Author Contributions
Conceptualization, R.S. and R.M.M.; methodology, R.S.; formal analysis, R.S. and N.M.; investigation, R.S. and R.M.M.; resources, R.S.; data curation, R.S. and R.M.M.; writing—original draft preparation, R.S.; writing—review and editing, R.S., N.M., J.T., N.K. and R.M.M.; supervision, N.M., J.T. and R.M.M. All authors have read and agreed to the published version of the manuscript.
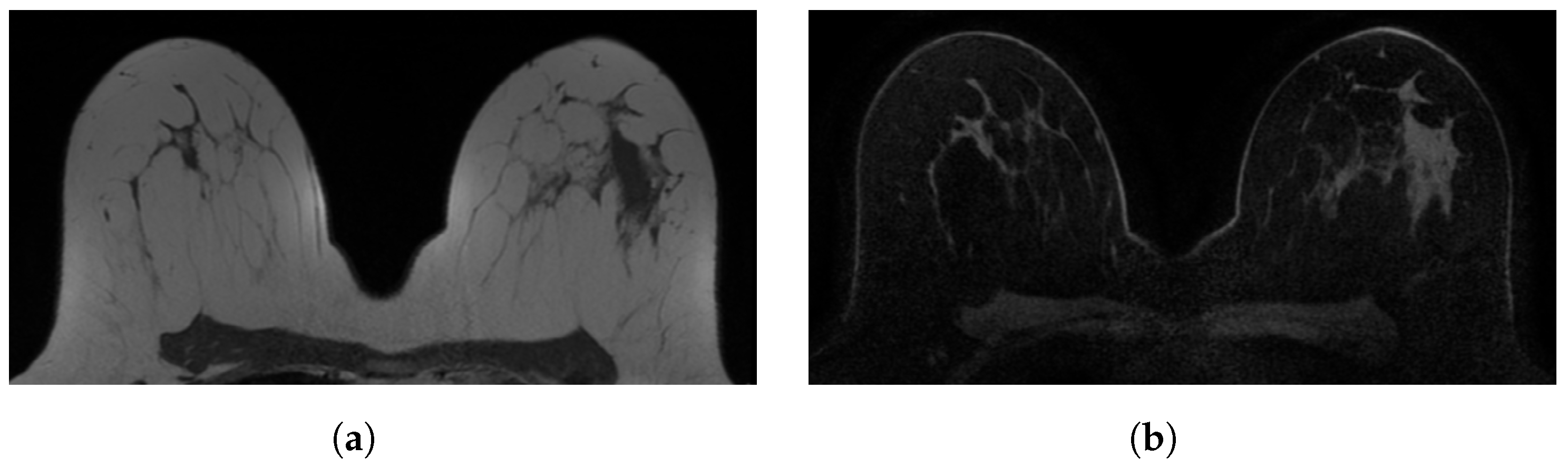
Figure 1.
Examples of in-phase and reconstructed water Dixon acquisitions from our internal dataset used to mimic acquisitions without (WOFS) and with (FS) fat suppression. (a) In-phase Dixon acquisition used as WOFS image. (b) Reconstructed water Dixon acquisition used as FS image.
Figure 1.
Examples of in-phase and reconstructed water Dixon acquisitions from our internal dataset used to mimic acquisitions without (WOFS) and with (FS) fat suppression. (a) In-phase Dixon acquisition used as WOFS image. (b) Reconstructed water Dixon acquisition used as FS image.
