Urine Flow Cytometry Parameter Cannot Safely Predict Contamination of Urine—A Cohort Study of a Swiss Emergency Department Using Machine Learning Techniques
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
2.2. Eligibility Criteria
2.3. Predictors: UFC Parameters
2.4. Two-Class Classification (Contamination or No Contamination)
2.5. Three-Class Classification of Urine Culture Growth
2.6. Data Collection
2.6.1. Urine Flow Cytometry
2.6.2. Urine Culture
2.7. Data Extraction
2.8. Statistical Analysis
Machine Learning Approach
2.9. Ethical Considerations
3. Results
3.1. Patient Characteristics
3.2. Descriptive Analysis
3.3. Prediction of Mixed Culture and the Role of Squamous Epithelial Cells (Two-Class Classification)
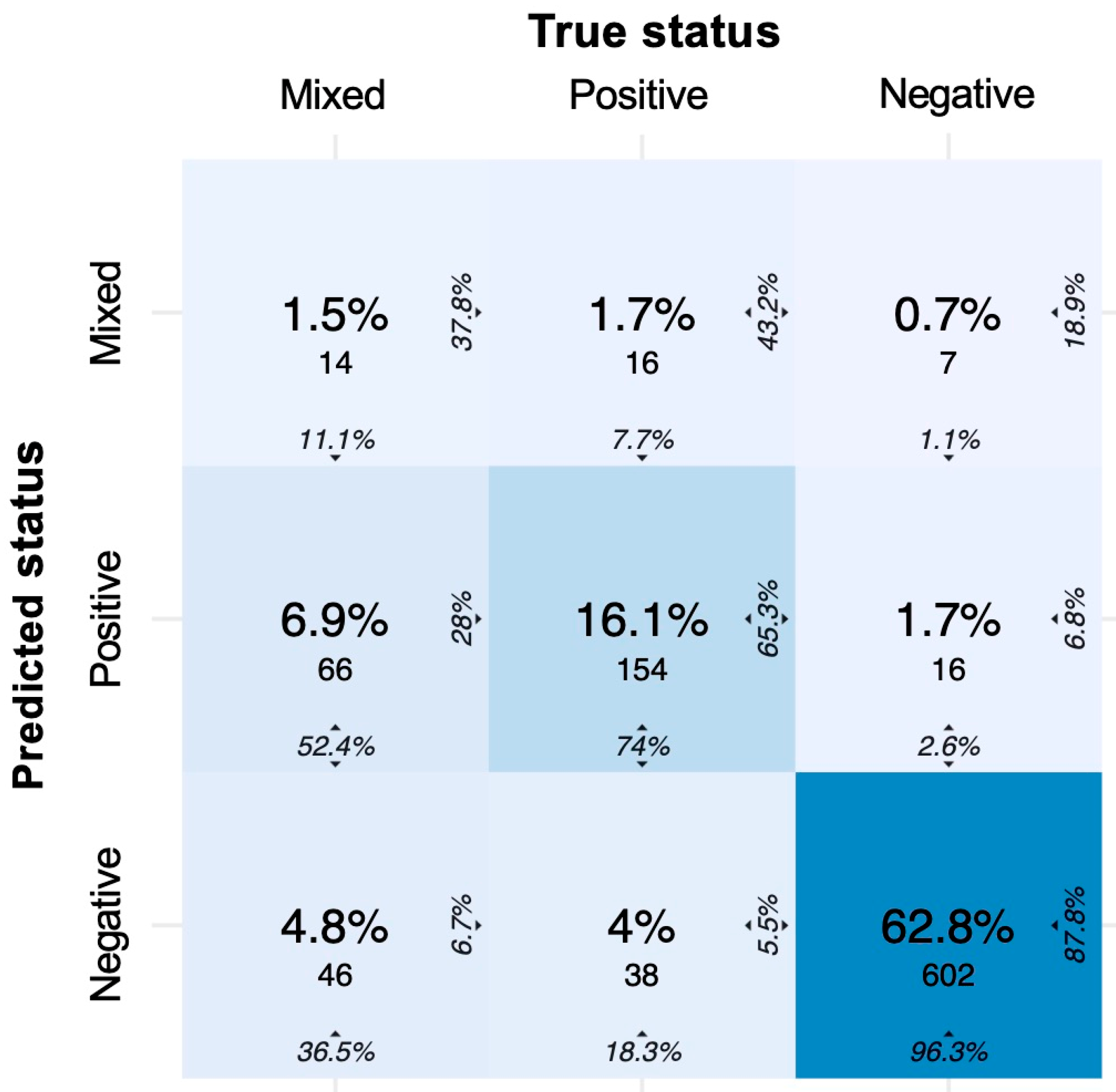
3.4. Three-Class Classification Using Machine Learning
4. Discussion
4.1. Statement of Principal Findings
4.2. Results in Context
4.3. Strengths and Weaknesses of the Study
4.4. Implications for Clinicians
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Foxman, B.; Barlow, R.; D’Arcy, H.; Gillespie, B.; Sobel, J.D. Urinary Tract Infection: Self-Reported Incidence and Associated Costs. Ann. Epidemiol. 2000, 10, 509–515. [Google Scholar] [CrossRef]
- Foxman, B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Disease-a-Month 2003, 49, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. Urinary Tract Infection Syndromes: Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clin. N. Am. 2014, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Geerlings, S.E. Clinical Presentations and Epidemiology of Urinary Tract Infections. Microbiol. Spectr. 2016, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Medina, M.; Castillo-Pino, E. An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol. 2019, 11, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, D.J.; Hicks, L.A.; Pavia, A.T.; Hersh, A.L. Antibiotic prescribing for adults in ambulatory care in the USA, 2007–09. J. Antimicrob. Chemother. 2014, 69, 234–240. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, M.; Shapiro, D.J.; Hersh, A.L.; Sanchez, G.V.; Hicks, L.A. Outpatient Antibiotic Prescribing Practices for Uncomplicated Urinary Tract Infection in Women in the United States, 2002–2011. Open Forum Infect. Dis. 2016, 3, ofw159. [Google Scholar] [CrossRef] [Green Version]
- Bell, B.G.; Schellevis, F.; Stobberingh, E.; Goossens, H.; Pringle, M. A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect. Dis. 2014, 14, 13. [Google Scholar] [CrossRef] [Green Version]
