Abstract
Background: FAM83H is important in teeth development; however, an increasing number of reports have indicated a role for it in human cancers. FAM83H is involved in cancer progression in association with various oncogenic molecules, including SCRIB. In the analysis of the public database, there was a significant association between FAM83H and SCRIB in colorectal carcinomas. However, studies evaluating the association of FAM83H and SCRIB in colorectal carcinoma have been limited. Methods: The clinicopathological significance of the immunohistochemical expression of FAM83H and SCRIB was evaluated in 222 colorectal carcinomas. Results: The expressions of FAM83H and SCRIB were significantly associated in colorectal carcinoma tissue. In univariate analysis, the nuclear expressions of FAM83H and SCRIB and the cytoplasmic expression of SCRIB were significantly associated with shorter survival of colorectal carcinomas. The nuclear expressions of FAM83H and SCRIB and the cytoplasmic expression of SCRIB were independent indicators of shorter cancer-specific survival in multivariate analysis. A co-expression pattern of nuclear FAM83H and cytoplasmic SCRIB predicted shorter cancer-specific survival (p < 0.001) and relapse-free survival (p = 0.032) in multivariate analysis. Conclusions: This study suggests that FAM83H and SCRIB might be used as prognostic markers of colorectal carcinomas and as potential therapeutic targets for colorectal carcinomas.
1. Introduction
FAM83H is primarily known for its importance in amelogenesis imperfecta and is involved in enamel formation during teeth development [1,2]. However, the role of FAM83H is not restricted to tooth development. It regulates cell biology in various human cells [3,4,5,6,7]. The expression of FAM83H was elevated in cancer tissue over its nonneoplastic counterpart tissue [4,5,8]. In addition, a higher expression of FAM83H is associated with a poor prognosis in human cancer patients [3,4,7,9]. In cancer of the liver [3], bone [6], kidney [9], and stomach [7], higher expressions of FAM83H predicted shorter survival. FAM83H is involved in the progression of human cancers in combination with various oncogenic signaling molecules. In conjunction with MYC and the Wnt/β-catenin pathway, FAM83H regulates cellular proliferation and invasiveness through the epithelial-to-mesenchymal transition (EMT) [3,6,10]. The role of FAM83H in EMT is suggested to be related to its localization in the cytoplasm and nuclei [7,11]. Deregulated localization of FAM83H in cancer cells has been reported in colorectal carcinoma (CRC) [11], hepatocellular carcinoma [3], gastric carcinomas [7], and gallbladder carcinoma [12].
SCRIB is a polarity protein essential in maintaining the epithelial junction and is deregulated in various pathologic conditions [13,14,15]. The abnormal expression pattern of SCRIB results in the alteration of cellular polarity [13,16]. Because of the deregulation of SCRIB in human cancers, SCRIB has been suggested to be a tumor suppressor [17]. A decreased expression of SCRIB in cancer-associated fibroblasts was associated with shorter survival of lung cancer patients [18]. However, an increasing number of reports have presented SCRIB as an important therapeutic target for cancers. In epithelial cells, the mislocalization of SCRIB resulted in the loss of the E-cadherin-mediated induction of EMT [16,19] and the activation of YAP1 signaling [20,21]. In the liver [22] and breast [23], the deregulation of SCRIB promoted tumor formation. In addition, a higher expression of SCRIB was associated with the shorter survival of ovarian and gastric cancer patients [7,24].
CRC is the third most common cancer, with the second-highest death rate [25]. Globally, 2.2 million new patients and 1.1 million deaths are predicted by 2030 [25]. The etiology and pathogenesis of CRC indicate that CRC is caused by multiple factors, such as lifestyle, genetic disorders, and the immune system [26]. Lifestyle changes are especially continuing to increase CRC incidence worldwide [26]. In addition, despite continuous improvement in the survival of CRC patients, the outcomes of CRCs vary widely according to cancer-specific molecular features and patient characteristics [27]. Therefore, it is important to find adequate biomarkers useful in treating CRCs. Recently, FAM83H and SCRIB have been presented as important regulators of human cancers [3,4,7,15,20], and FAM83H was closely associated with SCRIB in the progression of gastric carcinomas [7]. Furthermore, a search of the public database (GEPIA, http://gepia.cancer-pku.cn, accessed on 25 May 2022) indicated a significant association between SCRIB and FAM83H (Pearson’s correlation r = 0.74, p < 0.001) in CRC cancer [28]. However, there is no well-established study showing the relationship between FAM83H and SCRIB in CRCs. Therefore, this study aimed to investigate the expression and clinical significance of FAM83H and SCRIB in CRC patients.
2. Materials and Methods
2.1. Colorectal Carcinoma Patients
This study was approved by the institutional review board of Jeonbuk National University Hospital (IRB number, CUH 2021-10-034) and was performed in compliance with the Declaration of Helsinki. The paraffin-embedded tissue blocks, microscopic slides, and clinicopathologic information for this study were provided by the Biobank of Jeonbuk National University Hospital. Written informed consent was obtained by the Biobank of Jeonbuk National University Hospital. The patients included in this study were CRC patients who had received an operation between January 2018 and May 2019 at Jeonbuk National University Hospital, and their biospecimen were archived in the Biobank of Jeonbuk National University Hospital. Two hundred and twenty-two cases of CRCs were included in this study, and all cases were evaluated according to the latest WHO classification [26] and the American Joint Committee Cancer Staging System [29]. The patients who received neoadjuvant chemotherapy were not included in this study. Eighty-four patients received adjuvant chemotherapy principally based on the FOLFOX regimen (5-fluorouracil, leucovorin, and oxaliplatin) for twelve cycles over six months. The clinicopathological factors included in this study were the age of the patients, sex, site of the tumor, histologic grade, preoperative serum level of CEA (reference value; ~5.0 ng/mL), and CA19-9 (reference value; ~37 kU/L), TNM stage, T category of the stage, lymph node metastasis, and distant metastasis.
2.2. Immunohistochemical Staining and Scoring
The expressions of FAM83H and SCRIB in CRC tissue samples were evaluated with immunohistochemical staining. Immunohistochemical staining was performed on tissue microarray sections with 4 μm thickness. The tissue microarrays contained two 3.0 mm cores per case. Antigen retrieval of tissue sections was performed by boiling in pH 6.0 antigen retrieval solution (DAKO, Glostrup, Denmark) for 20 min. The tissue sections were incubated with primary antibodies for FAM83H (dilution: 1:100, Bethyl Laboratories, Montgomery, TX, USA) and SCRIB (dilution: 1:50, Santa Cruz Biotechnology, Santa Cruz, CA, USA). Then, the appropriate secondary antibodies were applied and samples counterstained with hematoxylin. The immunostained slides were evaluated by all authors by consensus under multi-viewing microscopy. The staining slides were scored according to staining intensity (0; no, 1; weak, 2; intermediate, 3; strong) and staining area scores (0; 0%, 1; 0~1%, 2; 2~10%, 3; 11~33%, 4; 34~66%, 5; 67~100%) [6,12,30]. The score of each tissue microarray core was determined by the sum of the intensity score and staining area score. Therefore, the scores ranged from zero to eight. After that, we used the sum of immunohistochemical staining scores from each core. The final immunohistochemical staining score ranged from zero to sixteen [3,9,24,31]
2.3. Statistical Analysis
The positivity for the immunohistochemical expression of FAM83H and SCRIB was determined by using a receiver operating characteristic (ROC) curve analysis [32]. The cut-off point for the immunohistochemical staining score was the point with the highest area under the curve (AUC) to predict the cancer-related deaths of patients [6,24,32]. The prognosis of CRC patients was evaluated for cancer-specific survival (CSS) and relapse-free survival (RFS) through September 2021. An event in CSS analysis was the death of a patient from CRC. An event in the RFS analysis was a relapse of CRC or the death of a patient from CRC. Statistical analysis was performed with the chi-square test, Kaplan–Meier survival analysis, and univariate and multivariate Cox proportional hazards regression analysis using SPSS software (IBM, version 25.0, Armonk, NY, USA). Statistical significance was determined by a p-value less than 0.05.
