The Need for Standardization in Next-Generation Sequencing Studies for Classic Hodgkin Lymphoma: A Systematic Review
Abstract
1. Introduction
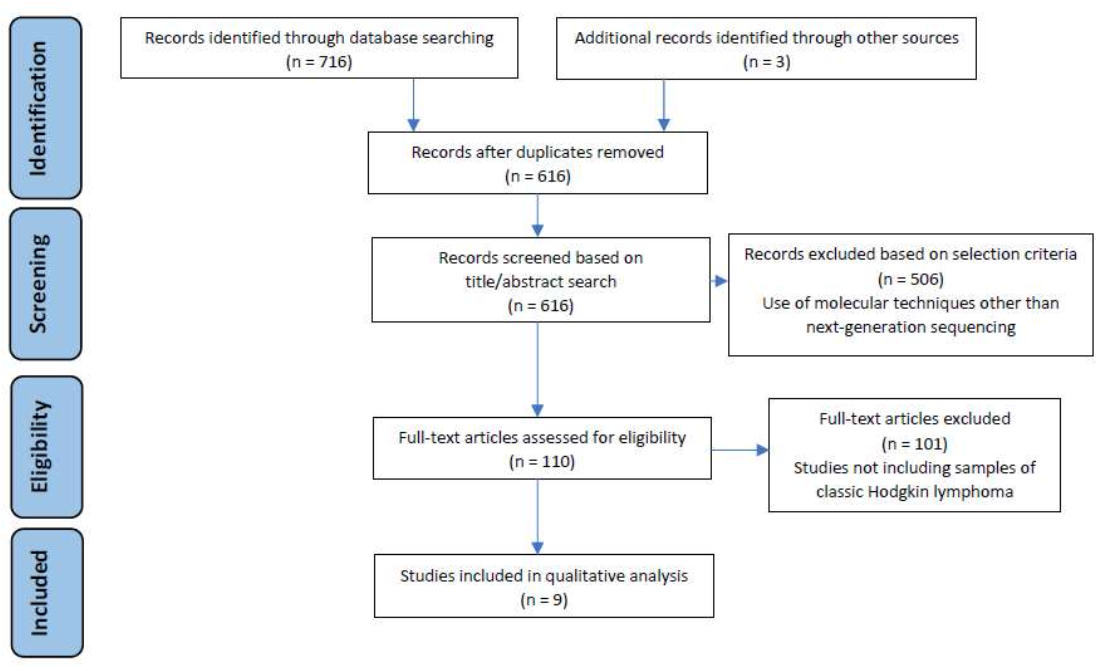
2. Materials and Methods
2.1. Search Strategy
2.2. Selection Criteria
2.3. Study Selection Process and Data Extraction
3. Results
3.1. Next-Generation Sequencing for the Assessment of Clonality in Classic Hodgkin Lymphoma
3.2. Next-Generation Sequencing and Liquid Biopsy: New Approaches for the Diagnosis and Follow-Up of Patients with Classic Hodgkin Lymphoma
3.3. Identification of High-Risk Mutational Profiles in Classic Hodgkin Lymphoma through Next-Generation Sequencing Methods
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hodgkin, T. On some Morbid Appearances of the Absorbent Glands and Spleen. Med. Chir. Trans. 1832, 17, 68–114. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, C. Über Eine Eigenartige Unter Dem Bilde Der Pseudoleukamie Verlaufende Tuberculose Des Lymphatischen Apparates. Z. Heilkd. 1898, 19, 21–90. [Google Scholar]
- Reed, D. On the Pathological Changes in Hodgkin’s Disease, with Special reference to its relation to tuberculosis. John Hopkins Hosp. Rep. 1902, 10, 133–196. [Google Scholar] [CrossRef]
- Vardhana, S.; Younes, A. The immune microenvironment in Hodgkin lymphoma: T cells, B cells and immune checkpoints. Haematologica 2016, 101, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Küppers, R.; Rajewsky, K.; Zhao, M.; Simons, G.; Laumann, R.; Fischer, R.; Hansmann, M.L. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show cloncal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc. Natl. Acad. Sci. USA 1994, 91, 10962–10966. [Google Scholar] [CrossRef] [PubMed]
- Mata, E.; Diaz-Lopez, A.; Martin-Moreno, A.; Sanchez-Beato, M.; Varela, I.; Mestre, M.J.; Garcia, J.F. Analysis of the mutational landscape of classic Hodgkin lymphoma identifies disease heterogeneity and potential therapeutic targets. Oncotarget 2017, 8, 111386. [Google Scholar] [CrossRef] [PubMed]
- Tiacci, E.; Ladewig, E.; Schiavoni, G.; Penson, A.; Fortini, E.; Pettirossi, V.; Wang, Y.; Rosseto, A.; Venanzi, A.; Vlasevska, S.; et al. Pervasive mutations of JAK/STAT pathways genes in classical Hodgkin lymphoma. Blood 2018, 131, 2454–2465. [Google Scholar] [CrossRef]
- Joos, S.; Küpper, M.; Ohl, S.; von Bonin, F.; Mechtersheimer, G.; Bentz, M.; Marynen, P.; Möller, P.; Pfreundschuh, M.; Trümper, L.; et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res. 2000, 60, 549–552. [Google Scholar]