- Kouri, T.T.; Gant, V.A.; Fogazzi, G.B.; Hofmann, W.; Hallander, H.O.; Guder, W.G. Towards European urinalysis guidelines: Introduction of a Project under European Confederation of Laboratory Medicine. Clin. Chim. Acta 2000, 297, 305–311. [Google Scholar] [CrossRef]
- Schmiemann, G.; Kniehl, E.; Gebhardt, K.; Matejczyk, M.M.; Hummers-Pradier, E. The Diagnosis of Urinary Tract Infection. Dtsch. Ärzteblatt Int. 2010, 107, 361–367. [Google Scholar] [CrossRef]
- Sheerin, N.S. Urinary tract infection. Medicine 2011, 39, 384–389. [Google Scholar] [CrossRef]
- Chu, C.M.; Lowder, J.L. Diagnosis and treatment of urinary tract infections across age groups. Am. J. Obstet. Gynecol. 2018, 219, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Gilboe, H.M.; Reiakvam, O.M.; Aasen, L.; Tjade, T.; Bjerner, J.; Ranheim, T.E.; Gaustad, P. Rapid diagnosis and reduced workload for urinary tract infection using flowcytometry combined with direct antibiotic susceptibility testing. PLoS ONE 2021, 16, e0254064. [Google Scholar] [CrossRef]
- Wagenlehner, F.M.E.; Hoyme, U.; Kaase, M.; Fünfstück, R.; Naber, K.G.; Schmiemann, G. Uncomplicated Urinary Tract Infections. Dtsch. Ärzteblatt Int. 2011, 108, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, L.E.; Gupta, K.; Bradley, S.F.; Colgan, R.; DeMuri, G.P.; Drekonja, D.; Eckert, L.O.; Geerlings, S.E.; Köves, B.; Hooton, T.M.; et al. Clinical Practice Guideline for the Management of Asymptomatic Bacteriuria: 2019 Update by the Infectious Diseases Society of Americaa. Clin. Infect. Dis. 2019, 68, e83–e110. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Seidenberg, R.; Schuh, S.K.; Exadaktylos, A.K.; Schechter, C.B.; Leichtle, A.B.; Hautz, W.E. The development and validation of different decision-making tools to predict urine culture growth out of urine flow cytometry parameter. PLoS ONE 2018, 13, e0193255. [Google Scholar] [CrossRef]
- Alenkaer, L.K.; Pedersen, L.; Szecsi, P.B.; Bjerrum, P.J. Evaluation of the sysmex UF-5000 fluorescence flow cytometer as a screening platform for ruling out urinary tract infections in elderly patients presenting at the Emergency Department. Scand. J. Clin. Lab. Investig. 2021, 81, 379–384. [Google Scholar] [CrossRef]
- Sharp, S.E. Laboratory Diagnosis of Urinary Tract Infections; ASM Press: Washington, DC, USA, 2009; ISBN 978-1-55581-517-2. [Google Scholar]
- Kim, S.Y.; Kim, Y.J.; Lee, S.M.; Hwang, S.H.; Kim, H.H.; Son, H.C.; Lee, E.Y. Evaluation of the Sysmex UF-100 Urine Cell Analyzer as a Screening Test to Reduce the Need for Urine Cultures for Community-Acquired Urinary Tract Infection. Am. J. Clin. Pathol. 2007, 128, 922–925. [Google Scholar] [CrossRef] [Green Version]