3. Results
3.1. Clinicopathologic Significance of Immunohistochemical Expressions of FAM83H and SCRIB in CRCs
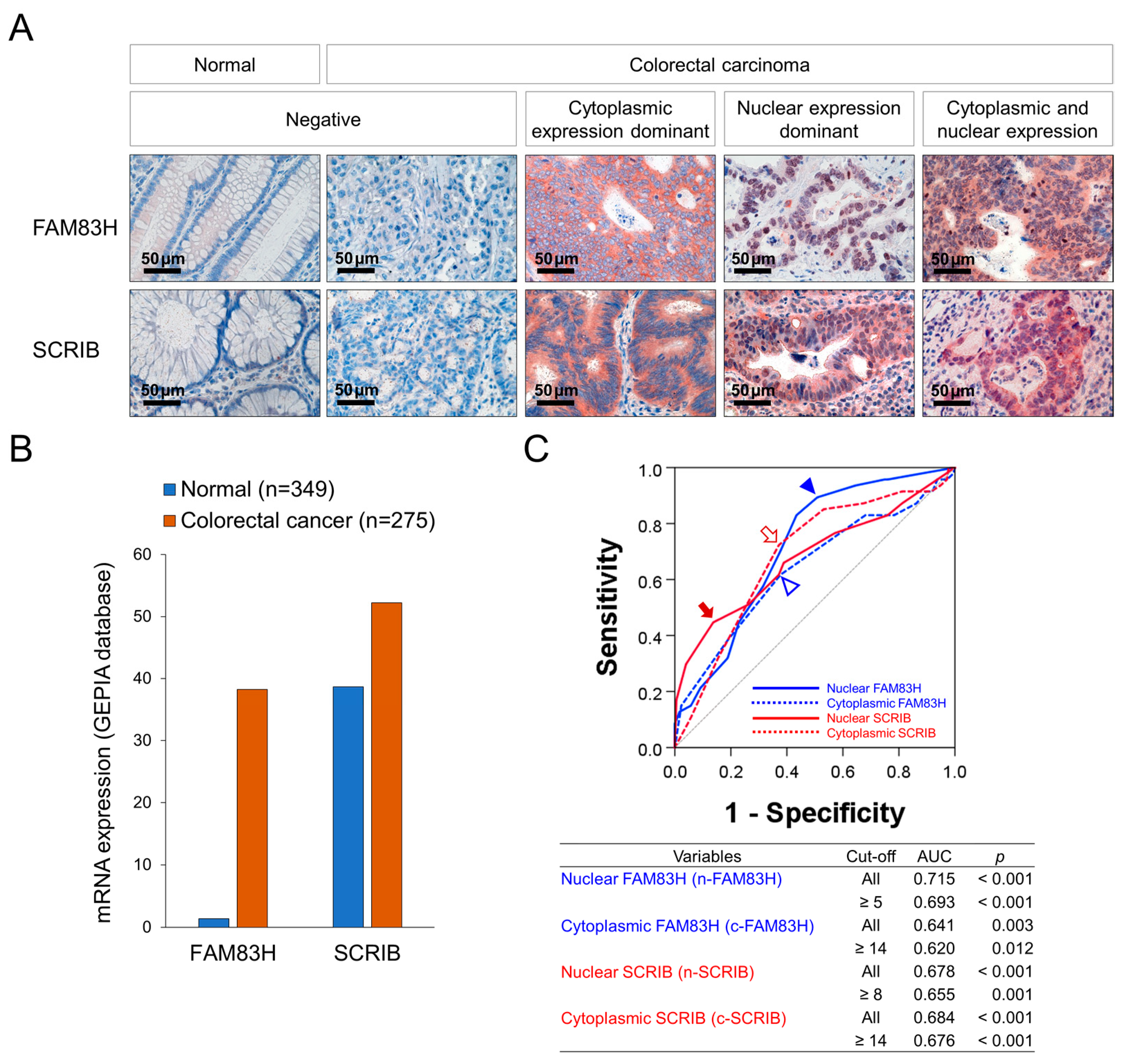
Immunohistochemically, representative examples of FAM83H and SCRIB expression in normal colonic and CRC tissue are presented in Figure 1A. The expression of the mRNA of FAM83H and SCRIB was higher in CRC tissue compared to normal conic tissue in the GEPIA2 database (Figure 1B) [28]. In our human CRC tissue, the expressions of FAM83H and SCRIB were primarily seen in the cytoplasm and nuclei of the cancer cells (Figure 1A). Therefore, we separately evaluated their nuclear and cytoplasmic expressions: nuclear expression of FAM83H (n-FAM83H), cytoplasmic expression of FAM83H (c-FAM83H), nuclear expression of SCRIB (n-SCRIB), and cytoplasmic expression of SCRIB (c-SCRIB). The immunohistochemical expressions of n-FAM83H, c-FAM83H, n-SCRIB, and c-SCRIB were classified into negative or positive groups according to their expression intensity and area. The cut-off points were determined as the points with the highest area under the curve in ROC curve analysis to predict the deaths of the patients (Figure 1C). The cut-off points for the expressions of n-FAM83H, c-FAM83H, n-SCRIB, and c-SCRIB were 5, 14, 8, and 14, respectively (Figure 1C). The cases with immunohistochemical staining scores equal to or greater than 5 for n-FAM83H, 14 for c-FAM83H, 8 for n-SCRIB, and 14 for c-SCRIB expression were considered positive (Figure 1B). With these cut-off values, n-FAM83H positivity was significantly associated with the histologic grade (p = 0.042), serum level of CA19-9 (p < 0.001), T category of stage (p = 0.033), distant metastasis (p = 0.003), and the expressions of c-SCRIB (p = 0.018) and n-SCRIB (p = 0.011) (Table 1). The expression of c-FAM83H was significantly associated with sex (p = 0.020) and c-SCRIB expression (p < 0.001) (Table 1). The positivity of n-SCRIB was significantly associated with the serum levels of CEA (p = 0.011) and CA19-9 (p = 0.006), stage (p = 0.003), lymph node metastasis (p = 0.005), distant metastasis (p < 0.001), and the expression of c-SCRIB (p < 0.001) (Table 1). The expression of c-SCRIB was significantly associated with distant metastasis (p = 0.013) (Table 1).
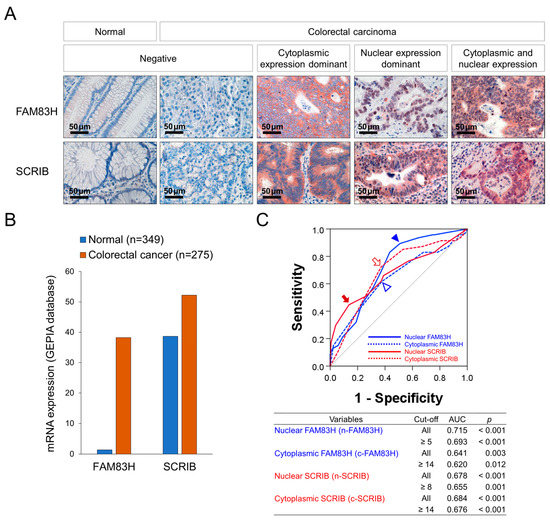
Figure 1.
Immunohistochemical staining for FAM83H and SCRIB and statistical analysis in CRCs. (A) Immunohistochemical expressions of FAM83H and SCRIB in normal and CRC tissue. FAM83H and SCRIB were expressed in the cytoplasm and nuclei of the CRC cells. (B) The expression of the mRNA of FAM83H and SCRIB in normal colonic tissue and colorectal carcinoma tissue from the GEPIA2 database (http://gepia2.cancer-pku.cn, accessed on 23 June 2022) (C) Receiver operating characteristic curve analysis was performed to determine the positivity of the nuclear expression of FAM83H (n-FAM83H), cytoplasmic expression of FAM83H (c-FAM83H), nuclear expression of SCRIB (n-SCRIB), and cytoplasmic expression of SCRIB (c-SCRIB). The blue arrowhead indicates the cut-off point for n-FAM83H expression, and the empty blue arrowhead indicates the cut-off point for c-SCRIB expression. The red arrow indicates the cut-off point for n-SCRIB expression, and the open red arrow indicates the cut-off point for c-SCRIB expression. The cut-off points were determined to predict the deaths of CRC patients, and the points have the highest area under the curve (AUC).