- Hartmann, S.; Martin-Subero, J.I.; Gesk, S.; Hüsken, J.; Gieding, M.; Nagel, I.; Riemke, J.; Chott, A.; Klapper, W.; Parrens, M.; et al. Detection of genomic imbalances in microdissected Hodgkin and Reed-Sternberg cells of classical Hodgkin’s lymphoma by array-based comparative genomic hybridization. Haematologica 2008, 93, 1318–1326. [Google Scholar] [CrossRef]
- Lake, A.; Shield, L.A.; Cordano, P.; Chui, D.T.; Osborne, J.; Crae, S.; Wilson, K.S.; Tosi, S.; Knight, S.J.; Gesk, S.; et al. Mutations of NFKBIA, encoding IkappaB alpha, are a recurrent finding in classical Hodgkin lymphoma but are not a unifying feature of non-EBV-associated cases. Int. J. Cancer. 2009, 125, 1334–1342. [Google Scholar] [CrossRef]
- Jungnickel, B.; Staratschek-Jox, A.; Bräuninger, A.; Spieker, T.; Wolf, J.; Diehl, V.; Hansmann, M.L.; Rajewsky, K.; Küppers, R. Clonal deleterious mutations in the IkappaBalpha gene in the malignant cells in Hodgkin’s lymphoma. J. Exp. Med. 2000, 191, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Monti, S.; Rodig, S.J.; Juszczynski, P.; Currie, T.; O’Donnell, E.; Chapuy, B.; Takeyama, K.; Neuberg, D.; Golub, T.R.; et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010, 116, 3268–3277. [Google Scholar] [CrossRef] [PubMed]
- Roemer, M.G.; Advani, R.H.; Ligon, A.H.; Natkunam, Y.; Redd, R.A.; Homer, H.; Connelly, C.F.; Sun, H.H.; Daadi, S.E.; Freeman, G.J.; et al. PD-L1 and PD-L2 Genetic alterations define classical Hodgkin lymphoma and predict outcome. J. Clin. Oncol. 2016, 34, 2690–2697. [Google Scholar] [CrossRef] [PubMed]
- Sujobert, P.; Le Bris, Y.; de Leval, L.; Gros, A.; Merlio, J.P.; Pastoret, C.; Huet, S.; Sarkozy, C.; Davi, F.; Callanan, M.; et al. The need for a Consensus Next-Generation Sequencing panel for Mature Lymphoid Malignancies. Hemasphere 2018, 3, e169. [Google Scholar] [CrossRef]
- Eichenauer, D.A.; Aleman, B.M.P.; André, M.; Federico, M.; Hutchings, M.; Illidge, T.; Engert, A.; Ladetto, M. ESMO Guidelines Committee. Hodgkin lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv19–iv29. [Google Scholar] [CrossRef]
- Hutton, B.; Salanti, G.; Caldwell, D.M.; Chaimani, A.; Schmid, C.H.; Cameron, C.; Ioannidis, J.P.; Straus, S.; Thorlund, K.; Jansen, J.P.; et al. The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: Checklist and explanations. Ann. Intern. Med. 2015, 162, 777–784. [Google Scholar] [CrossRef]
