Rapid One-Tube RPA-CRISPR/Cas12 Detection Platform for Methicillin-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Materials and Methods
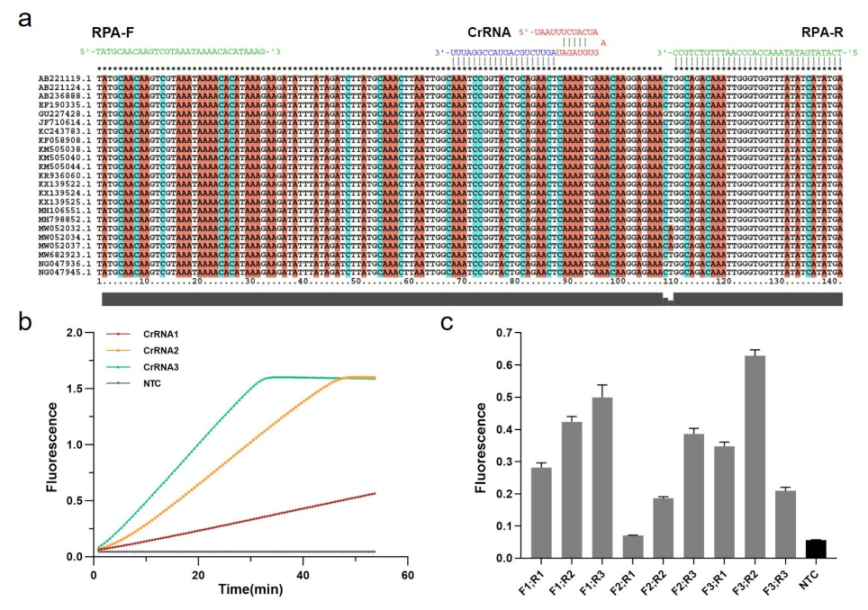
2.1. CrRNA Synthesis and Purification
2.2. Polymerase Chain Reaction (PCR) and Cas12a Fluorescence Detection
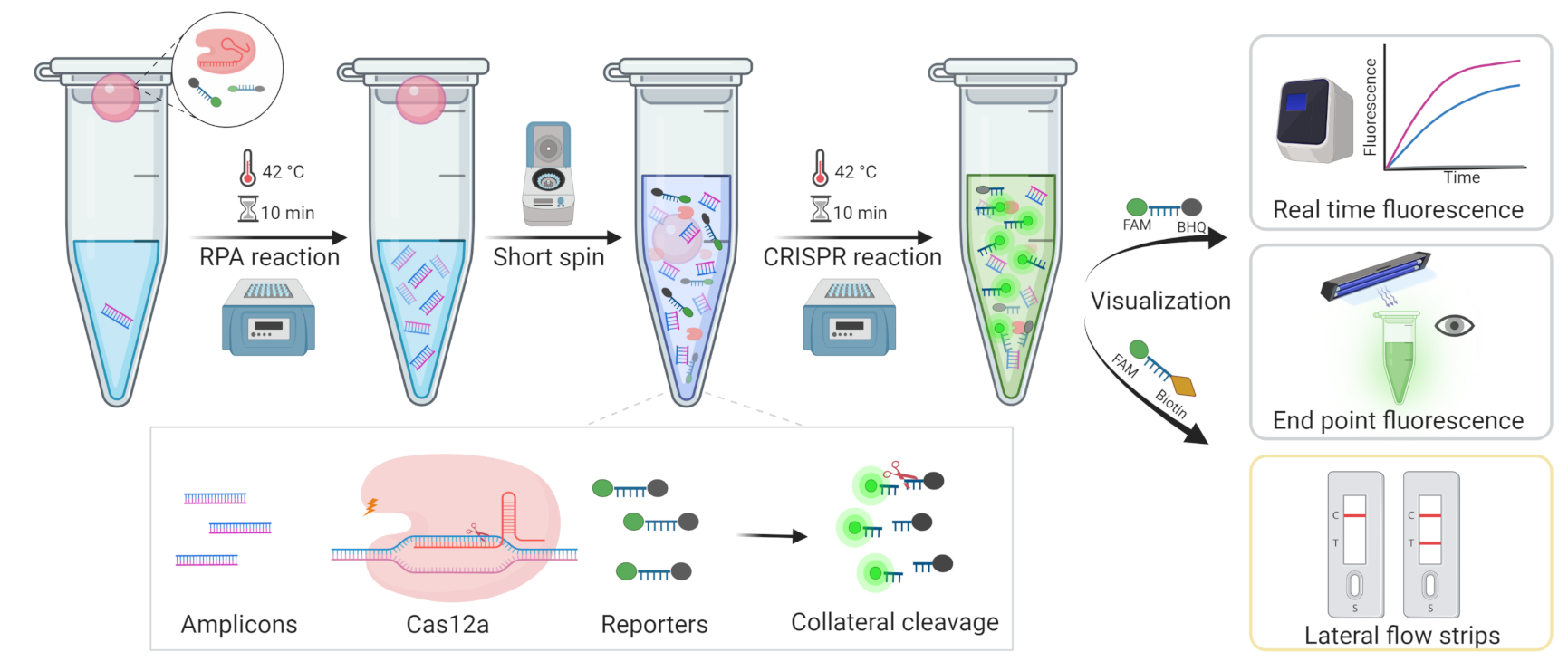
2.3. One Tube RPA-Cas12a Detection Platform
2.4. Visual End-Point Detection with a UV Light Illuminator
2.5. Lateral Flow Strip Assay
2.6. Bacterial Strains and Clinical MRSA Samples Preparation
2.7. Real-Time PCR Detection and Standard Curve
2.8. Statistical Analysis
3. Results
3.1. Development of One-Tube RPA-CRISPR/Cas12a Assay for MRSA Detection
3.2. Optimization of Experimental Conditions
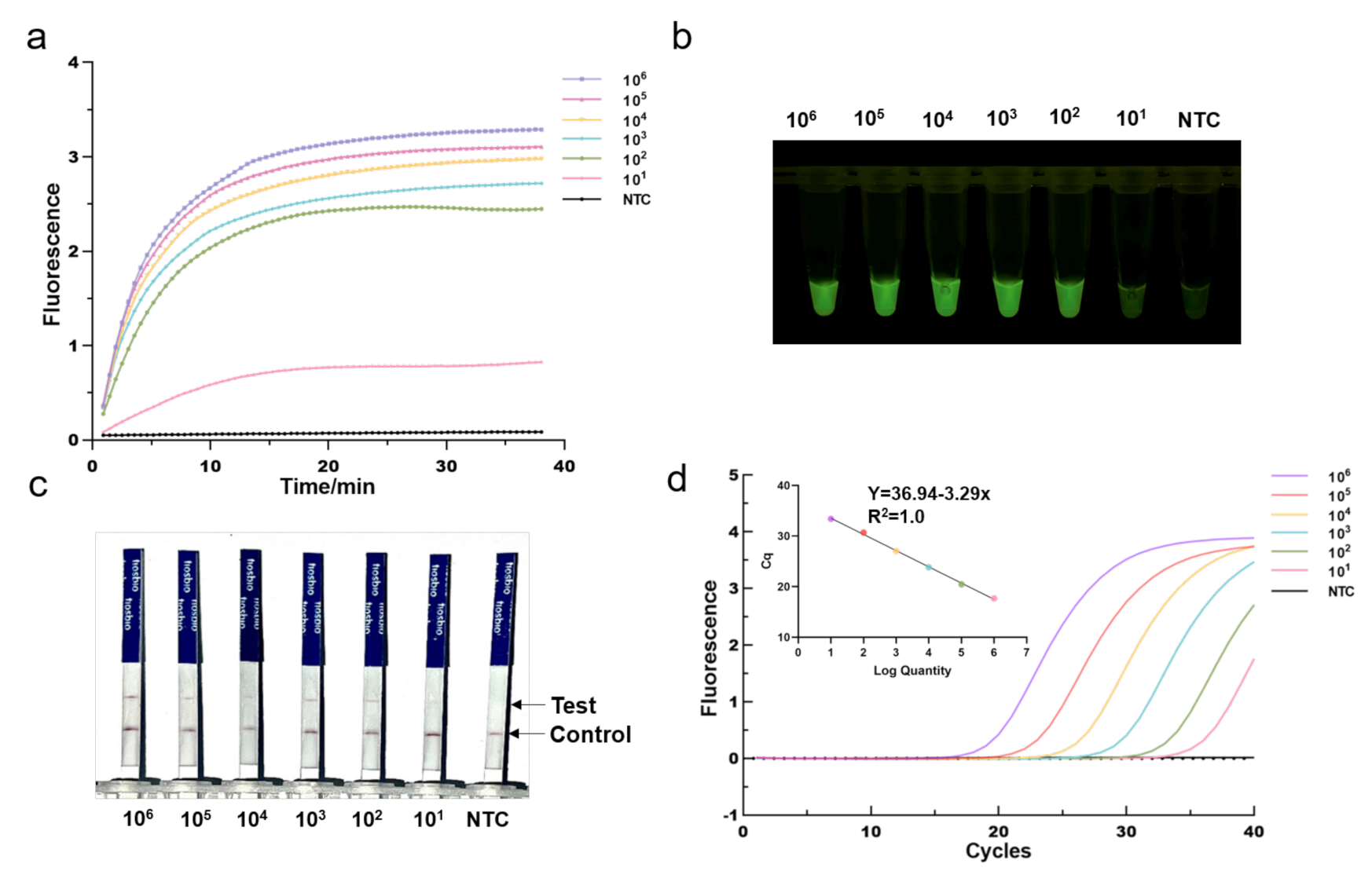
3.3. Sensitivity of the One-Tube RPA-CRISPR/Cas12a Detection
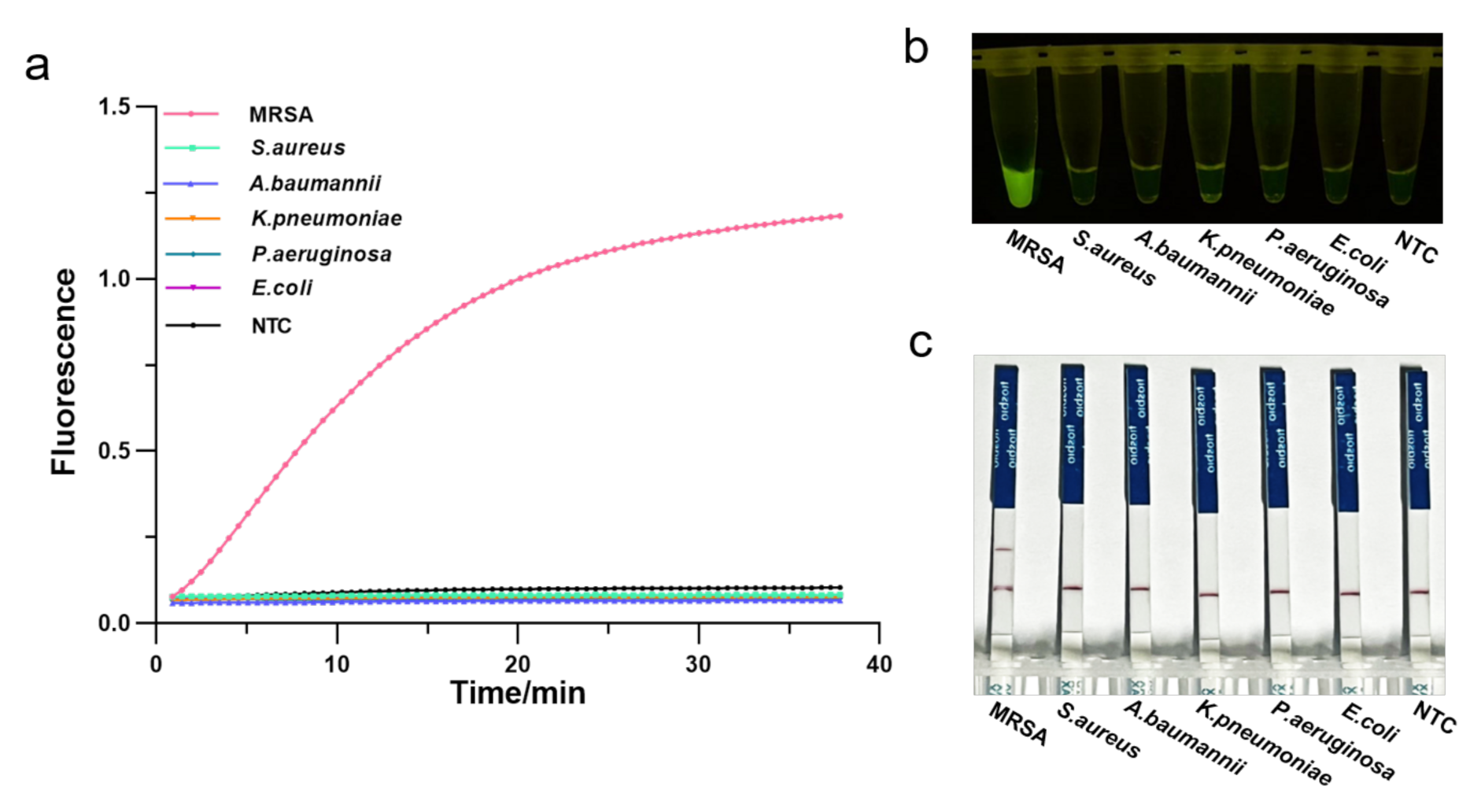
3.4. Specificity of the One-Tube RPA-CRISPR/Cas12a Detection
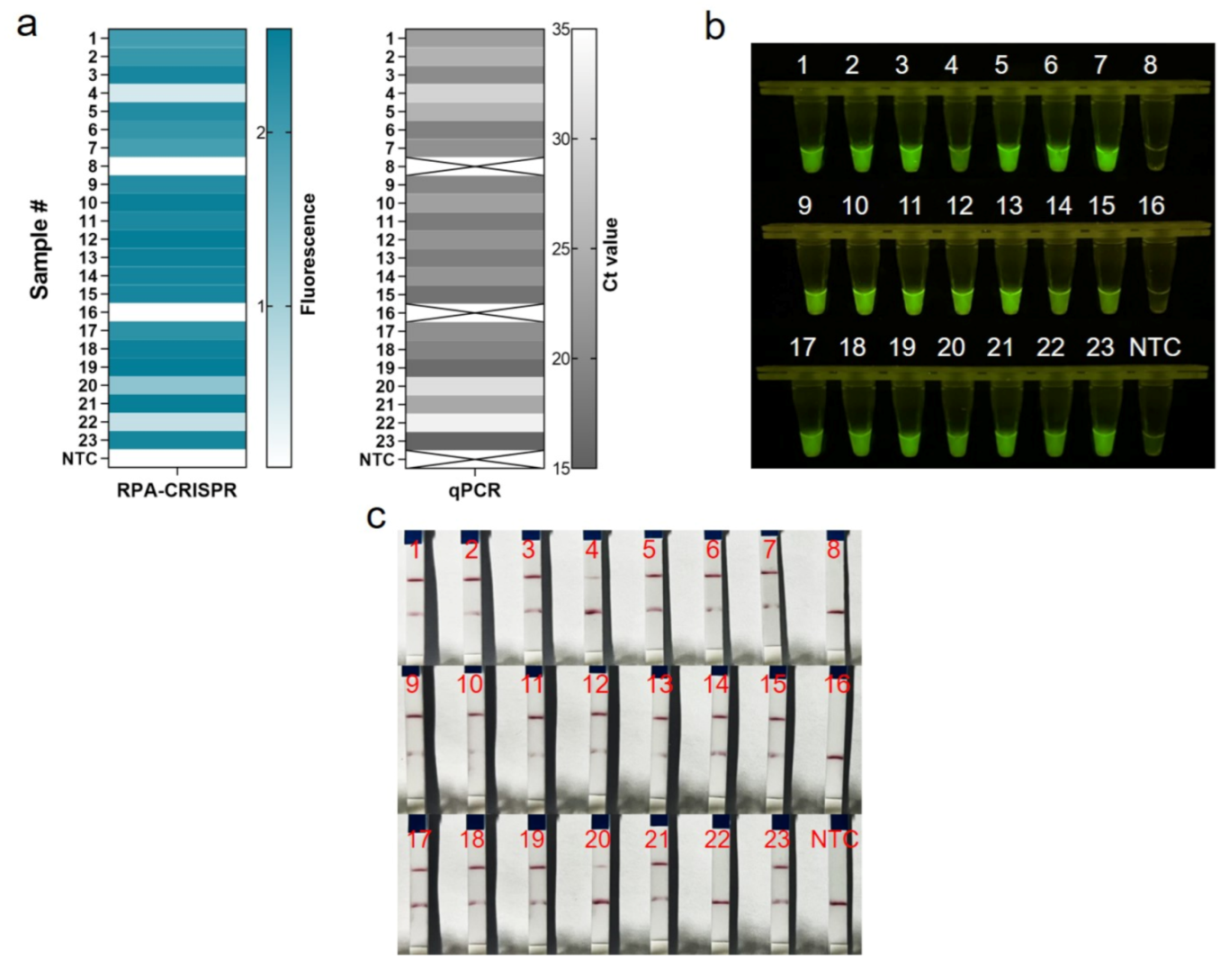
3.5. Validation of One-Tube RPA-CRISPR/Cas12a MRSA Detection Assay for Clinical Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Pulverer, G. Taxonomy of Staphylococcus aureus. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 1986, 262, 425–437. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Craft, K.M.; Nguyen, J.M.; Berg, L.J.; Townsend, S.D. Methicillin-resistant Staphylococcus aureus (MRSA): Antibiotic-resistance and the biofilm phenotype. MedChemComm 2019, 10, 1231–1241. [Google Scholar] [CrossRef]
- Sabath, L.D.; Finland, M. Inactivation of methicillin, oxacillin and ancillin by Staphylococcus aureus. Proc. Soc. Exp. Biol. Med. 1962, 111, 547–550. [Google Scholar] [CrossRef]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Stryjewski, M.E.; Corey, G.R. Methicillin-resistant Staphylococcus aureus: An evolving pathogen. Clin. Infect. Dis. 2014, 58, S10–S19. [Google Scholar] [CrossRef] [Green Version]
- Cascioferro, S.; Carbone, D.; Parrino, B.; Pecoraro, C.; Giovannetti, E.; Cirrincione, G.; Diana, P. Therapeutic Strategies to Counteract Antibiotic Resistance in MRSA Biofilm-Associated Infections. ChemMedChem 2021, 16, 65–80. [Google Scholar] [CrossRef]
- King, M.D.; Humphrey, B.J.; Wang, Y.F.; Kourbatova, E.V.; Ray, S.M.; Blumberg, H.M. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 2006, 144, 309–317. [Google Scholar] [CrossRef] [Green Version]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [Green Version]