Table 1.
Association between the expressions of FAM83H and SCRIB with clinicopathological characteristics of 122 CRCs.
3.2. Individual Expressions of FAM83H and SCRIB Were Associated with the Survival of CRC Patients
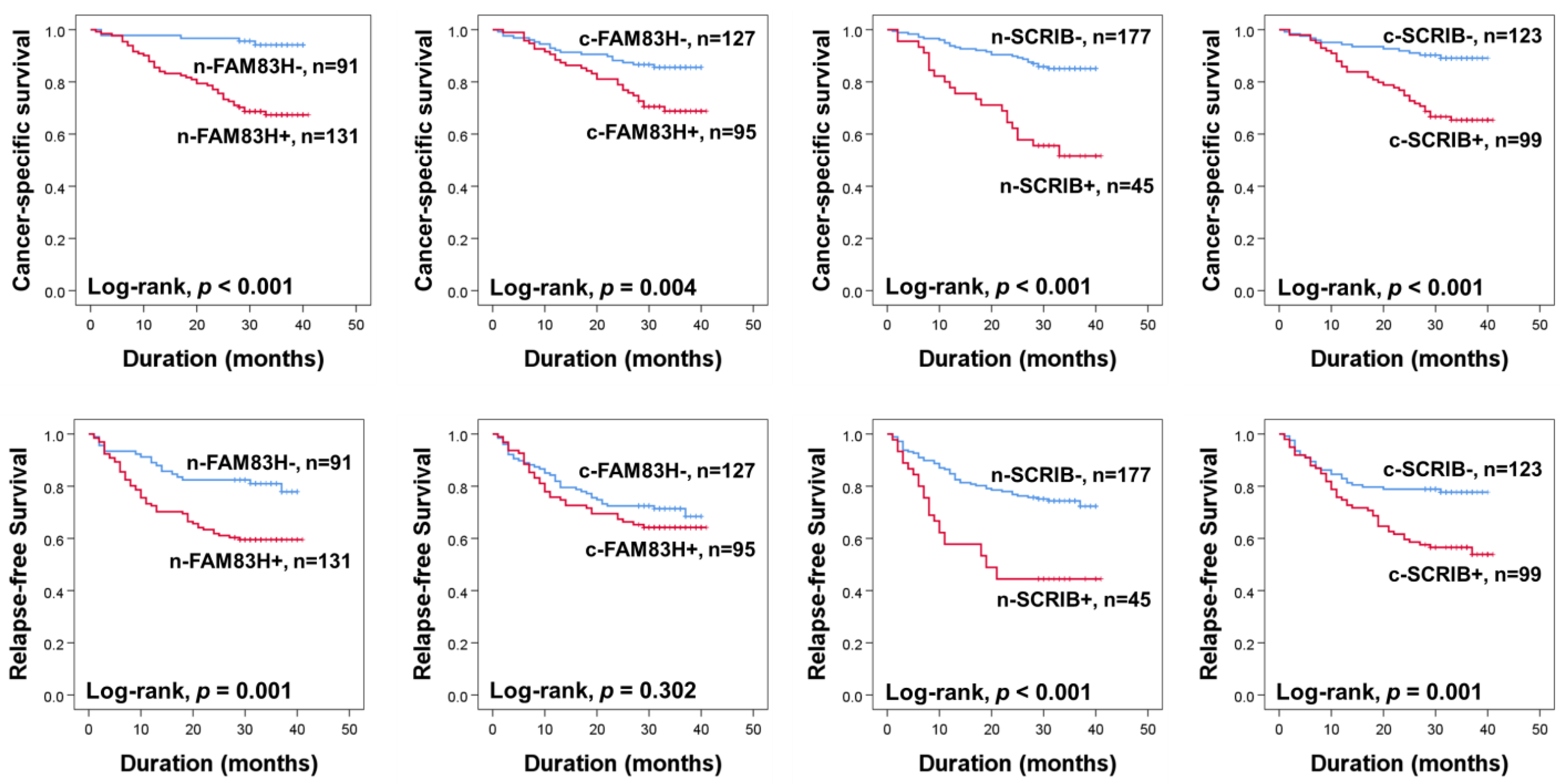
The prognoses of CRC patients were evaluated for CSS and RFS. In univariate analysis, the ages of the patients (CSS; p = 0.007, RFS; p = 0.062), elevated serum levels of CEA (CSS; p < 0.001, RFS; p < 0.001) and CA19-9 (CSS; p < 0.001, RFS; p < 0.001), cancer stage (CSS; p < 0.001, RFS; p < 0.001), T categories of stage (CSS; p < 0.001, RFS; p < 0.001), lymph node metastasis (CSS; p < 0.001, RFS; p < 0.001), distant metastasis at diagnosis (CSS; p < 0.001, RFS; p < 0.001), and the expressions of n-SCRIB (CSS; p < 0.001, RFS; p = 0.001), c-SCRIB (CSS; p < 0.001, RFS; p < 0.001), c-FAM83H (CSS; p = 0.006, RFS; p = 0.307), and n-FAM83H (CSS; p < 0.001, RFS; p < 0.001) were significantly associated with CSS or RFS (Table 2). The patients positive for n-FAM83H expression had a 6.793-fold (95% confidence interval (95% CI); 2.686–17.179) greater risk of death and a 2.356-fold (95% CI; 11.379–4.026) greater risk of relapse of tumor or death compared to the patients with an n-FAM83H negative tumor (Table 2). The positivity of c-FAM83H showed a 2.291-fold (95% CI; 1.272–4.126) greater risk in CSS (Table 2). The n-SCRIB positivity showed a 3.940-fold (95% CI; 2.213–7.014) greater risk in CSS and a 2.731-fold (95% CI; 1.675–4.453) greater risk in RFS (Table 2). The positivity of c-SCRIB showed a 3.597-fold (95% CI; 1.897–6.819) greater risk in CSS and a 2.175-fold (95% CI; 1.347–3.514) greater risk in RFS (Table 2). The Kaplan–Meier survival curves for CSS and RFS according to the expressions of n-FAM83H, c-FAM83H, n-SCRIB, and c-SCRIB are presented in Figure 2.

Table 2.
Univariate Cox proportional hazards regression analysis for cancer-specific survival and relapse-free survival in 122 CRCs.
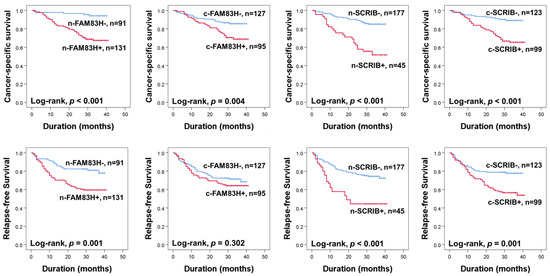
Figure 2.
Kaplan–Meier survival analysis for cancer-specific survival and relapse-free survival according to nuclear and cytoplasmic expressions of FAM83H and SCRIB in CRC patients. n-FAM83H, nuclear expression of FAM83H; c-FAM83H, cytoplasmic expression of FAM83H; n-SCRIB, nuclear expression of SCRIB; c-SCRIB, cytoplasmic expression of SCRIB.