- Trecourt, A.; Mauduit, C.; Szablewski, V.; Fontaine, J.; Balme, B.; Donzel, M.; Laurent, C.; Sesques, P.; Ghesquières, H.; Bachy, E.; et al. Plasticity of Mature B Cells between Follicular and Classic Hodgkin lymphomas: A series of 22 cases expanding the spectrum of transdifferentiation. Am. J. Surg. Pathol. 2020, 46, 58–70. [Google Scholar] [CrossRef]
- Alcoceba, M.; Garcia-Alvarez, M.; Chillon, M.C.; Jimenez, C.; Medina, A.; Anton, A.; Blanco, O.; Diaz, L.G.; Tamayo, P.; Gonzalez-Calle, V.; et al. Liquid biopsy: A non-invasive approach for Hodgkin lymphoma genotyping. Br. J. Haematol. 2021, 195, 542–551. [Google Scholar] [CrossRef]
- Van Bladel, D.A.G.; van den Brand, M.; Rijntjes, J.; Pamidimarri Naga, S.; Haacke, D.L.C.M.; Luijks, J.A.C.W.; Hebeda, K.M.; van Krieken, J.H.J.M.; Groenen, P.J.T.A.; Scheijen, B. Clonality assessment and detection of clonal diversity in classic Hodgkin lymphoma by next-generation sequencing of immunoglobulin gene rearrangements. Mod. Pathol. 2021, 1–10. [Google Scholar] [CrossRef]
- Camus, V.; Viennot, M.; Lequesne, J.; Viailly, P.J.; Bohers, E.; Bessi, L.; Marcq, B.; Etancelin, P.; Dubois, S.; Picquenot, J.M.; et al. Targeted genotyping of circulating tumor DNA for classical Hodgkin lymphoma monitoring: A prospective study. Haematologica 2021, 106, 154–162. [Google Scholar] [CrossRef]
- Mata, E.; Fernandez, S.; Astudillo, A.; Fernandez, R.; Garcia-Cosio, M.; Sanchez-Beato, M.; Provencio, M.; Estevez, M.; Montalban, C.; Piris, M.A.; et al. Genomic analyses of microdissected Hodgkin and Reed-Sternberg cells: Mutations in epigenetic regulators and p53 are frequent in refractory classic Hodgkin lymphoma. Blood Cancer J. 2019, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.S.; Vergilio, J.A.; Salhia, B.; Huang, H.J.; Oki, Y.; Garrido-Laguna, I.; Park, H.; Westin, J.R.; Meric-Bernstam, F.; Fabrizio, D.; et al. Comprehensive Genomic Profiling of Hodgkin lymphoma reveals recurrently mutated genes and increased mutation burden. Oncologist 2019, 24, 219–228. [Google Scholar] [CrossRef] [PubMed]
- European Network-Paediatric Hodgkin Lymphoma Study Group (EuroNet-PHL) Second International Inter-Group Study for Classical Hodgkin Lymphoma in Children and Adolescents. Available online: https://clinicaltrials.gov/ct2/show/NCT02684708 (accessed on 11 April 2022).