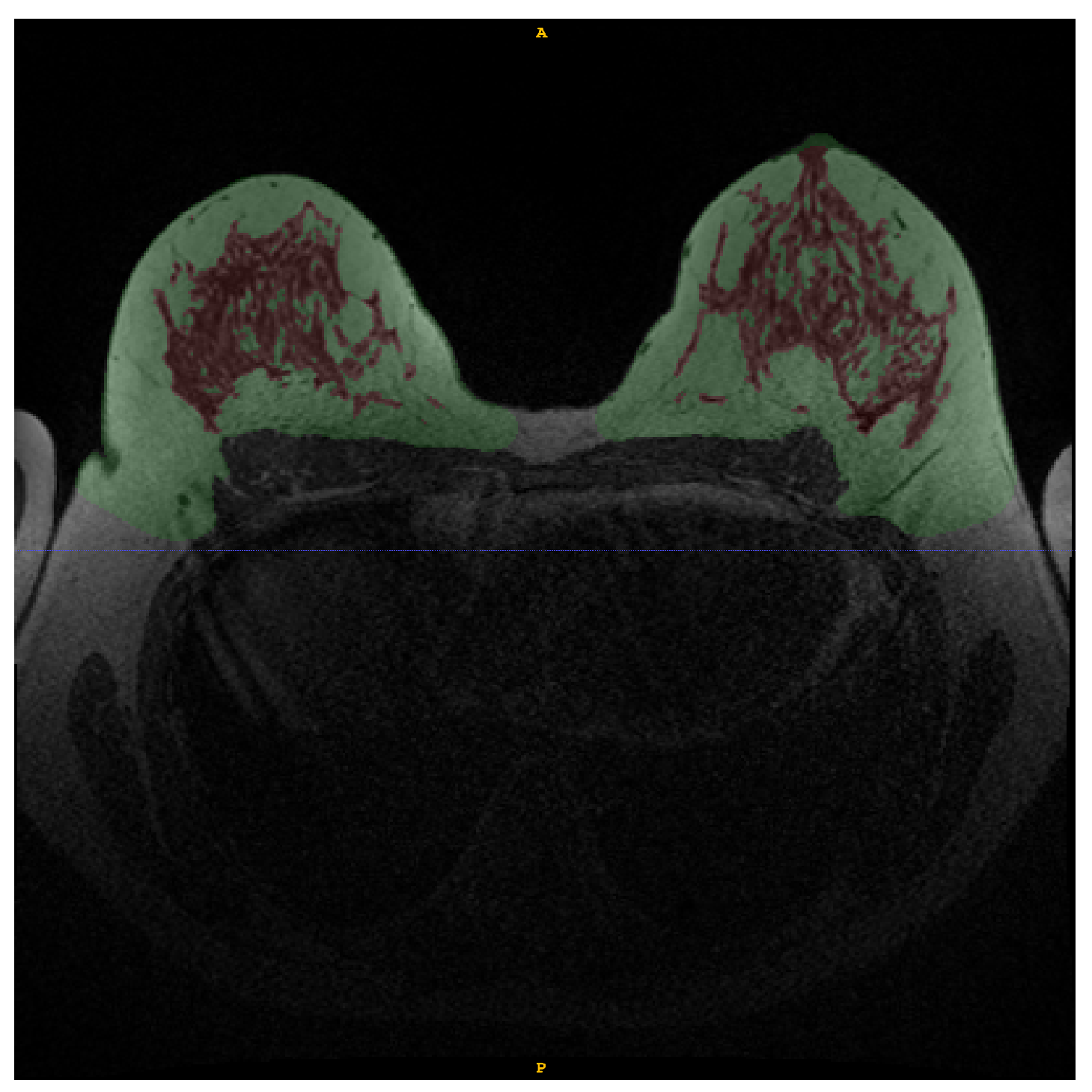
Figure 2.
Example of manual annotation of fat tissue (green) and FGT (red) in WOFS image.
Figure 2.
Example of manual annotation of fat tissue (green) and FGT (red) in WOFS image.
Figure 3.
Screenshot of reader study set-up. The Grand Challenge platform allows the set-up of a web-based reader study with a fully functional medical imaging viewer. In this reader study, we showed participants the same WOFS and FS case side-by-side, and they were asked to answer the questions on the left side of the screen, which, in this case, were multiple choice questions.
Figure 3.
Screenshot of reader study set-up. The Grand Challenge platform allows the set-up of a web-based reader study with a fully functional medical imaging viewer. In this reader study, we showed participants the same WOFS and FS case side-by-side, and they were asked to answer the questions on the left side of the screen, which, in this case, were multiple choice questions.