Multivariate analysis was performed with the factors significantly associated with CSS and RFS: preoperative serum levels of CEA and CA19-9, tumor stage, T category of tumor stage, lymph node metastasis, distant metastasis, and the expressions of n-FAM83H, n-SCRIB, and c-SCRIB. The factors significantly associated with CSS in multivariate analysis were CA19-9 (p = 0.004), T category of stage (p = 0.041), distant metastasis (p < 0.001), and the expressions of n-SCRIB (p = 0.042), c-SCRIB (p = 0.033), and n-FAM83H (p = 0.020) (Table 3). Distant metastasis was the factor significantly associated with RFS in multivariate analysis (p < 0.001) (Table 3). n-FAM83H positivity predicted a 3.170-fold (95% CI; 1.197–8.397) greater death risk than FAM83H-negative tumors (Table 3). c-SCRIB positivity showed a 2.087-fold (95% CI; 1.063–4.099) greater risk in CSS (Table 3). n-SCRIB-positive cancer patients had a 1.884-fold (95% CI; 1.022–3.474) greater risk of CSS (Table 3).

Table 3.
Multivariate Cox regression analysis for cancer-specific survival and relapse-free survival.
3.3. The Co-Expression Pattern of FAM83H and SCRIB Predicted the Survival of CRC Patients
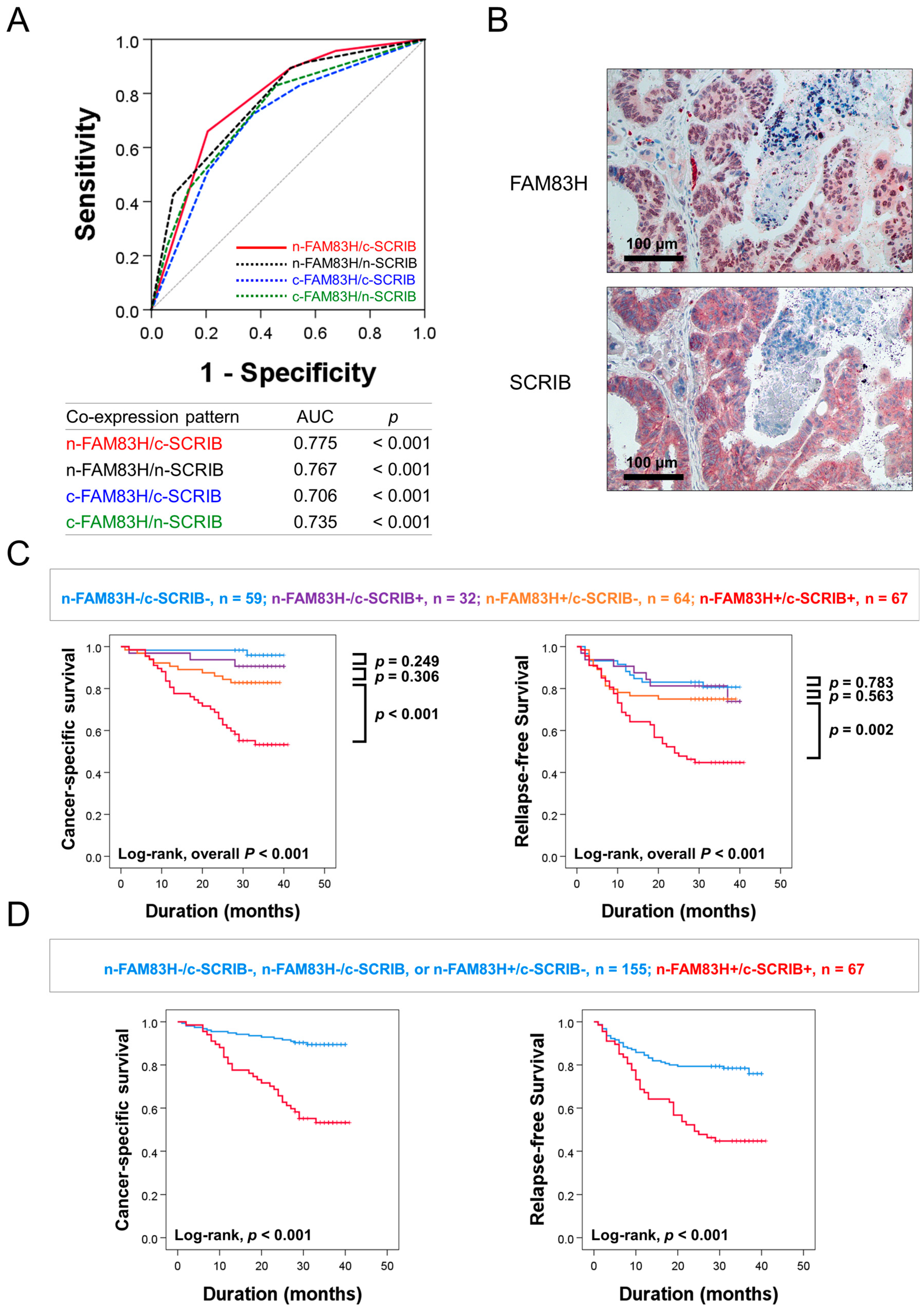
In this study, there were significant associations among the expressions of n-FAM83H, c-FAM83H, n-SCRIB, and c-SCRIB (Table 1). The relationship between FAM83H and SCRIB was also reported in gastric cancers [7]. Based on this relationship, we evaluated the clinical significance of the combined expression patterns of n-FAM83H, c-FAM83H, n-SCRIB, and c-SCRIB in CRCs: n-FAM83H/c-SCRIB, n-FAM83H/n-SCRIB, c-FAM83H/c-SCRIB, and c-FAM83H/n-SCRIB. Among the four types of combined expression patterns, the co-expression pattern of n-FAM83H/c-SCRIB had the largest AUC (AUC = 0.775, p < 0.001) (Figure 3A). Therefore, we first evaluated the prognostic significance of the co-expression pattern of n-FAM83H/c-SCRIB in CRC patients. Representative images of the co-expression pattern of n-FAM83H and c-SCRIB in the same sample are presented in Figure 3B. Based on n-FAM83H/c-SCRIB co-expression patterns, the CRCs were grouped into four subgroups: n-FAM83H−/c-SCRIB−, n = 59; n-FAM83H−/c-SCRIB+, n = 32; n-FAM83H+/c-SCRIB−, n = 64; n-FAM83H+/c-SCRIB+, n = 67. Of these four subgroups, the n-FAM83H−/c-SCRIB− subgroup had the longest survival and the n-FAM83H+/c-SCRIB+ subgroup had the shortest survival (CSS; Log-rank overall p < 0.001, RFS; Log-rank overall p < 0.001) (Figure 3C). The n-FAM83H−/c-SCRIB− subgroup had a 96% three-year CSS rate and an 81% three-year RFS rate (Table 4). In contrast, the n-FAM83H+/c-SCRIB+ subgroup had only a 53% three-year CSS rate and a 45% three-year RFS rate (Table 4). However, there were no significant prognostic differences among the n-FAM83H−/c-SCRIB−, n-FAM83H−/c-SCRIB+, and n-FAM83H+/c-SCRIB− subgroups (Figure 3C). Therefore, we further classified the CRC patients into two prognostic subgroups: (n-FAM83H−/c-SCRIB−, n-FAM83H−/c-SCRIB+, or n-FAM83H+/c-SCRIB−) and (n-FAM83H+/c-SCRIB+) subgroups (Figure 3D). This subgrouping into two prognostic subgroups was significantly associated with CSS and RFS in univariate (CSS; p < 0.001, RFS; p < 0.001) and multivariate analysis (CSS; p < 0.001, RFS; p = 0.032) (Table 5). In multivariate analysis, the n-FAM83H+/c-SCRIB+ subgroup had a 3.210-fold (95% CI; 1.683–6.122) greater risk of shorter CSS and a 1.710-fold (95% CI; 1.047–2.793) greater risk of shorter RFS (Table 5). In addition, when we evaluated the prognostic significance of the co-expression pattern of n-FAM83H/n-SCRIB, subgrouping into the two prognostic subgroups was significantly associated with CSS (p < 0.001) and RFS (p < 0.001) in univariate analysis (Table 5). In multivariate analysis, the n-FAM83H+/n-SCRIB+ subgroup had a 2.847-fold (95% CI; 1.537–5.272, p < 0.001) greater risk of shorter CSS (Table 5).