- Desch, A.K.; Hartung, K.; Botzen, A.; Brobeil, A.; Rummel, M.; Kurch, L.; Georgi, T.; Jox, T.; Bielack, S.; Burdach, S.; et al. Genotyping circulating tumor DNA of pediatric Hodgkin lymphoma. Leukemia 2020, 34, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Spina, V.; Bruscaggin, A.; Cuccaro, A.; Martini, M.; Di Trani, M.; Forestieri, G.; Manzoni, M.; Condoluci, A.; Arribas, A.; Terzi-Di-Bergamo, L.; et al. Circulating tumor DNA reveals genetics, clonal evolution and residual disease in classical Hodgkin lymphoma. Blood 2018, 131, 2413–2425. [Google Scholar] [CrossRef]
- Ten Berge, R.L.; Oudejans, J.J.; Dukers, D.F.; Meijer, J.W.; Ossenkoppele, G.J.; Meijer, C.J. Percentage of activated cytotoxic T-lymphocytes in anaplastic large cell lymphoma and Hodgkin’s disease: An independent biological prognostic marker. Leukemia 2001, 15, 458–464. [Google Scholar] [CrossRef][Green Version]
- Alvaro, T.; Lejeune, M.; Salvadó, M.T.; Bosch, R.; García, J.F.; Jaén, J.; Banham, A.H.; Roncador, G.; Montalbán, C.; Piris, M.A. Outcome in Hodgkin’s lymphoma can be predicted from the presence of accompanying cytotoxic and regulatory T cells. Clin. Cancer Res. 2005, 11, 1467–1473. [Google Scholar] [CrossRef]
- Steidl, C.; Lee, T.; Shah, S.P.; Farinha, P.; Han, G.; Nayar, T.; Delaney, A.; Jones, S.J.; Iqbal, J.; Weisenburger, D.D.; et al. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N. Engl. J. Med. 2010, 362, 875–885. [Google Scholar] [CrossRef]
- Kamper, P.; Bendix, K.; Hamilton-Dutoit, S.; Honoré, B.; Nyengaard, J.R.; d’Amore, F. Tumor-infiltrating macrophages correlate with adverse prognosis and Epstein-Barr virus status in classical Hodgkin’s lymphoma. Haematologica 2011, 96, 269–276. [Google Scholar] [CrossRef]
- Sánchez-Espiridión, B.; Martin-Moreno, A.M.; Montalbán, C.; Medeiros, L.J.; Vega, F.; Younes, A.; Piris, M.A.; Garcia, J.F. Immunohistochemical markers for tumor associated macrophages and survival in advanced classical Hodgkin’s lymphoma. Haematologica 2012, 97, 1080–1084. [Google Scholar] [CrossRef]
- Schmitz, R.; Hansmann, M.L.; Bohle, V.; Martin-Subero, J.I.; Hartmann, S.; Mechtersheimer, G.; Klapper, W.; Vater, I.; Giefing, M.; Gesk, S.; et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J. Exp. Med. 2009, 206, 981–989. [Google Scholar] [CrossRef]
- Reichel, J.; Chadburn, A.; Rubinstein, P.G.; Giulino-Roth, L.; Tam, W.; Liu, Y.; Gaiolla, R.; Eng, K.; Brody, J.; Inghurami, G.; et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 2015, 125, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Mohty, R.; Dulery, R.; Bazarbachi, A.H.; Savani, M.; Hamed, R.A.; Bazarbachi, A.; Mohty, M. Latest advances in the management of classical Hodgkin lymphoma: The era of novel therapies. Blood Cancer J. 2021, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Hasenclever, D.; Diehl, V.A. A prognostic score for advanced Hodgkin’s disease. International Prognostic Factors Project on Advanced Hodgkin’s disease. N. Engl. J. Med. 1998, 339, 1506–1514. [Google Scholar] [CrossRef]
- Engert, A.; Plütschow, A.; Eich, H.T.; Lohri, A.; Dörken, B.; Borchmann, P.; Berger, B.; Greil, R.; Willborn, K.C.; Wilhem, M. Reduced treatment intensity in patients with early-stage Hodgkin’s lymphoma. N. Engl. J. Med. 2010, 363, 640–652. [Google Scholar] [CrossRef] [PubMed]