- Pickens, C.I.; Qi, C.; Postelnick, M.; Paonessa, J.; Donnelly, H.K.; Wunderink, R.G. Association between a rapid diagnostic test to detect methicillin-resistant Staphylococcus Aureus pneumonia and decreased vancomycin use in a medical intensive care unit over a 30-month period. Infect. Control Hosp. Epidemiol. 2021, 42, 1385–1387. [Google Scholar] [CrossRef]
- Zimmerman, C.E.; Stamper, P.D.; Bryant, L.; Farley, J.; Golova, J.; Holmberg, R.; Howard, T.; Linger, Y.; Meyers, K.; Perov, A.; et al. Development of a simple, low-density array to detect methicillin-resistant Staphylococcus aureus and mecA dropouts in nasal swabs. J. Microbiol. Methods 2012, 91, 366–376. [Google Scholar] [CrossRef]
- Francois, P.; Pittet, D.; Bento, M.; Pepey, B.; Vaudaux, P.; Lew, D.; Schrenzel, J. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 2003, 41, 254–260. [Google Scholar] [CrossRef] [Green Version]
- Peterson, L.R.; Liesenfeld, O.; Woods, C.W.; Allen, S.D.; Pombo, D.; Patel, P.A.; Mehta, M.S.; Nicholson, B.; Fuller, D.; Onderdonk, A. Multicenter evaluation of the LightCycler methicillin-resistant Staphylococcus aureus (MRSA) advanced test as a rapid method for detection of MRSA in nasal surveillance swabs. Clin. Microbiol. 2010, 48, 1661–1666. [Google Scholar] [CrossRef] [Green Version]