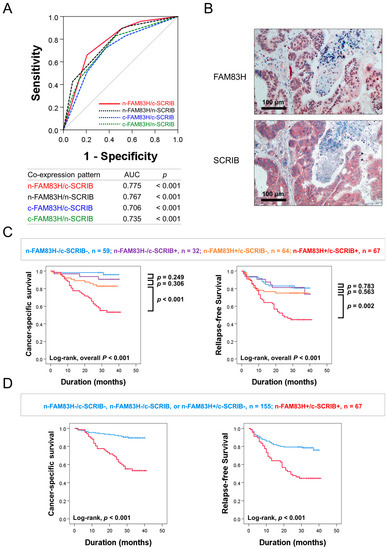
Figure 3.
Statistical analysis and Kaplan–Meier survival analysis according to the combined expression pattern of nuclear and cytoplasmic expressions of FAM83H and SCRIB. (A) Receiver operating characteristic curve analysis to predict the death of CRC patients according to combined expression patterns of nuclear and cytoplasmic expressions of FAM83H and SCRIB: n-FAM83H/c-SCRIB, n-FAM83H/n-SCRIB, c-FAM83H/c-SCRIB, and c-FAM83H/n-SCRIB. (B) Representative images of the co-expression patterns of n-FAM83H and c-SCRIB in the same CRC sample. (C) Kaplan–Meier survival analysis in four prognostic subgroups of CRCs according to the combined expression pattern of n-FAM83H and c-SCRIB: (n-FAM83H−/c-SCRIB−), (n-FAM83H−/c-SCRIB+), (n-FAM83H+/c-SCRIB−), and (n-FAM83H+/c-SCRIB+) subgroups. (D) Survival analysis in two prognostic subgroups of CRCs: (n-FAM83H−/c-SCRIB−, n-FAM83H−/c-SCRIB+, or n-FAM83H+/c-SCRIB−) and (n-FAM83H+/c-SCRIB+) subgroups.

Table 4.
One- and three-year cancer-specific survival and relapse-free survival according to co-expression patterns of n-FAM83H and c-SCRIB.

Table 5.
Univariate and multivariate Cox regression analysis for cancer-specific survival and relapse-free survival according to the co-expression patterns of FAM83H and SCRIB in CRCs.
3.4. Individual and Co-Expression Patterns of FAM83H and SCRIB Were Associated with Survival in the Subpopulation of CRCs According to Therapeutic Treatment
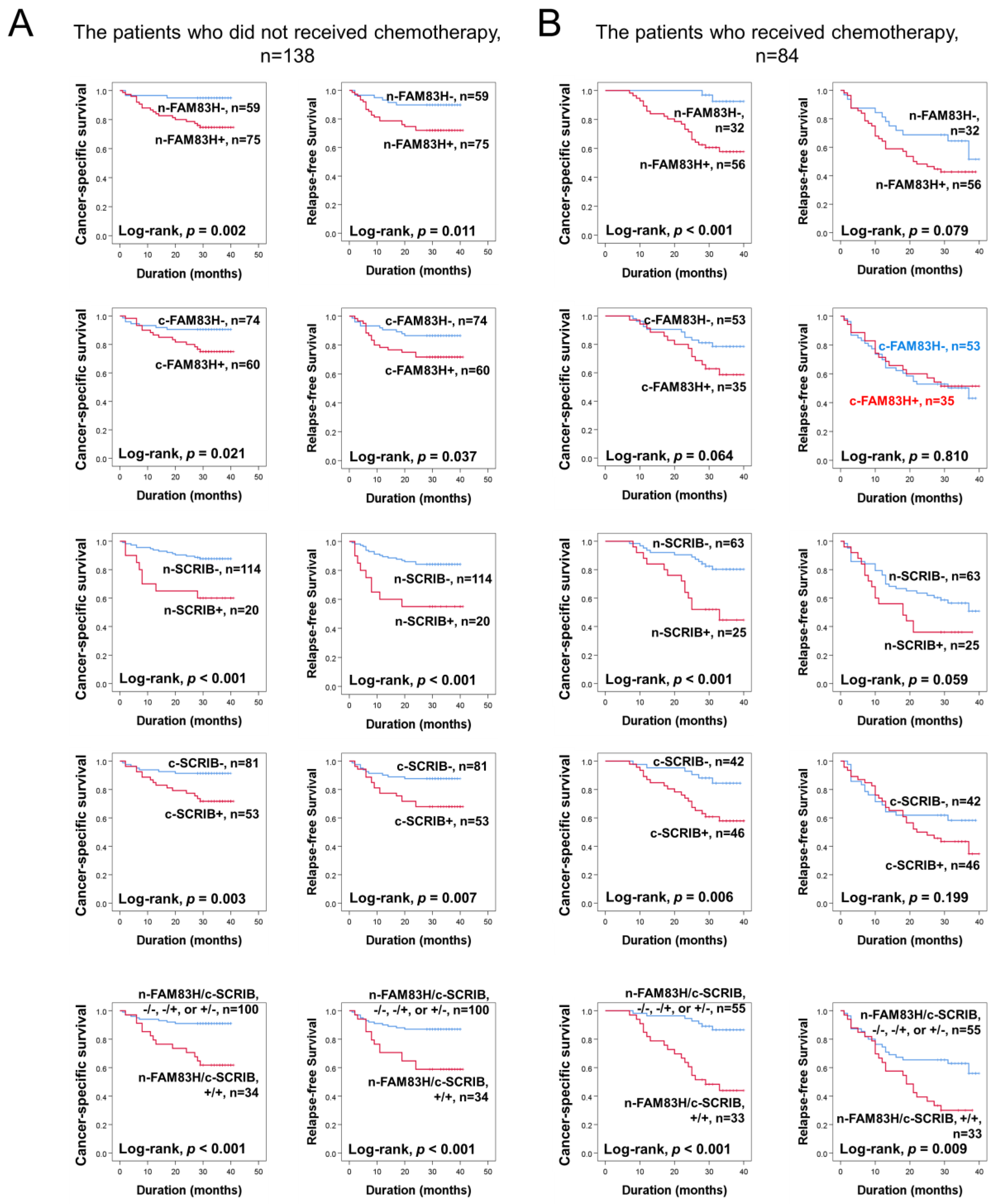
Based on the prognostic significance of FAM83H and SCRIB expressions in CRC, we further evaluated their prognostic significance in the subpopulation of CRCs according to adjuvant chemotherapy. In the 138 patients who did not receive adjuvant chemotherapy, the expressions of n-FAM83H (CSS; p = 0.002, RFS; p = 0.011), c-FAM83H (CSS; p = 0.021, RFS; p = 0.037), n-SCRIB (CSS; p < 0.001, RFS; p < 0.001), and c-SCRIB (CSS; p = 0.003, RFS; p = 0.007) and the co-expression pattern of n-FAM83H/c-SCRIB (CSS; p < 0.001, RFS; p < 0.001) were significantly associated with CSS and RFS (Figure 4A). In the 84 patients who received adjuvant chemotherapy, the expressions of n-FAM83H (p < 0.001), n-SCRIB (p < 0.001), and c-SCRIB (p = 0.006) and the co-expression pattern of n-FAM83H/c-SCRIB (p < 0.001) were significantly associated with CSS (Figure 4B). However, only the co-expression pattern of n-FAM83H/c-SCRIB (p = 0.009) was significantly associated with RFS (Figure 4B).