- André, M.P.E.; Girinsky, T.; Federico, M.; Reman, O.; Fortpied, C.; Gotti, M.; Casasnovas, O.; Brice, P.; van der Maazen, R.; Re, A. Early positron emission tomotraphy response-adapted treatment in stage I and II Hodgkin lymphoma: Final results of the randomized EORTC/LYSA/FILH10 trial. J. Clin. Oncol. 2017, 35, 1786–1794. [Google Scholar] [CrossRef]
- Weniger, M.A.; Melzner, I.; Menz, C.K.; Wegener, S.; Bucur, A.J.; Dorsch, K.; Mattfeldt, T.; Barth, T.F.; Möller, P. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006, 25, 2679–2684. [Google Scholar] [CrossRef]
- Jaffe, E.S. The elusive Reed-Sternberg cell. N. Engl. J. Med. 1989, 320, 529–531. [Google Scholar] [CrossRef]
- Küppers, R.; Zhao, M.; Hansmann, M.L.; Rajewsky, K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J. 1993, 12, 4955–4967. [Google Scholar] [CrossRef]
- Weniger, M.A.; Küppers, R. Molecular biology of Hodgkin lymphoma. Leukemia 2021, 35, 968–981. [Google Scholar] [CrossRef]
- Schmid, C.; Pan, L.; Diss, T.; Isaacson, P.G. Expression of B-cell antigens by Hodgkin’s and Reed-Sternberg cells. Am. J. Pathol. 1991, 139, 701–707. [Google Scholar]
- Hsu, S.M.; Yang, K.; Jaffe, E.S. Phenotypic expression of Hodgkin’s and Reed-Sternberg cells in Hodgkin’s disease. Am. J. Pathol. 1985, 118, 209–217. [Google Scholar]
- Kanzler, H.; Küppers, R.; Hansmann, M.L.; Rajewsky, K. Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J. Exp. Med. 1996, 184, 1495–1505. [Google Scholar] [CrossRef] [PubMed]
- Ushmorov, A.; Ritz, O.; Hummel, M.; Leithäuser, F.; Möller, P.; Stein, H.; Wirth, T. Epigenetic silencing of the immunoglobulin heavy-chain gene in classical Hodgkin lymphoma-derived cell lines contributes to the loss of immunoglobulin expression. Blood 2004, 104, 3326–3334. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, A.; Oker, E.; Bentink, S.; Lenze, D.; Stein, H.; Hummel, M. Histone acetylation and DNA demethylation of B cells result in a Hodgkin-like phenotype. Leukemia 2008, 22, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Stein, H.; Marafioti, T.; Foss, H.D.; Laumen, H.; Hummel, M.; Anagnostopoulos, I.; Wirth, T.; Demel, G.; Falini, B. Down-regulation of BOB.1/OBF.1 and Oct2 in classical Hodgkin disease but not in lymphocyte predominant Hodgkin disease correlates with immunoglobulin transcription. Blood 2001, 97, 496–501. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Ferreri, A.J.; Govi, S.; Pileri, S.A.; Savage, K.J. Anaplastic large cell lymphoma, ALK-positive. Crit. Rev. Oncol. Hematol. 2012, 83, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Van den Brand, M.; Rijntjes, J.; Möbs, M.; Steinhilber, J.; van der Klift, M.Y.; Heezen, K.C.; Kroeze, L.I.; Reigl, T.; Porc, J.; Darzentas, N.; et al. Euroclonality-NGS working group. Next-Generation sequencing-based clonality assessment of Ig gene rearrangements: A multicenter validation study by EuroClonality-NGS. J. Mol. Diagn. 2021, 23, 1105–1115. [Google Scholar] [CrossRef]
- Schneider, S.; Crescenzi, B.; Schneider, M.; Ascani, S.; Hartmann, S.; Hansmann, M.L.; Falini, B.; Mecucci, C.; Tiacci, E.; Küppers, R. Subclonal evoluation of a classical Hodgkin lymphoma from a germinal center B-cell derived mantle cell lymphoma. Int. J. Cancer 2014, 134, 832–843. [Google Scholar] [CrossRef]
