Rajoria, G.; Liu, Z.; Klimek, S.; Zoino, R.; Yousefi, K.; Sharma, R.; De Marzo, A.M.; et al. Analytic, preanalytic, and clinical validation of p53 ihc for detection of tp53 missense mutation in prostate cancer. Clin. Cancer Res. 2017, 23, 4693–4703. [Google Scholar] [CrossRef]
- Pardo, F.S.; Hsu, D.W.; Zeheb, R.; Efird, J.T.; Okunieff, P.G.; Malkin, D.M. Mutant, wild type, or overall p53 expression: Freedom from clinical progression in tumours of astrocytic lineage. Brit. J. Cancer 2004, 91, 1678–1686. [Google Scholar] [CrossRef]
- Takami, H.; Yoshida, A.; Fukushima, S.; Arita, H.; Matsushita, Y.; Nakamura, T.; Ohno, M.; Miyakita, Y.; Shibui, S.; Narita, Y.; et al. Revisiting tp53 mutations and immunohistochemistry-a comparative study in 157 diffuse gliomas. Brain Pathol. 2015, 25, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Yu, X.D.; Li, J.; Zhang, Z.Y.; Hou, J.; Li, F. Prognostic significance of p53 expression in patients with esophageal cancer: A meta-analysis. BMC Cancer 2016, 16, 373. [Google Scholar] [CrossRef] [PubMed]
- Cruz, I.; Snijders, P.J.F.; Van Houten, V.; Vosjan, M.; Van der Waal, I.; Meijer, C.J.L.M. Specific p53 immunostaining patterns are associated with smoking habits in patients with oral squamous cell carcinomas. J. Clin. Pathol. 2002, 55, 834–840. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hodgson, A.; Xu, B.; Downes, M.R. P53 immunohistochemistry in high-grade urothelial carcinoma of the bladder is prognostically significant. Histopathology 2017, 71, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Kelsey, K.T.; Hirao, T.; Schned, A.; Hirao, S.; Devi-Ashok, T.; Nelson, H.H.; Andrew, A.; Karagas, M.R. A population-based study of immunohistochemical detection of p53 alteration in bladder cancer. Brit. J. Cancer 2004, 90, 1572–1576. [Google Scholar] [CrossRef]