- Gajdacs, M. The Continuing Threat of Methicillin-Resistant Staphylococcus aureus. Antibiotics 2019, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, S.Y.; Wang, J.; Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37, 730–743. [Google Scholar] [CrossRef]
- Van Dongen, J.E.; Berendsen, J.T.W.; Steenbergen, R.D.M.; Wolthuis, R.M.F.; Eijkel, J.C.T.; Segerink, L.I. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens. Bioelectron. 2020, 166, 112445. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, Q.X.; Liu, J.K.; Nie, X.Q.; Zhao, G.P.; Wang, J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018, 28, 491–493. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Li, S.Y.; Cheng, Q.X.; Wang, J.M.; Li, X.Y.; Zhang, Z.L.; Gao, S.; Cao, R.B.; Zhao, G.P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.Q.; Ke, Y.Q.; Liu, W.J.; Sun, Y.Q.; Ding, X.T. A One-Pot Toolbox Based on Cas12a/crRNA Enables Rapid Foodborne Pathogen Detection at Attomolar Level. ACS Sens. 2020, 5, 1427–1435. [Google Scholar] [CrossRef]
- Chen, Y.; Mei, Y.; Zhao, X.; Jiang, X. Reagents-Loaded, Automated Assay that Integrates Recombinase-Aided Amplification and Cas12a Nucleic Acid Detection for a Point-of-Care Test. Anal. Chem. 2020, 92, 14846–14852. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Li, Z.Y.; Zhao, H.; Cooper, K.; Liu, C.C. Dynamic Aqueous Multiphase Reaction System for One-Pot CRISPR-Cas12a-Based Ultrasensitive and Quantitative Molecular Diagnosis. Anal. Chem. 2020, 92, 8561–8568. [Google Scholar] [CrossRef]