- Foo, W.C.; Huang, Q.; Sebastian, S.; Hutchinson, C.B.; Burchette, J.; Wang, E. Concurrent classical Hodgkin lymphoma and plasmablastic lymphoma in a patient with chronic lymphocytic leukemia/small lymphocytic lymphoma treated with fludarabine: A dimorphic presentation of iatrogenic immunodeficiency-associated lymphoproliferative disorder with evidence suggestive of multiclonal transformability of B cells by Epstein-Barr virus. Hum. Pathol. 2010, 41, 1802–1808. [Google Scholar] [CrossRef]
| Results | Database | Search Terms |
|---|---|---|
| 434 | Web of Science | “Hodgkin lymphoma”, “Hodgkin disease”, “Next Generation Sequencing”, “NGS” and “Molecular biology” |
| 282 | MEDLINE |
| Clinical Usefulness | Major Findings | Bioinformatic Analysis | Sequencing Chemistry | Origin of Tumor DNA | Sample Size and Clinical Features | Goal of the NGS Experiment | Study |
|---|---|---|---|---|---|---|---|
| Adjusted chemotherapy depending on the mutational profile | ARID1A and KTM2D commonly mutated in FL and cHL. There is a secondary clonal evolution after transdifferentiation | LymphoTrack and Vidijil software | Illumina, San Diego, CA, USA | FFPE and fresh frozen tissue | 3 sequential lymphomas for clonality and 5 cases for targeted NGS | To evaluate transdifferentiation between cHL and follicular lymphoma (FL) | Trecourt et al., (2021) [17] |
| ctDNA as a feasible strategy for genotyping and monitoring | Variants in SOCS1 (28%), IGLL5 (36%), TNFAIP3 (23%), GNA13 (23%) and STAT6 (21%). Poor prognosis features correlated with ctDNA concentration | VarScan2 and DGCaller algorithms for variant calling. RefSeq database for functional annotation | Illumina, San Diego, CA, USA | ctDNA | 60 cases of newly diagnosed cHL | Identify cHL somatic variants | Alcoceba et al., (2021) [18] |
| NGS as a sensitive and specific assay for clonality analysis | Clonality detection rates: fresh frozen: NGS (88%) vs. BIOMED-2 (63%); FFPE tissue: NGS (56%) vs. BIOMED-2 (20%) | ARResT/Interrogate pipeline | Ion TorrentTM, Thermo Fisher, Waltham, MA, USA | FFPE and fresh frozen tissue | Duplicated analysis (PCR and NGS) of 16 primary cHL cases | Compare NGS and BIOMED-2/EuroClonality for IG gene rearrangement | Van Bladel et al., (2021) [19] |
| ctDNA as a valid tool for genotyping and response assessment | Variants in SOCS1 (50%), B2M (33.3%), TNFAIP3 (31.7%), STAT6 (23.3%) and ITPKB (23.3%). ctDNA concentration correlated with metabolic tumor volume (MTV) | Software builder for base calling, alignment and quality control (Torrent Suite) | Ion TorrentTM, Thermo Fisher, Waltham, MA, USA | ctDNA | 60 cases of newly diagnosed cHL | Evaluate liquid biopsy as a new strategy for diagnosis and tailored treatment | Camus et al., (2021) [20] |
| Drugs targeting epigenetic modulators could be of interest in refractory cHL | Frequent mutations in epigenetic regulators as EP300 (41.6%) and CREBBP (33.3%) | Torrent Suite, Integrative Genomics Viewer (IGV) and PROVEAN and CONDEL algorithms | Ion TorrentTM, Thermo Fisher Scientific, NY, USA | FFPE | 12 cHL refractory patients (paired samples from diagnosis and relapse) | Identify genomic variants in refractory cHL | Mata et al., (2019) [21] |
