The Roles of TP53 and FGFR2 in Progress Made Treating Endometrial Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Microsatellite Stability
2.3. Molecular Analysis
2.4. Statistical Methods
3. Results
3.1. Clinical Characteristics of the Study Population
3.2. Microsatellite Stability Status
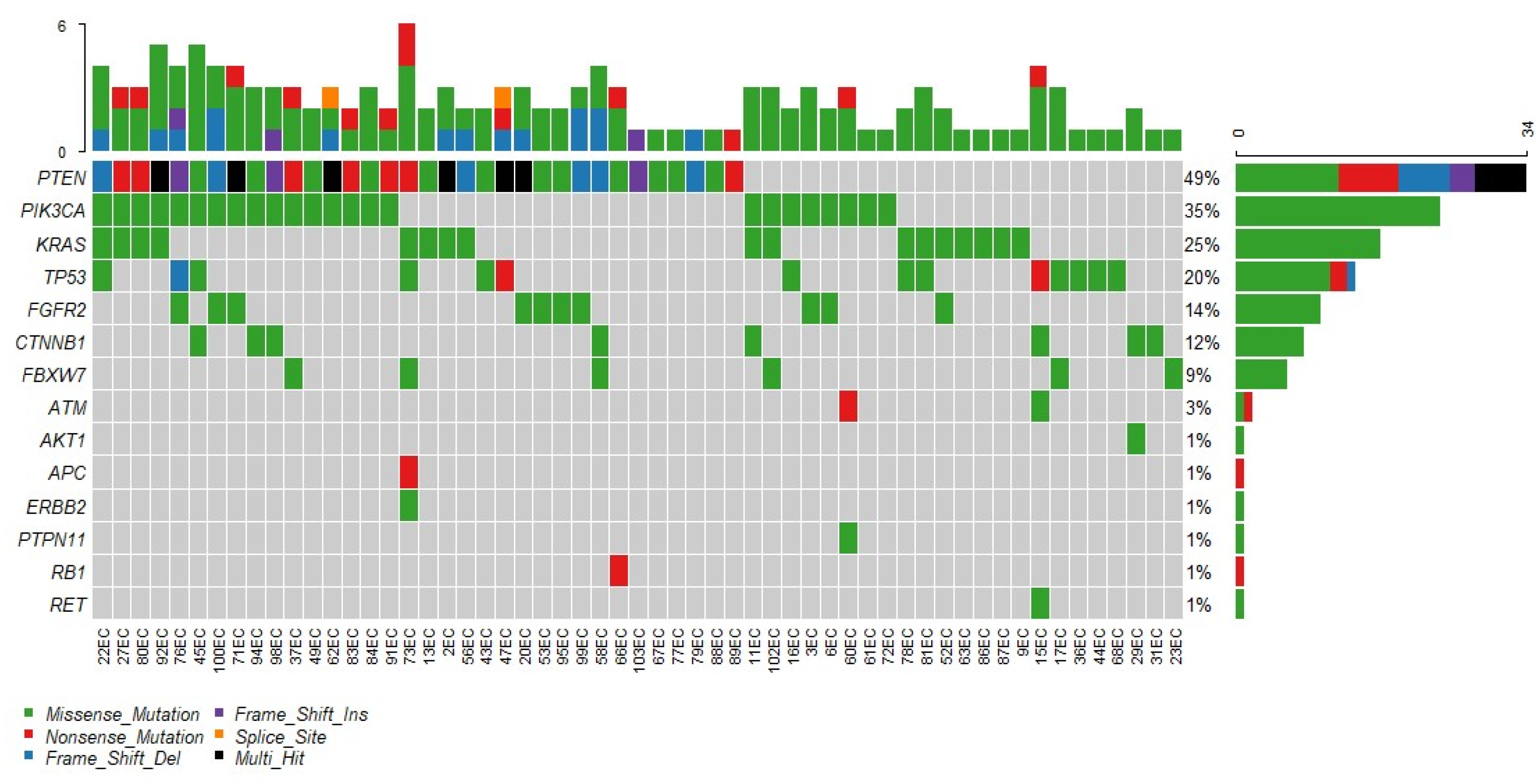
3.3. Genotyping
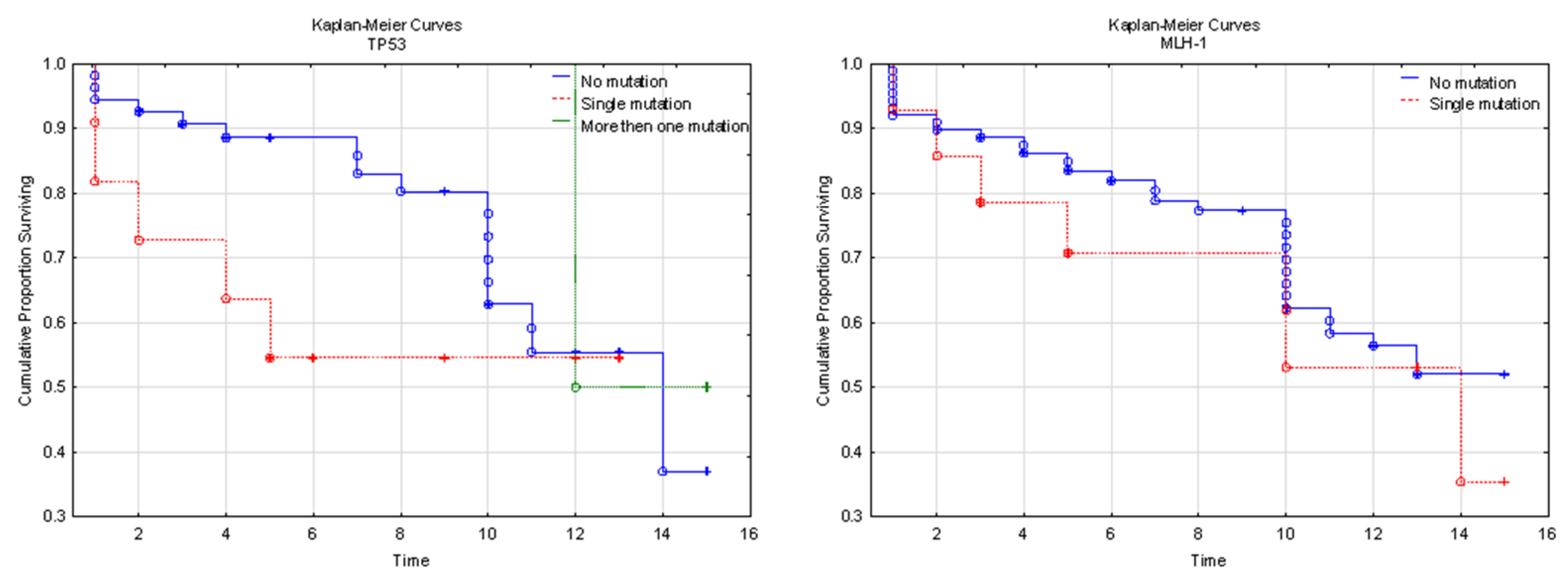
3.4. Genetic Background and Clinical Implications
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lortet-Tieulent, J.; Ferlay, J.; Bray, F.; Jemal, A. International Patterns and Trends in Endometrial Cancer Incidence, 1978–2013. JNCI J. Natl. Cancer Inst. 2017, 110, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; McAlpine, J.N. New Classification of Endometrial Cancers: The Development and Potential Applications of Genomic-Based Classification in Research and Clinical Care. Gynecol. Oncol. Res. Pract. 2016, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Cancer Society. Facts & Figures 2019; American Cancer Society: Atlanta, GA, USA, 2019. [Google Scholar]
- Jiang, X.; Tang, H.; Chen, T. Epidemiology of gynecologic cancers in China. J. Gynecol. Oncol. 2018, 29, e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henley, S.J.; Miller, J.W.; Dowling, N.F.; Benard, V.B.; Richardson, L.C. Uterine Cancer Incidence and Mortality—United States, 1999–2016. MMWR. Morb. Mortal. Wkly. Rep. 2018, 67, 1333–1338. [Google Scholar] [CrossRef] [Green Version]
- Clarke, M.A.; Long, B.J.; Del Mar Morillo, A.; Arbyn, M.; Bakkum-Gamez, J.N.; Wentzensen, N. Association of Endometrial Cancer Risk with Postmenopausal Bleeding in Women. JAMA Intern. Med. 2018, 178, 1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosman-Dziewiszek, P.; Dziegiel, P.; Zabel, M. Disturbance of gene expression in endometrial cancer as therapy aim. Ginekol. Pol. 2011, 82, 276–280. [Google Scholar]