- Manoni, F.; Fornasiero, L.; Ercolin, M.; Tinello, A.; Ferrian, M.; Hoffer, P.; Valverde, S.; Gessoni, G. Cutoff values for bacteria and leukocytes for urine flow cytometer Sysmex UF-1000i in urinary tract infections. Diagn. Microbiol. Infect. Dis. 2009, 65, 103–107. [Google Scholar] [CrossRef]
- Brilha, S.; Proença, H.; Cristino, J.M.; Hänscheid, T. Use of flow cytometry (Sysmex® UF-100) to screen for positive urine cultures: In search for the ideal cut-off. Clin. Chem. Lab. Med. (CCLM) 2010, 48, 289–292. [Google Scholar] [CrossRef] [Green Version]
- De Rosa, R.; Grosso, S.; Bruschetta, G.; Avolio, M.; Stano, P.; Modolo, M.L.; Camporese, A. Evaluation of the Sysmex UF1000i flow cytometer for ruling out bacterial urinary tract infection. Clin. Chim. Acta 2010, 411, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Xu, D.; Shao, W.; Lu, Y. Evaluation of the Sysmex UF-1000i for the Diagnosis of Urinary Tract Infection. Am. J. Clin. Pathol. 2010, 133, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Frazee, B.W.; Enriquez, K.; Ng, V.; Alter, H. Abnormal Urinalysis Results Are Common, Regardless of Specimen Collection Technique, in Women Without Urinary Tract Infections. J. Emerg. Med. 2015, 48, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.W.; Tenney, J.H.; Hoopes, J.M.; Muncie, H.L.; Anthony, W.C. A Prospective Microbiologic Study of Bacteriuria in Patients with Chronic Indwelling Urethral Catheters. J. Infect. Dis. 1982, 146, 719–723. [Google Scholar] [CrossRef]
- Podbielski, A.; Abele-Horn, M.; Becker, K.; Kniehl, E.; Mauch, H.; Ruessmann, H.; Schubert, S.; Zimmermann, S. Harnwegsinfektionen. In MiQ—Qualitätsstandards in der mikrobiologisch-infektiologischen Diagnostik; Elsevier Urban & Fischer: Munich, Germany, 2020; ISBN 978-3-437-41570-8. [Google Scholar]
- Guelman, L.; Hsieh, C.D.; Pine, K.; Lough, M.E. Optimizing Antibiotic Management For Acute Simple Cystitis in Women: An Evidence-Based Roadmap for the Primary Care Ambulatory Setting. Urol. Nurs. 2019, 39, 283–292. [Google Scholar] [CrossRef]
- Wright, R.A.; Euwer, R.; Scholes, E.N.; Miles, S.R. Accuracy of standard urinalysis in predicting culture results. J. Natl. Med. Assoc. 1986, 78, 43–48. [Google Scholar]
- Smith, P.; Morris, A.; Reller, L.B. Predicting Urine Culture Results by Dipstick Testing and Phase Contrast Microscopy. Pathology 2003, 35, 161–165. [Google Scholar]
- Mohr, N.M.; Harland, K.K.; Crabb, V.; Mutnick, R.; Baumgartner, D.; Spinosi, S.; Haarstad, M.; Ahmed, A.; Schweizer, M.; Faine, B. Urinary squamous epithelial cells do not accurately predict urine culture contamination, but may predict urinalysis performance in predicting bacteriuria. Acad. Emerg. Med. 2016, 23, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.-J.; Wang, Q.-Q.; Zhang, J.-R.; Xu, Y.-L.; Zhang, W.-W.; Chen, Y.; Gu, M.-L.; Hu, Z.-D.; Deng, A.-M. Systematic review and meta-analysis of flow cytometry in urinary tract infection screening. Clin. Chim. Acta 2013, 424, 90–95. [Google Scholar] [CrossRef]