- Tokajian, S. New epidemiology of Staphylococcus aureus infections in the Middle East. Clin. Microbiol. Infect. 2014, 20, 624–628. [Google Scholar] [CrossRef] [Green Version]
- Hulme, J. Recent Advances in the Detection of Methicillin Resistant Staphylococcus aureus (MRSA). Biochip J. 2017, 11, 89–100. [Google Scholar] [CrossRef]
- Wei, J. Accurate and sensitive analysis of Staphylococcus aureus through CRISPR-Cas12a based recycling signal amplification cascades for early diagnosis of skin and soft tissue infections. J. Microbiol. Methods 2021, 183, 106167. [Google Scholar] [CrossRef]
- Xu, L.Q.; Dai, Q.Q.; Shi, Z.Y.; Liu, X.T.; Gao, L.; Wang, Z.Z.; Zhu, X.Y.; Li, Z. Accurate MRSA identification through dual-functional aptamer and CRISPR-Cas12a assisted rolling circle amplification. J. Microbiol. Methods 2020, 173, 105917. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [Green Version]
- Aman, R.; Mahas, A.; Mahfouz, M. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 2020, 9, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.B.; Zeng, H.J.; Liu, X.F.; Jiang, W.; Wang, Y.; Ouyang, W.B.; Tang, X.M. RPA-Cas12a-FS: A frontline nucleic acid rapid detection system for food safety based on CRISPR-Cas12a combined with recombinase polymerase amplification. Food Chem. 2021, 334, 127608. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Long, J.; Yuan, M.; Zheng, X.; Shen, Y.; Jin, Y.; Yang, H.; Li, H.; Chen, S.; Duan, G. CRISPR/Cas12-Based Ultra-Sensitive and Specific Point-of-Care Detection of HBV. Int. J. Mol. Sci. 2021, 22, 4842. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Deng, X.D.; Yu, G.X.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.M.; Zhang, Y.; Xie, K.B. Evaluation of CRISPR/Cas12a-based DNA detection for fast pathogen diagnosis and GMO test in rice. Mol. Breed. 2020, 40, 11. [Google Scholar] [CrossRef]
- Wang, B.; Wang, R.; Wang, D.Q.; Wu, J.; Li, J.X.; Wang, J.; Liu, H.H.; Wang, Y.M. Cas12aVDet: A CRISPR/Cas12a-Based Platform for Rapid and Visual Nucleic Acid Detection. Anal. Chem. 2019, 91, 12156–12161. [Google Scholar] [CrossRef]
- Kim, T.H.; Park, J.; Kim, C.J.; Cho, Y.K. Fully integrated lab-on-a-disc for nucleic acid analysis of food-borne pathogens. Anal. Chem. 2014, 86, 3841–3848. [Google Scholar] [CrossRef]

| qPCR | Coincidence Rate (CR) | |||
|---|---|---|---|---|
| Positive | Negative | |||
| RPA-Cas12a fluorescence-based | Positive | 21 | 0 | 100% |