Figure 4.
Bland–Altman plot to compare the breast density (BD) calculated on ground truth manual annotation and BD calculated from breast and FGT segmentations generated by the network trained with WOFS + FS acquisitions. (a) Bland–Altman plot for WOFS test cases. (b) Bland–Altman plot for FS test cases.
Figure 4.
Bland–Altman plot to compare the breast density (BD) calculated on ground truth manual annotation and BD calculated from breast and FGT segmentations generated by the network trained with WOFS + FS acquisitions. (a) Bland–Altman plot for WOFS test cases. (b) Bland–Altman plot for FS test cases.
Figure 5.
Internal test set results (n = 9 patients, 18 acquisitions (9 WOFS, 9 FS)). Reader study confusion matrix comparing the ratings (1—Poor, 2—Fair, 3—Good, 4—Excellent) from the two readers.
Figure 5.
Internal test set results (n = 9 patients, 18 acquisitions (9 WOFS, 9 FS)). Reader study confusion matrix comparing the ratings (1—Poor, 2—Fair, 3—Good, 4—Excellent) from the two readers.
Figure 6.
External test set results (n = 30 patients, 60 acquisitions (30 WOFS, 30 FS)). Reader study confusion matrix comparing the ratings (1—Poor, 2—Fair, 3—Good, 4—Excellent) from the two readers.
Figure 6.
External test set results (n = 30 patients, 60 acquisitions (30 WOFS, 30 FS)). Reader study confusion matrix comparing the ratings (1—Poor, 2—Fair, 3—Good, 4—Excellent) from the two readers.
Figure 7.
Example slices of FGT segmentations from external test set acquisitions rated either poor or fair during the reader study assessment.
Figure 7.
Example slices of FGT segmentations from external test set acquisitions rated either poor or fair during the reader study assessment.
Table 1.
Breast DCE-MRI acquisition parameters for internal dataset.
Table 1.
Breast DCE-MRI acquisition parameters for internal dataset.
| Acquisition Parameter | |
|---|
| Repetition Time (ms) | 4–5.68 |
| Echo Time (ms) | 2–2.46 |
| Flip Angle | 15–20 |
| Magnetic Field Strength | 3.0 |
| Slice Thickness (mm) | 1–2.5 |
| Pixel Spacing (mm) | (0.8 × 0.8)–(0.98 × 0.98) |
Table 2.
MRI scanner information from the randomly selected subset of the external validation dataset.
Table 2.
MRI scanner information from the randomly selected subset of the external validation dataset.
| Manufacturer | Magnetic Field Strength (Tesla) | Model | Total (n = 30) |
|---|
| GE Medical Systems | 1.5 | Optima MR450w | 5 |
| | | Signa HDx | 3 |
| | | Signa HDxt | 2 |
| | 3.0 | Signa HDx | 2 |
| | | Signa HDxt | 3 |
| Siemens | 1.5 | Avanto | 5 |
| | 3.0 | Skyra | 5 |
| | | TrioTim | 5 |
Table 3.
Internal test set results (n = 9 patients, 18 acquisitions (9 WOFS, 9 FS)). Breast and FGT Dice Similarity Coefficients (DSC) and 95% Confidence Intervals (CI) from network trained only without fat suppression (WOFS only) acquisitions and without and with fat suppression acquisitions (WOFS + FS).
Table 3.
Internal test set results (n = 9 patients, 18 acquisitions (9 WOFS, 9 FS)). Breast and FGT Dice Similarity Coefficients (DSC) and 95% Confidence Intervals (CI) from network trained only without fat suppression (WOFS only) acquisitions and without and with fat suppression acquisitions (WOFS + FS).
| Network Trained on | Test | Breast DSC [95% CI] | FGT DSC [95% CI] |
|---|
| WOFS only | WOFS | 0.96 [0.94, 0.97] | 0.92 [0.89, 0.95] |
| FS | 0.10 [0.08, 0.12] | 0.15 [0.11, 0.19] |
| WOFS + FS | WOFS | 0.96 [0.95, 0.97] | 0.91 [0.87, 0.94] |
| FS | 0.95 [0.94, 0.96] | 0.86 [0.82, 0.91] |
Table 4.