- Walter, F.G.; Gibly, R.L.; Knopp, R.K.; Roe, D.J. Squamous cells as predictors of bacterial contamination in urine samples. Ann. Emerg. Med. 1998, 31, 455–458. [Google Scholar] [CrossRef]
- Yang, C.-C.; Yang, S.S.-D.; Hung, H.-C.; Chiang, I.-N.; Peng, C.C.-H.; Chang, S.-J. Rapid differentiation of cocci/mixed bacteria from rods in voided urine culture of women with uncomplicated urinary tract infections. J. Clin. Lab. Anal. 2017, 31, e22071. [Google Scholar] [CrossRef] [PubMed]
- Bekeris, L.G.; Jones, B.A.; Walsh, M.K.; Wagar, E.A. Urine Culture Contamination: A College of American Pathologists Q-Probes Study of 127 Laboratories. Arch. Pathol. Lab. Med. 2008, 132, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Lough, M.E.; Shradar, E.; Hsieh, C.; Hedlin, H. Contamination in Adult Midstream Clean-Catch Urine Cultures in the Emergency Department: A Randomized Controlled Trial. J. Emerg. Nurs. 2019, 45, 488–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowe, T.A.; Juthani-Mehta, M. Urinary tract infection in older adults. Aging Health 2013, 9, 519–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Zare, M.; Vehreschild, M.J.; Wagenlehner, F. Management of uncomplicated recurrent urinary tract infections. Br. J. Urol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Tandogdu, Z.; Wagenlehner, F.M. Global epidemiology of urinary tract infections. Curr. Opin. Infect. Dis. 2016, 29, 73–79. [Google Scholar] [CrossRef]
- Delcaru, C.; Alexandru, I.; Podgoreanu, P.; Grosu, M.; Stavropoulos, E.; Chifiriuc, M.C.; Lazar, V. Microbial Biofilms in Urinary Tract Infections and Prostatitis: Etiology, Pathogenicity, and Combating strategies. Pathogens 2016, 5, 65. [Google Scholar] [CrossRef] [Green Version]
- Sen, A.; Kaul, A.; Kaul, R. Estrogen receptors in human bladder cells regulate innate cytokine responses to differentially modulate uropathogenic E. coli colonization. Immunobiology 2021, 226, 152020. [Google Scholar] [CrossRef]
- Lüthje, P.; Brauner, H.; Ramos, N.L.; Övregaard, A.; Gläser, R.; Hirschberg, A.L.; Aspenström, P.; Brauner, A. Estrogen Supports Urothelial Defense Mechanisms. Sci. Transl. Med. 2013, 5, 190ra80. [Google Scholar] [CrossRef]