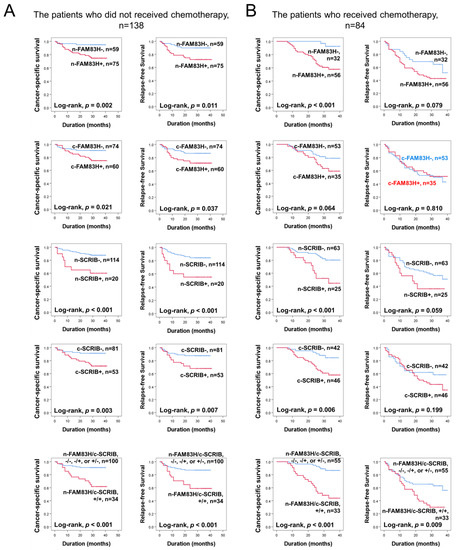
Figure 4.
Survival analysis according to individual expressions of nuclear FAM83H (n-FAM83H), cytoplasmic FAM83H (c-FAM83H), nuclear SCRIB (n-SCRIB), cytoplasmic FAM83H (c-SCRIB), and co-expression patterns of nuclear FAM83H and cytoplasmic SCRIB (n-FAM83H/c-SCRIB) in the subgroups of CRC patients who received or did not receive adjuvant chemotherapy. (A) Kaplan–Meier survival curves in the 138 patients who did not receive chemotherapy. (B) Kaplan–Meier survival curves in the 84 patients who received adjuvant chemotherapy.
4. Discussion
In this study, we have shown that the expressions of FAM83H and SCRIB were associated with advanced clinicopathologic characteristics of CRC patients. The expression of n-FAM83H was associated with a higher histologic grade, increased serum levels of CA19-9, a higher T category, and distant metastasis. In addition, higher expressions of n-FAM83H and c-FAM83H were associated with a shorter CSS of CRC patients in univariate analysis. In multivariate analysis, the expression of n-FAM83H predicted shorter CSS in CRC patients. Consistently, a higher expression of FAM83H in cancer tissue was reported in cancers of the breast, colon, liver, lung, ovary, pancreas, stomach, and uterus [4,5,8]. FAM83H positivity was associated with a higher uterine cancer stage [8] and pancreatic ductal carcinoma [33]. In line with our results, a higher expression of FAM83H was associated with the poor prognosis of cancer patients with tumors of the bladder [31], bone [6], gallbladder [12], kidney [9], liver [3], lung [34], stomach [7], and uterus [4]. However, in contrast to our results, there have been controversial reports on the significance of the expression of FAM83H in human cancers. The expression of FAM83H was decreased in cutaneous squamous cell carcinomas compared to normal tissue [35], and a higher expression of FAM83H was associated with a favorable prognosis in gastric adenocarcinomas [36]. Therefore, despite the prognostic significance of FAM83H expression in CRC patients, further study is needed to clarify the clinical relevance of FAM893H expression in CRC.
In addition to the prognostic significance of n-FAM83H expression, the expression of SCRIB was also significantly associated with shorter survival of CRC patients. The expression of n-SCRIB was significantly associated with higher serum levels of CEA and CA19-9, a higher tumor stage, lymph node metastasis, and distant metastasis. In addition, higher expressions of n-SCRIB and c-SCRIB were associated with a shorter CSS of CRC patients in univariate and multivariate analysis. In line with our results, a higher expression of SCRIB in human cancers has been reported in CRC [20,37], hepatocellular carcinoma [22], breast cancer [15], and prostate cancer [15]. Furthermore, a higher expression of SCRIB also predicted shorter survival of ovarian carcinoma [24] and gastric carcinoma [7]. However, controversially, the expression of SCRIB was decreased compared to normal tissue in oropharyngeal squamous cell carcinoma, breast cancer, uterine cervical cancer, lung cancer, and lymphoma [15,38]. Therefore, although our results suggest that the expression of SCRIB might be used as a prognostic indicator for CRC patients, further study is needed to determine the clinical significance of SCRIB expression.
In this study, there was a significant association between the expressions of FAM83H and SCRIB, as well as their co-expression patterns, which were independent prognostic indicators of CSS and RFS in CRC patients. The molecular relationship between FAM83H and SCRIB is evident in data from the public database [28] and is further supported by a study on the relationship between FAM83H and SCRIB in gastric carcinomas [7]. In gastric carcinomas, FAM83H is involved in the regulation of the expression of SCRIB and forms the FAM83H-SCRIB complex, and FAM83H-SCRIB stabilizes β-catenin to induce EMT and the proliferation of cancer cells [7]. FAM83H overexpression especially activated the tumor growth and pulmonary metastasis of gastric cancer cells, which was attenuated with the knock-down of SCRIB in vivo [7]. These results suggest that the FAM83H-mediated activation of SCRIB might be an important mechanism in FAM83H-mediated oncogenesis. This relationship between FAM83H and SCRIB might be explained by the shorter survival of CRC patients with an n-FAM83H+/c-SCRIB+ phenotype. In gastric carcinomas, patients with positive expressions of both n-FAM83H and n-SCRIB showed the shortest overall survival and RFS [7]. In addition to the molecular association of FAM83H and SCRIB in human cancers, FAM83H forms a more extensive molecular network in the progression of human cancers. In hepatocellular carcinomas, oncogene MYC induces FAM83H transcription, and the increased expression of FAM83H stimulates the growth and invasiveness of cells [3]. In osteosarcoma cells, FAM83H activated the canonical Wnt/β-catenin pathway by stabilizing β-catenin from proteasome-mediated ubiquitination [6]. A higher expression of FAM83H in KHOS/NP osteosarcoma cells increased in vivo tumor growth and pulmonary metastasis [6]. In clear cell renal cell carcinomas, the co-expression of FAM83H and pannexin-2 predicted the shortest overall survival and RFS in multivariate analysis [9]. In addition, FAM83H is involved in the proliferation of cancer cells by activating PI3K/AKT pathway in pancreatic cancer [10] and uterine cervical cancer [5]. Therefore, it is suggested that FAM83H is involved in the progression of human cancers by forming a complex network with the molecules associated with cancer progression and SCRIB in CRCs.
With regard to the prognostic significance of FAM83H and SCRIB expression in CRC patients, an interesting finding in this study is that the co-expression pattern of n-FAM83H and c-SCRIB was associated with the CSS and RFS in the patients who received adjuvant chemotherapy. This result suggests that higher expressions of FAM83H and/or SCRIB would be associated with resistance to chemotherapy. In addition, the positivity of n-FAM83H or n-SCRIB was associated with a higher tumor stage, lymph node metastasis, or distant metastasis at diagnosis. Invasive and metastatic potential is an important clinical feature of EMT, and EMT has been presented as an important phenotype of resistance to conventional anti-cancer therapies [39,40]. These results suggest the possibility that FAM83H/SCRIB might be involved in resistance to anti-cancer chemotherapy through the induction of EMT. Similarly, a higher expression of n-FAM83H and n-SCRIB was associated with higher tumor stage and lymph node metastasis of gastric carcinomas [7]. EMT phenotype, loss of E-cadherin with the induction of N-cadherin and Snail has been induced with the overexpression of FAM83H and SCRIB in gastric cancer cells and ovarian cancer cells [7,24]. A higher expression of FAM83H was associated with the induction of EMT by stimulating the PI3K pathway in pancreatic cancer cells [10]. In ovarian carcinomas, the positivity of n-SCRIB was significantly associated with platinum resistance and has been suggested as an independent indicator of poor prognosis for ovarian carcinoma patients who received adjuvant chemotherapy [24]. In in vivo experiments, the overexpression of FAM83H increased the pulmonary metastasis of NCI-N87 gastric cancer cells [7] and KHOS/NP osteosarcoma cells [6]. Regarding therapeutic application targeting the FAM83H-SCRIB pathway, the knock-down of FAM83H and/or SCRIB inhibited the growth of various human cancer cells, such as hepatocellular carcinoma [3], gastric carcinoma [7], ovarian carcinoma [24], and osteosarcoma [6]. Therefore, the FAM83H-SCRIB pathway might be a therapeutic target for human cancers, especially for the poorly prognostic subgroup highly expressing FAM83H and SCRIB. However, further study, especially on the precise mechanism regarding the role of FAM83H-SCRIB in conventional anti-cancer therapies, is needed to select a population of patients who could potentially benefit from anti-FAM83H/SCRIB-targeted therapy.