- Kryniewska, K.; Sznurkowski, J.J. Key problems in the treatment of endometrial cancer in the light of ESMO-ESGO-ESTRO Consensus of December 2015. Curr. Gynecol. Oncol. 2016, 14, 145–155. [Google Scholar] [CrossRef]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Murali, R.; Soslow, R.A.; Weigelt, B. Classification of endometrial carcinoma: More than two types. Lancet Oncol. 2014, 15, e268–e278. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, D.; Gong, C.; Zhang, F.; He, J.; Zhang, W.; Zhao, Y.; Sun, J. Prognostic role of hormone receptors in endometrial cancer: A systematic review and meta-analysis. World J. Surg. Oncol. 2015, 13, 208. [Google Scholar] [CrossRef] [Green Version]
- Buza, N.; Roque, D.M.; Santin, A.D. HER2/neu in Endometrial Cancer: A Promising Therapeutic Target with Diagnostic Challenges. Arch. Pathol. Lab. Med. 2014, 138, 343–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urick, M.E.; Bell, D.W. Clinical actionability of molecular targets in endometrial cancer. Nat. Rev. Cancer 2019, 19, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research; Schultz, N. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Vermij, L.; Smit, V.; Nout, R.; Bosse, T. Incorporation of molecular characteristics into endometrial cancer management. Histopathology 2019, 76, 52–63. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [Green Version]
- Evrard, C.; Alexandre, J. Predictive and Prognostic Value of Microsatellite Instability in Gynecologic Cancer (Endometrial and Ovarian). Cancers 2021, 13, 2434. [Google Scholar] [CrossRef]
- Siemanowski, J.; Schömig-Markiefka, B.; Buhl, T.; Haak, A.; Siebolts, U.; Dietmaier, W.; Arens, N.; Pauly, N.; Ataseven, B.; Büttner, R.; et al. Managing Difficulties of Microsatellite Instability Testing in Endometrial Cancer-Limitations and Advantages of Four Different PCR-Based Approaches. Cancers 2021, 13, 1268. [Google Scholar] [CrossRef]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours, 8th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2016. [Google Scholar]
- Brooks, R.A.; Fleming, G.F.; Lastra, R.R.; Lee, N.K.; Moroney, J.W.; Son, C.H.; Tatebe, K.; Veneris, J.L. Current recommendations and recent progress in endometrial cancer. CA A Cancer J. Clin. 2019, 69, 258–279. [Google Scholar] [CrossRef]