- Sonnex, C. Influence of ovarian hormones on urogenital infection. Sex. Transm. Infect. 1998, 74, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuh, S.K.; Seidenberg, R.; Arampatzis, S.; Leichtle, A.B.; Hautz, W.; Exadaktylos, A.K.; Schechter, C.B.; Müller, M. Diagnosis of Urinary Tract Infections by Urine Flow Cytometry: Adjusted Cut-Off Values in Different Clinical Presentations. Dis. Markers 2019, 2019, 5853486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurlbut, T.A., III; Littenberg, B. The Diagnostic Accuracy of Rapid Dipstick Tests to Predict Urinary Tract Infection. Am. J. Clin. Pathol. 1991, 96, 582–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bent, S.; Nallamothu, B.K.; Simel, D.L.; Fihn, S.D.; Saint, S. Does This Woman Have an Acute Uncomplicated Urinary Tract Infection? JAMA: J. Am. Med. Assoc. 2002, 287, 2701–2710. [Google Scholar] [CrossRef]
- Stamm, W.E.; Counts, G.W.; Running, K.R.; Fihn, S.; Turck, M.; Holmes, K.K. Diagnosis of Coliform Infection in Acutely Dysuric Women. N. Engl. J. Med. 1982, 307, 463–468. [Google Scholar] [CrossRef]
- Hooton, T.M. The current management strategies for community-acquired urinary tract infection. Infect. Dis. Clin. N. Am. 2003, 17, 303–332. [Google Scholar] [CrossRef]
- Nicolle, L.E. Uncomplicated Urinary Tract Infection in Adults Including Uncomplicated Pyelonephritis. Urol. Clin. N. Am. 2008, 35, 1–12. [Google Scholar] [CrossRef]





| Total (n = 3835) | Training Set (n = 2876) | Validation Set (n = 959) | ||||
|---|---|---|---|---|---|---|
| Age [years], med (IQR) | 67 | (51–78) | 67 | (50.5–78) | 66 | (51–78) |
| Gender, n (%) | ||||||
| Male | 2084 | (54.3) | 1548 | (53.8) | 536 | (55.9) |
| Female | 1751 | (45.7) | 1328 | (46.2) | 423 | (44.1) |
| Urine sample, n (%) | ||||||
| Single-use catheter urine | 493 | (12.9) | 363 | (12.6) | 130 | (13.6) |
| Midstream urine | 2206 | (57.5) | 1639 | (57.0) | 567 | (59.1) |
| Spontaneous urine | 1136 | (29.6) | 874 | (30.4) | 262 | (27.3) |
| Bacteria in UFC/µL, med (IQR) | 150 | (19–2814) | 149 | (18–2901) | 150 | (21–2711) |
| Leucocytes in UFC/µL, med (IQR) | 17 | (4–167) | 17 | (4–170) | 18 | (4–155) |
| Squamous epithelial cells in UFC/µL, med (IQR) | 2 | (0–6) | 2 | (0–6) | 2 | (0–7) |
| Urine culture growth, n (%) | ||||||
| Negative culture | ||||||
| Positive culture | 2501 | (65.2) | 1876 | (65.2) | 625 | (65.2) |
| Escherichia coli | 552 | (14.4) | 420 | (14.6) | 132 | (13.8) |
| Klebsiella pneumoniae | 75 | (2.0) | 49 | (1.7) | 26 | (2.7) |
| Enterococcus faecalis | 18 | (0.5) | 12 | (0.4) | 6 | (0.6) |
| Aerococcus urinae | 16 | (0.4) | 12 | (0.4) | 4 | (0.4) |
| Staphylococcus aureus | 15 | (0.4) | 12 | (0.4) | 3 | (0.3) |
| Klebsiella oxytoca | 14 | (0.4) | 9 | (0.3) | 5 | (0.5) |
| Lactobacillus species | 14 | (0.4) | 9 | (0.3) | 5 | (0.5) |