Our results suggest that the expressions of FAM83H and SCRIB might be used as prognostic markers for CRC patients. With regard to the evaluation of immunohistochemically stained slides, scoring according to the subcellular localization of selected markers is an important point. In evaluating the subcellular localization of FAM83H and SCRIB, FAM83H expression is expected in the cytoplasmic membrane and the cytosol [11,41,42], and SCRIB expression is expected in the plasma membrane [13,14]. However, as shown in Figure 1A, the expressions of FAM83H and SCRIB are primarily seen in the cytoplasm and nuclei. Furthermore, the nuclear expressions of FAM83H and SCRIB were independent indicators of shorter CSS in CRC patients. Consistently, n-FAM83H expression was an indicator of the poor prognosis of hepatocellular carcinoma [3], gastric carcinoma [7], clear cell renal cell carcinoma [9], and osteosarcoma [6]. In gastric carcinoma and ovarian carcinomas, positivity for n-SCRIB was also significantly associated with the shorter survival of patients [7,24]. The prognostic significance of the nuclear expressions of FAM83H and SCRIB might be related to their roles in cancer progression in conjunction with nuclear proteins important in cancer progression, such as MYC and β-catenin [3,7,24]. In CRC, the nuclear localization of FAM83H was present in a minor population of cancers, but its localization to nuclei has been suggested to be involved in cancer progression with the interaction of SON and casein kinase 1α [11,42]. Therefore, our results suggest that careful evaluation of the subcellular localization of FAM83H and SCRIB is important for the FAM83H and SCRIB immunostaining of cancer tissue. However, further study is needed to clarify the significance of the expressions of FAM83H and SCRIB in predicting the prognosis of human cancers.
5. Conclusions
In conclusion, the results show that individual and combined expression patterns of FAM83H and SCRIB are significantly associated with shorter CSS and RFS in CRC patients. The combined expression pattern of n-FAM83H and c-SCRIB was especially significantly associated with the shorter survival of CRC patients who received adjuvant chemotherapy. Therefore, FAM83H and SCRIB might be used as prognostic markers for CRC patients and as potential therapeutic targets for the poor prognostic population of CRC patients who have tumors that highly express FAM83H and SCRIB.
Author Contributions
T.Y.J., H.I.L., M.S.P., M.Y.S. and K.Y.J. participated in the study design. T.Y.J., H.I.L., M.S.P., M.Y.S. and K.Y.J. performed the experiment. T.Y.J., H.I.L., M.S.P., M.Y.S. and K.Y.J. were involved in the data collection and data interpretation. T.Y.J., H.I.L., M.S.P., M.Y.S. and K.Y.J. participated in the statistical analyses. T.Y.J., H.I.L., M.S.P., M.Y.S. and K.Y.J. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Medical Research Center Program (2017R1A5A2015061) through the National Research Foundation (NRF), which is funded by the Korean government (MSIP).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Jeonbuk National University Hospital (IRB number: CUH 2019-11-041, date of approval: 1 November 2021) and was performed in compliance with the Declaration of Helsinki. The paraffin-embedded tissue blocks, microscopic slides, and clinicopathologic information for this study were provided by the Biobank of Jeonbuk National University Hospital.
Informed Consent Statement
Written informed consent was obtained from the patients by the Biobank of Jeonbuk National University Hospital to publish this paper.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Acknowledgments
This study was performed with the support of a research program in the Medical Research class of Jeonbuk National University Medical School. We thank Ae Ri Ahn (Department of Pathology, Jeonbuk National University Hospital) for reviewing the clinicopathological data and supporting the scoring of immunohistochemical staining slides. We thank D.B. Leveson-Gower, who provided medical writing services. The biospecimens and clinicopathologic information for this study were provided by the Biobank of Jeonbuk National University Hospital, a member of the National Biobank of Korea, which is supported by the Ministry of Health, Welfare, and Family Affairs. All samples derived from the Korea Biobank Network were obtained with informed consent under institutional review board-approved protocols.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hart, P.S.; Becerik, S.; Cogulu, D.; Emingil, G.; Ozdemir-Ozenen, D.; Han, S.T.; Sulima, P.P.; Firatli, E.; Hart, T.C. Novel FAM83H mutations in Turkish families with autosomal dominant hypocalcified amelogenesis imperfecta. Clin. Genet. 2009, 75, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.W.; Lee, S.K.; Lee, Z.H.; Park, J.C.; Lee, K.E.; Lee, M.H.; Park, J.T.; Seo, B.M.; Hu, J.C.; Simmer, J.P. FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am. J. Hum. Genet. 2008, 82, 489–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.M.; Park, S.H.; Bae, J.S.; Noh, S.J.; Tao, G.Z.; Kim, J.R.; Kwon, K.S.; Park, H.S.; Park, B.H.; Lee, H.; et al. FAM83H is involved in the progression of hepatocellular carcinoma and is regulated by MYC. Sci. Rep. 2017, 7, 3274. [Google Scholar] [CrossRef]
- Snijders, A.M.; Lee, S.Y.; Hang, B.; Hao, W.; Bissell, M.J.; Mao, J.H. FAM83 family oncogenes are broadly involved in human cancers: An integrative multi-omics approach. Mol. Oncol. 2017, 11, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Li, H.F.; Hu, Y.J.; Jiang, M.J.; Liu, Q.S.; Zhou, J. Family with Sequence Similarity 83 Member H Promotes the Viability and Metastasis of Cervical Cancer Cells and Indicates a Poor Prognosis. Yonsei Med. J. 2019, 60, 611–618. [Google Scholar] [CrossRef]
- Kim, K.M.; Hussein, U.K.; Park, S.H.; Kang, M.A.; Moon, Y.J.; Zhang, Z.; Song, Y.; Park, H.S.; Bae, J.S.; Park, B.H.; et al. FAM83H is involved in stabilization of beta-catenin and progression of osteosarcomas. J. Exp. Clin. Cancer Res. 2019, 38, 267. [Google Scholar] [CrossRef] [Green Version]
- Hussein, U.K.; Ha, S.H.; Ahmed, A.G.; Kim, K.M.; Park, S.H.; Kim, C.Y.; Kwon, K.S.; Zhang, Z.; Lee, S.A.; Park, H.S.; et al. FAM83H and SCRIB stabilize beta-catenin and stimulate progression of gastric carcinoma. Aging 2020, 12, 11812–11834. [Google Scholar] [CrossRef]
- Lin, S.; Du, J.; Hao, J.; Luo, X.; Wu, H.; Zhang, H.; Zhao, X.; Xu, L.; Wang, B. Identification of Prognostic Biomarkers Among FAM83 Family Genes in Human Ovarian Cancer Through Bioinformatic Analysis and Experimental Verification. Cancer Manag. Res. 2021, 13, 8611–8627. [Google Scholar] [CrossRef]
- Kim, K.M.; Hussein, U.K.; Bae, J.S.; Park, S.H.; Kwon, K.S.; Ha, S.H.; Park, H.S.; Lee, H.; Chung, M.J.; Moon, W.S.; et al. The Expression Patterns of FAM83H and PANX2 Are Associated With Shorter Survival of Clear Cell Renal Cell Carcinoma Patients. Front. Oncol. 2019, 9, 14. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, H.; Zhang, C.; Hou, B. FAM83H overexpression predicts worse prognosis and correlates with less CD8(+) T cells infiltration and Ras-PI3K-Akt-mTOR signaling pathway in pancreatic cancer. Clin. Transl. Oncol. 2020, 22, 2244–2252. [Google Scholar] [CrossRef]