- Hussein, Y.R.; Soslow, R.A. Molecular insights into the classification of high-grade endometrial carcinoma. Pathology 2018, 50, 151–161. [Google Scholar] [CrossRef] [Green Version]
- Imboden, S.; Nastic, D.; Ghaderi, M.; Rydberg, F.; Rau, T.T.; Mueller, M.D.; Epstein, E.; Carlson, J.W. Phenotype of POLE-mutated endometrial cancer. PLoS ONE 2019, 14, e0214318. [Google Scholar] [CrossRef] [Green Version]
- Alkushi, A.; Abdul-Rahman, Z.H.; Lim, P.; Schulzer, M.; Coldman, A.; Kalloger, S.E.; Miller, D.; Gilks, C.B. Description of a Novel System for Grading of Endometrial Carcinoma and Comparison with Existing Grading Systems. Am. J. Surg. Pathol. 2005, 29, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Scholten, A.N.; Smit, V.T.H.B.M.; Beerman, H.; van Putten, W.L.J.; Creutzberg, C.L. Prognostic significance and interobserver variability of histologic grading systems for endometrial carcinoma. Cancer 2004, 100, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Soslow, R.A.; Tornos, C.; Park, K.J.; Malpica, A.; Matias-Guiu, X.; Oliva, E.; Parkash, V.; Carlson, J.; McCluggage, W.G.; Gilks, C.B. Endometrial Carcinoma Diagnosis use of figo grading and genomic subcategories in clinical practice: Recommendations of the International Society of Gynecological Pathologists. Int. J. Gynecol. Pathol. 2019, 38, S64–S74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köbel, M.; Ronnett, B.M.; Singh, N.; Soslow, R.A.; Gilks, C.B.; McCluggage, W.G. Interpretation of P53 Immunohistochemistry in Endometrial Carcinomas: Toward Increased Reproducibility. Int. J. Gynecol. Pathol. 2019, 38 (Suppl. 1), S123–S131. [Google Scholar] [CrossRef]
- Nakamura, M.; Obata, T.; Daikoku, T.; Fujiwara, H. The Association and Significance of p53 in Gynecologic Cancers: The Potential of Targeted Therapy. Int J. Mol. Sci. 2019, 20, 5482. [Google Scholar] [CrossRef] [Green Version]
- Subramaniam, K.S.; Tham, S.T.; Mohamed, Z.; Woo, Y.L.; Mat Adenan, N.A.; Chung, I. Cancer-Associated Fibroblasts Promote Proliferation of Endometrial Cancer Cells. PLoS ONE 2013, 8, e68923. [Google Scholar] [CrossRef]