- Yemelyanova, A.; Vang, R.; Kshirsagar, M.; Lu, D.; Marks, M.A.; Shih, I.M.; Kurman, R.J. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for tp53 mutations in ovarian carcinoma: An immunohistochemical and nucleotide sequencing analysis. Mod. Pathol. 2011, 24, 1248–1253. [Google Scholar] [CrossRef]
- Kobel, M.; Kang, E.Y. The many uses of p53 immunohistochemistry in gynecological pathology: Proceedings of the isgyp companion society session at the 2020 uscap annual9 meeting. Int. J. Gynecol. Pathol. 2021, 40, 32–40. [Google Scholar] [CrossRef]
- Cole, A.J.; Dwight, T.; Gill, A.J.; Dickson, K.A.; Zhu, Y.; Clarkson, A.; Gard, G.B.; Maidens, J.; Valmadre, S.; Clifton-Bligh, R.; et al. Assessing mutant p53 in primary high-grade serous ovarian cancer using immunohistochemistry and massively parallel sequencing. Sci. Rep. 2016, 6, 26191. [Google Scholar] [CrossRef]
- Chene, P. The role of tetramerization in p53 function. Oncogene 2001, 20, 2611–2617. [Google Scholar] [CrossRef]

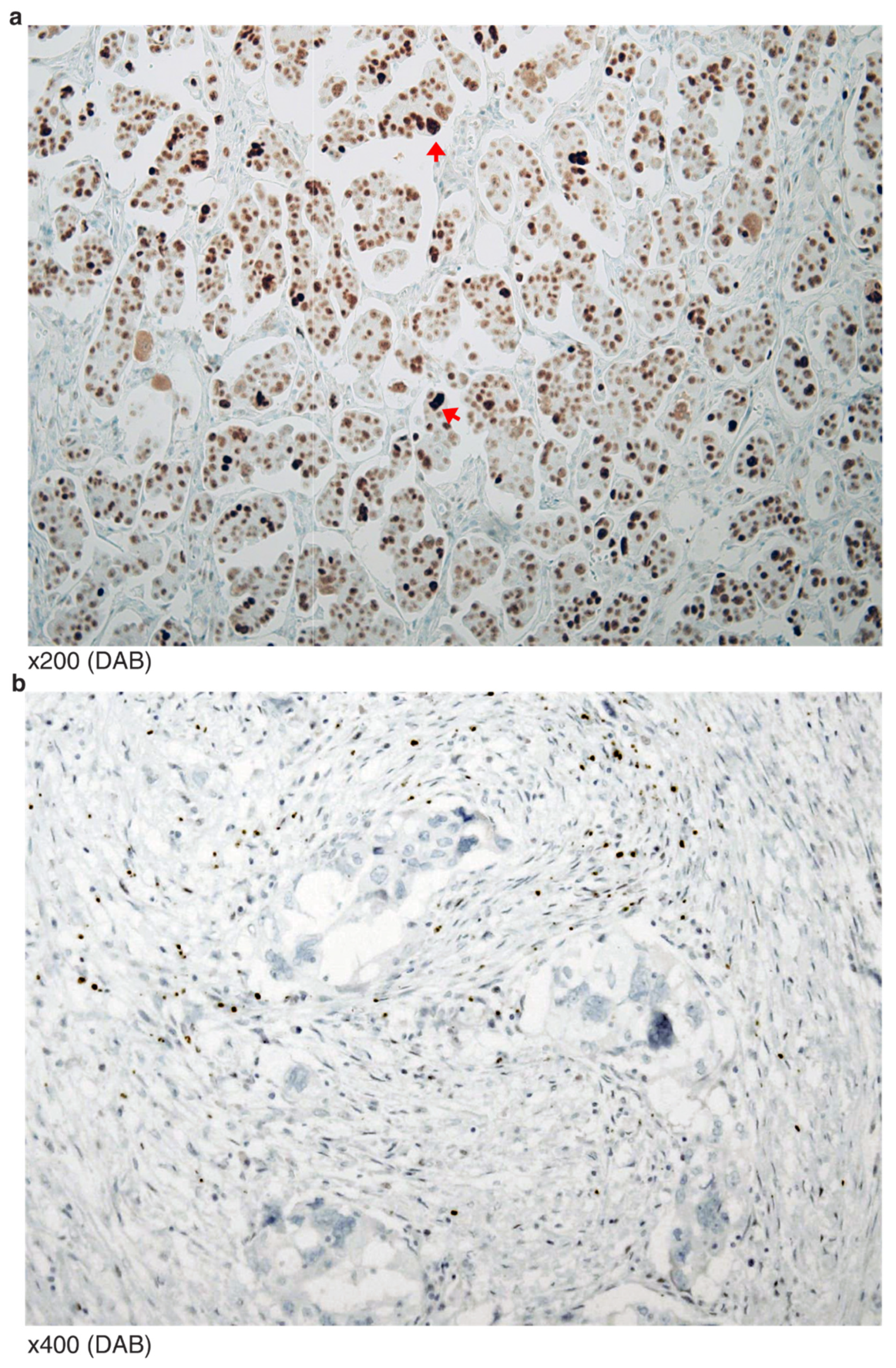


| N Samples * (N DBD) † | MM | NM | Frameshift | In-Frame | SP | WT | Total | ||
|---|---|---|---|---|---|---|---|---|---|
| Del | Ins | Del | Ins | ||||||
| HGSCa | 130 (128) | 32 (20) | 27 (19) | 6 (5) | 3 (3) | 4 (0) | 202 (175) | ||
| LGSCa | 1 (1) | 1 (1) | |||||||
| Carcino-sarcoma | 7 (7) | 2 (1) | 9 (8) | ||||||
| Endometrioid carcinoma | 4 (4) | 3 (1) | 1 (1) | 1 (0) | 9 (6) | ||||
| Mucinous carcinoma | 5 (5) | 1 (1) | 2 (1) | 8 (7) | |||||
| Clear cell carcinoma | 3 (3) | 3 (3) | |||||||
| Metastatic carcinoma | 3 (3) | 1 (0) | 4 (3) | ||||||
| Total | 153 (151) | 35 (22) | 33 (21) | 6 (5) | 3 (3) | 1 (1) | 4 (0) | 1 (0) | 236 (203) |
| Type | AA Change | Domain | Mutation Type | IHC Staining | Tumor % | VAF |
|---|---|---|---|---|---|---|
| endometrioid | V73W fs*50 | TD | FSD | WT | 30 | 10.3 |
| clear cell | G105R | DBD | MM | WT | 80 | 49.88 |
| HGSCa | S183* | DBD | NM | “All” | 70 | 42.67 |
| HGSCa | F212_H214del | DBD | IFD | “All” | 70 | 13.62 |