| Negative | 0 | 2 | ||
| Total | 21 | 2 | ||
| RPA-Cas12a lateral flow strip | Positive | 20 | 0 | 95.7% |
| Negative | 1 | 2 | ||
| Total | 21 | 2 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Shi, Z.; Hu, A.; Cui, J.; Yang, K.; Liu, Y.; Deng, G.; Zhu, C.; Zhu, L. Rapid One-Tube RPA-CRISPR/Cas12 Detection Platform for Methicillin-Resistant Staphylococcus aureus. Diagnostics 2022, 12, 829. https://doi.org/10.3390/diagnostics12040829
Li Y, Shi Z, Hu A, Cui J, Yang K, Liu Y, Deng G, Zhu C, Zhu L. Rapid One-Tube RPA-CRISPR/Cas12 Detection Platform for Methicillin-Resistant Staphylococcus aureus. Diagnostics. 2022; 12(4):829. https://doi.org/10.3390/diagnostics12040829
Chicago/Turabian StyleLi, Yanan, Zhonglin Shi, Anzhong Hu, Junsheng Cui, Ke Yang, Yong Liu, Guoqing Deng, Cancan Zhu, and Ling Zhu. 2022. "Rapid One-Tube RPA-CRISPR/Cas12 Detection Platform for Methicillin-Resistant Staphylococcus aureus" Diagnostics 12, no. 4: 829. https://doi.org/10.3390/diagnostics12040829
APA StyleLi, Y., Shi, Z., Hu, A., Cui, J., Yang, K., Liu, Y., Deng, G., Zhu, C., & Zhu, L. (2022). Rapid One-Tube RPA-CRISPR/Cas12 Detection Platform for Methicillin-Resistant Staphylococcus aureus. Diagnostics, 12(4), 829. https://doi.org/10.3390/diagnostics12040829