| Pseudomonas aeruginosa | 11 | (0.3) | 5 | (0.2) | 6 | (0.6) |
| Other | 102 | (2.7) | 81 | (2.8) | 21 | (2.2) |
| Mixed culture | 517 | (13.5) | 391 | (13.6) | 126 | (13.1) |
| Squamous Epithelial Cells Group/µL 1 | Mixed Culture, n (%) | No Mixed Culture, n (%) | Odds Ratio (95% CI) | |||
|---|---|---|---|---|---|---|
| 0–0.1 | 2 | (1.6) | 64 | (7.7) | 1.00 | (baseline) |
| >0.1–0.5 | 6 | (4.8) | 154 | (18.5) | 1.25 | (0.25–6.34) |
| >0.5–1.9 | 27 | (21.4) | 254 | (30.5) | 3.40 | (0.79–14.68) |
| >1.9–6.3 | 27 | (21.4) | 173 | (20.8) | 4.99 | (1.15–21.61) |
| >6.3–17.5 | 32 | (25.4) | 113 | (13.6) | 9.06 | (2.10–39.06) |
| >17.5 | 32 | (25.4) | 75 | (9.0) | 13.65 | (3.15–59.20) |
| Prediction of Mixed Cultures | (%) |
| Sensitivity (SE) | 11.1 |
| Specificity (SP) | 97.2 |
| Positive Predictive Value (PPV) | 37.8 |
| Negative Predictive Value (NPV) | 87.9 |
| Prediction of Positive Cultures | (%) |
| Sensitivity (SE) | 74.0 |
| Specificity (SP) | 89.1 |
| Positive Predictive Value (PPV) | 65.3 |
| Negative Predictive Value (NPV) | 92.5 |
| Prediction of Negative Cultures | (%) |
| Sensitivity (SE) | 96.3 |
| Specificity (SP) | 74.9 |
| Positive Predictive Value (PPV) | 87.8 |
| Negative Predictive Value (NPV) | 91.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, M.; Sägesser, N.; Keller, P.M.; Arampatzis, S.; Steffens, B.; Ehrhard, S.; Leichtle, A.B. Urine Flow Cytometry Parameter Cannot Safely Predict Contamination of Urine—A Cohort Study of a Swiss Emergency Department Using Machine Learning Techniques. Diagnostics 2022, 12, 1008. https://doi.org/10.3390/diagnostics12041008
Müller M, Sägesser N, Keller PM, Arampatzis S, Steffens B, Ehrhard S, Leichtle AB. Urine Flow Cytometry Parameter Cannot Safely Predict Contamination of Urine—A Cohort Study of a Swiss Emergency Department Using Machine Learning Techniques. Diagnostics. 2022; 12(4):1008. https://doi.org/10.3390/diagnostics12041008
Chicago/Turabian StyleMüller, Martin, Nadine Sägesser, Peter M. Keller, Spyridon Arampatzis, Benedict Steffens, Simone Ehrhard, and Alexander B. Leichtle. 2022. "Urine Flow Cytometry Parameter Cannot Safely Predict Contamination of Urine—A Cohort Study of a Swiss Emergency Department Using Machine Learning Techniques" Diagnostics 12, no. 4: 1008. https://doi.org/10.3390/diagnostics12041008
APA StyleMüller, M., Sägesser, N., Keller, P. M., Arampatzis, S., Steffens, B., Ehrhard, S., & Leichtle, A. B. (2022). Urine Flow Cytometry Parameter Cannot Safely Predict Contamination of Urine—A Cohort Study of a Swiss Emergency Department Using Machine Learning Techniques. Diagnostics, 12(4), 1008. https://doi.org/10.3390/diagnostics12041008