| endometrioid | S261_G262dup | DBD | IFI | WT | 40 | 16.71 |
| HGSCa | G262del | DBD | IFD | “All” | 80 | 32.92 |
| HGSCa | R273H | DBD | MM | “None” | 90 | 0.75 |
| HGSCa | L289F fs*57 | DBD | FSI | Cyto | 70 | 23.66 |
| HGSCa | P301Q fs*44 | NLS | FSD | WT | 60 | 27.28 |
| HGSCa | P301Q fs*44 | NLS | FSD | “All” | 90 | 82.71 |
| HGSCa | R306* | NLS | NM | Cyto | 80 | 0.86 |
| HGSCa | R306* | NLS | NM | WT + Cyto | 80 | 0.60 |
| HGSCa | R342E fs*3 | OD | FSI | “All” | 30 | 36.69 |
| HGSCa | R342* | OD | NM | “All” + Cyto | 90 | 37.4 |
| HGSCa | R342* | OD | NM | “All” | 50 | 7.08 |
| HGSCa | R342* | OD | NM | “All” | 60 | 44.4 |
| HGSCa | E343* | OD | NM | WT | 90 | 0.31 |
| HGSCa | E346* | OD | NM | “All” | 90 | 0.35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, E.; Han, H.; Choi, S.-E.; Park, H.; Woo, H.-Y.; Jang, M.; Shim, H.-S.; Hwang, S.; Kang, H.; Cho, N.-H. p53 Immunohistochemistry and Mutation Types Mismatching in High-Grade Serous Ovarian Cancer. Diagnostics 2022, 12, 579. https://doi.org/10.3390/diagnostics12030579
Park E, Han H, Choi S-E, Park H, Woo H-Y, Jang M, Shim H-S, Hwang S, Kang H, Cho N-H. p53 Immunohistochemistry and Mutation Types Mismatching in High-Grade Serous Ovarian Cancer. Diagnostics. 2022; 12(3):579. https://doi.org/10.3390/diagnostics12030579
Chicago/Turabian StylePark, Eunhyang, Hyunho Han, Sung-Eun Choi, Hyunjin Park, Ha-Young Woo, Mi Jang, Hyo-Sup Shim, Sohyun Hwang, Haeyoun Kang, and Nam-Hoon Cho. 2022. "p53 Immunohistochemistry and Mutation Types Mismatching in High-Grade Serous Ovarian Cancer" Diagnostics 12, no. 3: 579. https://doi.org/10.3390/diagnostics12030579
APA StylePark, E., Han, H., Choi, S.-E., Park, H., Woo, H.-Y., Jang, M., Shim, H.-S., Hwang, S., Kang, H., & Cho, N.-H. (2022). p53 Immunohistochemistry and Mutation Types Mismatching in High-Grade Serous Ovarian Cancer. Diagnostics, 12(3), 579. https://doi.org/10.3390/diagnostics12030579