- Kuga, T.; Kume, H.; Adachi, J.; Kawasaki, N.; Shimizu, M.; Hoshino, I.; Matsubara, H.; Saito, Y.; Nakayama, Y.; Tomonaga, T. Casein kinase 1 is recruited to nuclear speckles by FAM83H and SON. Sci. Rep. 2016, 6, 34472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, S.W.; Ahn, A.R.; Ha, S.H.; Hussein, U.K.; Do Yang, J.; Kim, K.M.; Park, H.S.; Park, S.H.; Yu, H.C.; Jang, K.Y. Expression of FAM83H and ZNF16 are associated with shorter survival of patients with gallbladder carcinoma. Diagn. Pathol. 2020, 15, 63. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Muthuswamy, S.K. Polarity protein alterations in carcinoma: A focus on emerging roles for polarity regulators. Curr. Opin. Genet. Dev. 2010, 20, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsum, I.A.; Martin, C.; Humbert, P.O. Scribble regulates an EMT polarity pathway through modulation of MAPK-ERK signaling to mediate junction formation. J. Cell. Sci. 2013, 126, 3990–3999. [Google Scholar] [CrossRef] [Green Version]
- Santoni, M.J.; Kashyap, R.; Camoin, L.; Borg, J.P. The Scribble family in cancer: Twentieth anniversary. Oncogene 2020, 39, 7019–7033. [Google Scholar] [CrossRef]
- Martin-Belmonte, F.; Perez-Moreno, M. Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer 2011, 12, 23–38. [Google Scholar] [CrossRef]
- Moreno-Bueno, G.; Portillo, F.; Cano, A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 2008, 27, 6958–6969. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, H.; Bian, Y.; An, J.; Duan, X.; Wan, J.; Yao, X.; Du, C.; Ni, C.; Zhu, L.; et al. Low SCRIB expression in fibroblasts promotes invasion of lung cancer cells. Life Sci. 2020, 256, 117955. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Shen, H.; Huang, C.; Wu, J.; Li, J.; Hu, T.; Wang, Z.; Zhang, H.; Shao, Y.; Fu, Z. SCRIB Promotes Proliferation and Metastasis by Targeting Hippo/YAP Signalling in Colorectal Cancer. Front. Cell Dev. Biol. 2021, 9, 656359. [Google Scholar] [CrossRef]
- Zhao, D.; Yin, Z.; Soellner, M.B.; Martin, B.R. Scribble sub-cellular localization modulates recruitment of YES1 to regulate YAP1 phosphorylation. Cell Chem. Biol. 2021, 28, 1235–1241.e5. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Meyer, A.S.; Weiler, S.M.E.; Rupp, C.; Toth, M.; Sticht, C.; Singer, S.; Thomann, S.; Roessler, S.; Schorpp-Kistner, M.; et al. Cytoplasmic localization of the cell polarity factor scribble supports liver tumor formation and tumor cell invasiveness. Hepatology 2018, 67, 1842–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, L.; Rosenberg, A.; Bergami, K.C.; Yu, M.; Xuan, Z.; Jaffe, A.B.; Allred, C.; Muthuswamy, S.K. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 2008, 135, 865–878. [Google Scholar] [CrossRef] [Green Version]
- Hussein, U.K.; Ahmed, A.G.; Choi, W.K.; Kim, K.M.; Park, S.H.; Park, H.S.; Noh, S.J.; Lee, H.; Chung, M.J.; Moon, W.S.; et al. SCRIB Is Involved in the Progression of Ovarian Carcinomas in Association with the Factors Linked to Epithelial-to-Mesenchymal Transition and Predicts Shorter Survival of Diagnosed Patients. Biomolecules 2021, 11, 405. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Digestive System Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2019. [Google Scholar]
- Grothey, A.; Sargent, D.J. Adjuvant Therapy for Colon Cancer: Small Steps Toward Precision Medicine. JAMA Oncol. 2016, 2, 1133–1134. [Google Scholar] [CrossRef]
- Li, C.; Tang, Z.; Zhang, W.; Ye, Z.; Liu, F. GEPIA2021: Integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021, 49, W242–W246. [Google Scholar] [CrossRef]
- Amin, M.B.; American Joint Committee on Cancer; American Cancer Society. AJCC Cancer Staging Manual, 8th ed.; American Joint Committee on Cancer; Springer: Chicago, IL, USA, 2017. [Google Scholar]
- Allred, D.; Harvey, J.M.; Berardo, M.; Clark, G.M. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod. Pathol. 1998, 11, 155–168. [Google Scholar]
- Ahn, A.R.; Noh, S.J.; Hussein, U.K.; Park, H.S.; Chung, M.J.; Lee, H.; Moon, W.S.; Kang, M.J.; Kim, H.J.; Lee, N.R.; et al. FAM83H and Nectin1 expression are related with survival and relapse of bladder urothelial carcinoma patients. BMC Urol. 2021, 21, 143. [Google Scholar] [CrossRef]
- De Long, E.R.; De Long, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Ma, Z.; Zhou, Z.; Zhuang, H.; Li, Z.; Ma, Z.; Huang, B.; Liu, C.; Gong, Y.; Zou, Y.; Zheng, Z.; et al. Identification of Prognostic and Therapeutic Biomarkers among FAM83 Family Members for Pancreatic Ductal Adenocarcinoma. Dis. Markers 2021, 2021, 6682697. [Google Scholar] [CrossRef]
- Gan, J.; Li, Y.; Meng, Q. Systematic Analysis of Expression Profiles and Prognostic Significance for FAM83 Family in Non-small-Cell Lung Cancer. Front. Mol. Biosci. 2020, 7, 572406. [Google Scholar] [CrossRef]
- Tokuchi, K.; Kitamura, S.; Maeda, T.; Watanabe, M.; Hatakeyama, S.; Kano, S.; Tanaka, S.; Ujiie, H.; Yanagi, T. Loss of FAM83H promotes cell migration and invasion in cutaneous squamous cell carcinoma via impaired keratin distribution. J. Dermatol. Sci. 2021, 104, 112–121. [Google Scholar] [CrossRef]
- Zhang, T.; Lai, S.; Cai, Y.; Huang, Z.; Li, Y.; Chen, S.; Zhang, Z.; Ye, Z.; Lai, X.; Zhai, E.; et al. Comprehensive Analysis and Identification of Prognostic Biomarkers and Therapeutic Targets Among FAM83 Family Members for Gastric Cancer. Front. Cell Dev. Biol. 2021, 9, 719613. [Google Scholar] [CrossRef]
- Shen, H.; Meng, Y.; Hu, T.; Li, S.; Du, M.; Xin, J.; Gu, D.; Wang, M.; Fu, Z. Genetic variants in Hippo signalling pathway-related genes affect the risk of colorectal cancer. Arch. Toxicol. 2021, 95, 271–281. [Google Scholar] [CrossRef]
- Lulic, L.; Jakovcevic, A.; Manojlovic, L.; Dediol, E.; Banks, L.; Tomaic, V. Human DLG1 and SCRIB Are Distinctly Regulated Independently of HPV-16 during the Progression of Oropharyngeal Squamous Cell Carcinomas: A Preliminary Analysis. Cancers 2021, 13, 4461. [Google Scholar] [CrossRef]
- Cho, E.S.; Kang, H.E.; Kim, N.H.; Yook, J.I. Therapeutic implications of cancer epithelial-mesenchymal transition (EMT). Arch. Pharm. Res. 2019, 42, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Du, B.; Shim, J.S. Targeting Epithelial-Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules 2016, 21, 965. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Estrella, M.R.; Hu, Y.Y.; Chan, H.L.; Zhang, H.D.; Kim, J.W.; Simmer, J.P.; Hu, J.C. Fam83h is associated with intracellular vesicles and ADHCAI. J. Dent. Res. 2009, 88, 991–996. [Google Scholar] [CrossRef]
- Kuga, T.; Kume, H.; Kawasaki, N.; Sato, M.; Adachi, J.; Shiromizu, T.; Hoshino, I.; Nishimori, T.; Matsubara, H.; Tomonaga, T. A novel mechanism of keratin cytoskeleton organization through casein kinase Iα and FAM83H in colorectal cancer. J. Cell Sci. 2013, 126, 4721–4731. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).