| Modifications in the variant allele frequency of XPO1 in ctDNA correlates with clinical outcomes | Variants in TP53 (22%), B2M (22%), XPO1 (18%), TNFAIP3 (14%) and SOCS1 (10%) in biopsied tissues. XPO1 was detected in 31% of ctDNA | Not detailed | Illumina, San Diego, CA, USA | FFPE tissue and ctDNA | 63 cHL patients (clinical features not specified) | Describe the mutational profile of cHL by CGP | Liang et al., (2019) [22] |
| ctDNA quantification as a useful tool for monitoring pediatric HL patients | SOCS1 (80%), IGLL5 (33%) and TNFAIP3 (32%). Pretherapy ctDNA load was statistically significant and correlated with MTV (p = 0.0059) | Enrichment v3.0.0 and Variant Studio v3.0 For variant calling: Exome Varian, Server and ExAc | Illumina, San Diego, CA, USA | ctDNA | 96 newly diagnosed pediatric patients enrolled in the EuroNet-PHL-C2 trial [23] | Use NGS on ctDNA from cHL pediatric patients | Desch et al., (2019) [24] |
| ctDNA mirrors genetic landscape of isolated HRS cells ctDNA may serve to personalize therapeutic decisions | Mutations identified in ctDNA and biopsies were highly concordant (87.50%) Treatment pressure induced differential patterns of clonal selection | BWA software and SAM tool. VarScan2 and Integrative Genome Viewer software (IGV) | Illumina, San Diego, CA, USA | FFPE tissue and ctDNA | 80 cHL new diagnoses and 32 refractory patients | Identify the genetics of cHL in different clinical phases as well as its modifications on treatment | Spina et al., (2018) [25] |
| Drugs against members of JAK/STAT, NF-kB and BCR could be rationally used in cHL | Variants in EP300 (12.3%), CSFR2B (12.3%), BTK (10.5%) and STAT6 (10.5%). Frequent mutations involving the BCR pathway | Torrent Suite, Integrative Genomics Viewer (IGV), RAMSES, PROVEAN and Alamut algorithms | Ion TorrentTM, Thermo Fisher Scientific, NY, USA | FFPE tissue and cHL-derived cell lines | 57 cHL samples and 6 cHL-derived cell lines | Describe the mutational landscape of cHL | Mata et al., (2017) [6] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santisteban-Espejo, A.; Bernal-Florindo, I.; Perez-Requena, J.; Atienza-Cuevas, L.; Moran-Sanchez, J.; Fernandez-Valle, M.d.C.; Romero-Garcia, R.; Garcia-Rojo, M. The Need for Standardization in Next-Generation Sequencing Studies for Classic Hodgkin Lymphoma: A Systematic Review. Diagnostics 2022, 12, 963. https://doi.org/10.3390/diagnostics12040963
Santisteban-Espejo A, Bernal-Florindo I, Perez-Requena J, Atienza-Cuevas L, Moran-Sanchez J, Fernandez-Valle MdC, Romero-Garcia R, Garcia-Rojo M. The Need for Standardization in Next-Generation Sequencing Studies for Classic Hodgkin Lymphoma: A Systematic Review. Diagnostics. 2022; 12(4):963. https://doi.org/10.3390/diagnostics12040963
Chicago/Turabian StyleSantisteban-Espejo, Antonio, Irene Bernal-Florindo, Jose Perez-Requena, Lidia Atienza-Cuevas, Julia Moran-Sanchez, María del Carmen Fernandez-Valle, Raquel Romero-Garcia, and Marcial Garcia-Rojo. 2022. "The Need for Standardization in Next-Generation Sequencing Studies for Classic Hodgkin Lymphoma: A Systematic Review" Diagnostics 12, no. 4: 963. https://doi.org/10.3390/diagnostics12040963
APA StyleSantisteban-Espejo, A., Bernal-Florindo, I., Perez-Requena, J., Atienza-Cuevas, L., Moran-Sanchez, J., Fernandez-Valle, M. d. C., Romero-Garcia, R., & Garcia-Rojo, M. (2022). The Need for Standardization in Next-Generation Sequencing Studies for Classic Hodgkin Lymphoma: A Systematic Review. Diagnostics, 12(4), 963. https://doi.org/10.3390/diagnostics12040963