- Alexa, M.; Hasenburg, A.; Battista, M.J. The TCGA Molecular Classification of Endometrial Cancer and Its Possible Impact on Adjuvant Treatment Decisions. Cancers 2021, 13, 1478. [Google Scholar] [CrossRef]



| N | Average | Median | SD | Minimum | Maximum | |
|---|---|---|---|---|---|---|
| Age (years) | 103 | 71.08 | 70 | 10.98 | 48.00 | 99.00 |
| <50 | 2 | 49 | 49 | 1.41 | 48 | 50 |
| 51–60 | 16 | 56.69 | 57 | 2.70 | 52 | 60 |
| 61–70 | 34 | 65.59 | 65.5 | 2.88 | 61 | 70 |
| 71–80 | 30 | 75.43 | 75 | 3.17 | 71 | 80 |
| 81–90 | 15 | 83.93 | 83 | 2.22 | 81 | 89 |
| >90 | 6 | 94 | 93.5 | 2.68 | 91 | 99 |
| BMI | 103 | 34.07 | 34.20 | 2.44 | 23.00 | 38.50 |
| 18.5–24.99 | 2 | 23.9 | 23.9 | 1.27 | 23 | 24.8 |
| 25–29.99 | 1 | 29.8 | 29.8 | - | 29.8 | 29.8 |
| 30–34.99 | 61 | 33.06 | 33 | 1.20 | 30 | 34.9 |
| 35–39.99 | 39 | 36.29 | 36.2 | 1.01 | 35 | 38.5 |
| Follow up time (years) | 103 | 9.97 | 11 | 4.48 | 1 | 16 |
| Rtg-therapy | 59 | 4938.98 | 4600.00 | 1132.59 | 4500.00 | 9200.00 |
| OS (years) | 102 | 8.23 | 9.00 | 4.42 | 1.00 | 15.00 |
| RFS (years) | 6 | 4.67 | 3.00 | 4.23 | 1.00 | 10.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adamczyk-Gruszka, O.; Horecka-Lewitowicz, A.; Strzelecka, A.; Wawszczak-Kasza, M.; Gruszka, J.; Lewitowicz, P. The Roles of TP53 and FGFR2 in Progress Made Treating Endometrial Cancer. Diagnostics 2022, 12, 1737. https://doi.org/10.3390/diagnostics12071737
Adamczyk-Gruszka O, Horecka-Lewitowicz A, Strzelecka A, Wawszczak-Kasza M, Gruszka J, Lewitowicz P. The Roles of TP53 and FGFR2 in Progress Made Treating Endometrial Cancer. Diagnostics. 2022; 12(7):1737. https://doi.org/10.3390/diagnostics12071737
Chicago/Turabian StyleAdamczyk-Gruszka, Olga, Agata Horecka-Lewitowicz, Agnieszka Strzelecka, Monika Wawszczak-Kasza, Jakub Gruszka, and Piotr Lewitowicz. 2022. "The Roles of TP53 and FGFR2 in Progress Made Treating Endometrial Cancer" Diagnostics 12, no. 7: 1737. https://doi.org/10.3390/diagnostics12071737