Internal test set results (n = 9 patients, 18 acquisitions (9 WOFS, 9 FS)). Reader study average rating (1—Poor, 2—Fair, 3—Good, 4—Excellent) and 95% Confidence Intervals (CI) per reader.
Table 4.
Internal test set results (n = 9 patients, 18 acquisitions (9 WOFS, 9 FS)). Reader study average rating (1—Poor, 2—Fair, 3—Good, 4—Excellent) and 95% Confidence Intervals (CI) per reader.
| | | Breast | | FGT | |
|---|
| | Acquisitions Type | Avg. Rating | 95% CI | Avg. Rating | 95% CI |
| Reader 1 | WOFS | 3.89 | [3.67, 4.0] | 3.89 | [3.67, 4.0] |
| FS | 3.89 | [3.67, 4.0] | 3.78 | [3.44, 4.0] |
| Reader 2 | WOFS | 4.0 | [4.0, 4.0] | 4.0 | [4.0, 4.0] |
| FS | 4.0 | [4.0, 4.0] | 4.0 | [4.0, 4.0] |
Table 5.
Internal test set results (n = 9 patients, 18 acquisitions (9 WOFS, 9 FS)). Reader study overall average rating (1—Poor, 2—Fair, 3—Good, 4—Excellent) and percentage agreement between readers.
Table 5.
Internal test set results (n = 9 patients, 18 acquisitions (9 WOFS, 9 FS)). Reader study overall average rating (1—Poor, 2—Fair, 3—Good, 4—Excellent) and percentage agreement between readers.
| | Breast | | FGT | |
|---|
| Acquisitions Type | Avg. Rating | % Agreement | Avg. Rating | % Agreement |
| WOFS | 3.94 | 89% | 3.94 | 89% |
| FS | 3.94 | 78% | 3.88 | 89% |
Table 6.
External test set results (n = 30 patients, 60 acquisitions (30 WOFS, 30 FS)). Reader study average rating (1—Poor, 2—Fair, 3—Good, 4—Excellent) and 95% Confidence Intervals (CI) per reader.
Table 6.
External test set results (n = 30 patients, 60 acquisitions (30 WOFS, 30 FS)). Reader study average rating (1—Poor, 2—Fair, 3—Good, 4—Excellent) and 95% Confidence Intervals (CI) per reader.
| | | Breast | | FGT | |
|---|
| | Acquisitions Type | Avg. Rating | 95% CI | Avg. Rating | 95% CI |
| Reader 1 | WOFS | 3.7 | [3.53, 3.87] | 3.77 | [3.6, 3.9] |
| FS | 3.53 | [3.33, 3.73] | 3.43 | [3.17, 3.67] |
| Reader 2 | WOFS | 4.0 | [4.0, 4.0] | 3.93 | [3.83, 4.0] |
| FS | 4.0 | [4.0, 4.0] | 3.5 | [3.23, 3.73] |
Table 7.
External test set results (n = 30 patients, 60 acquisitions (30 WOFS, 30 FS)). Reader study overall average rating (1—Poor, 2—Fair, 3—Good, 4—Excellent) and percentage agreement between readers.
Table 7.
External test set results (n = 30 patients, 60 acquisitions (30 WOFS, 30 FS)). Reader study overall average rating (1—Poor, 2—Fair, 3—Good, 4—Excellent) and percentage agreement between readers.
| | Breast | | FGT | |
|---|
| Acquisitions Type | Avg. Rating | % Agreement | Avg. Rating | % Agreement |
| WOFS | 3.85 | 70% | 3.85 | 70% |
| FS | 3.77 | 57% | 3.47 | 73% |