Deep-Risk: Deep Learning-Based Mortality Risk Predictive Models for COVID-19
Abstract
:1. Introduction
- Developing three deep learning predictive models with different architectures to predict the mortality risk for COVID-19 patients.
- Using the clinical features of COVID-19 patients to predict their chances of survival.
- Converting the original clinical dataset into images that would be used by one of the proposed predictive models and subsequently analyzing its performance.
- Conducting a comparative analysis with some previous studies that used well-known ML methods on the same dataset.
- Comparing our proposed work to previous studies that employed the deep learning CNN model with various meta-heuristic methods and datasets.
2. Background and Related Works on Predicting Mortality Risk and Severity
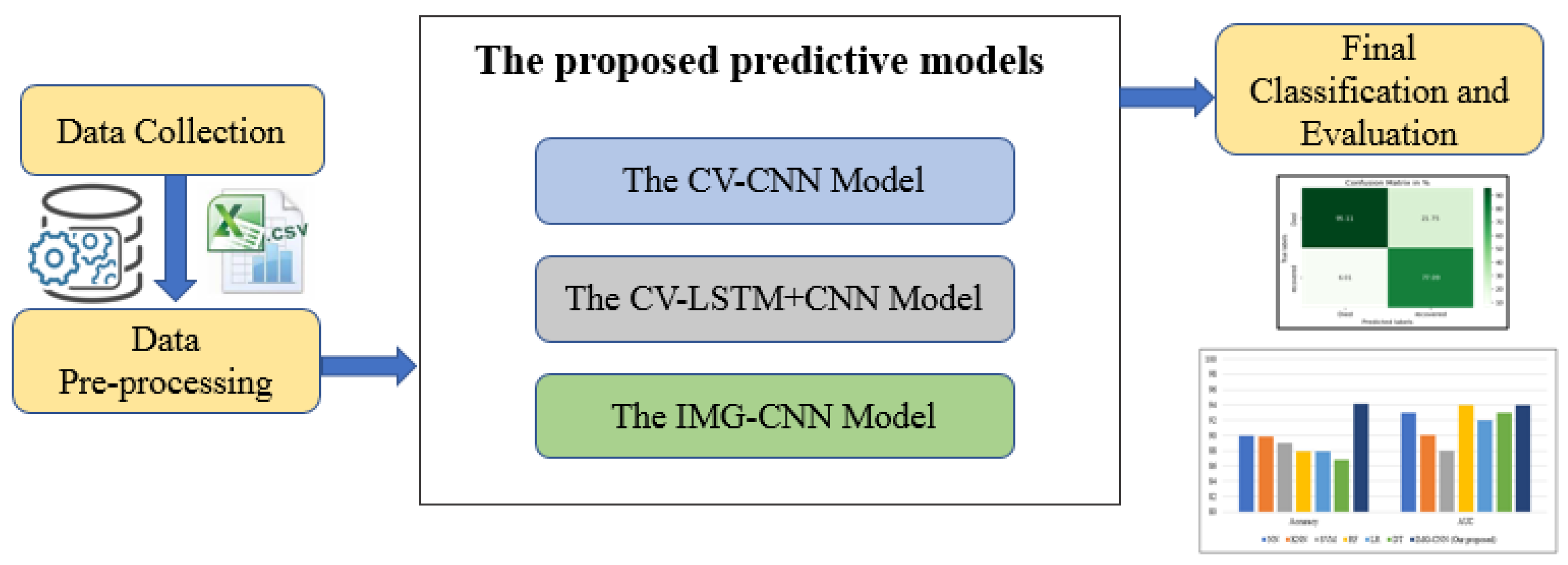
3. Materials and Methods
3.1. Datasets for the Study
3.2. Dataset Pre-Processing
3.3. The Proposed Deep Learning Predictive Models
3.3.1. The First Predictive Model: CV-CNN
3.3.2. The Second Predictive Model: CV- LSTM + CNN
3.3.3. The Third Predictive Model: IMG-CNN
4. Results
4.1. Experimental Parameters
4.2. Performance Metrics
4.3. Deep-Risk: Deep Learning-Based Risk Mortality Prediction System Results
4.3.1. CV-CNN Model
4.3.2. CV-LSTM + CNN Model
4.3.3. IMG-CNN Model
4.4. Proposed Models Comparison and Results Discussion
5. Comparative Analysis with State-Of the-Art Work
5.1. Dataset-Based Comparison with State-Of the-Art Work
5.2. Deep Learning-Based Comparison with State-Of the-Art Work
6. Conclusions and Future Work
- Apply different large datasets and deeply analyze the effectiveness of converting these datasets into images;
- Try to improve the performance of the predictive models by using other deep learning methods.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pourhomayoun, M.; Shakibi, M. Predicting mortality risk in patients with COVID-19 using machine learning to help medical decision-making. Smart Health 2021, 20, 100178. [Google Scholar] [CrossRef] [PubMed]
- Aldweesh, A.; Derhab, A.; Emam, A.Z. Deep learning approaches for anomaly-based intrusion detection systems: A survey, taxonomy, and open issues. Knowl.-Based Syst. 2020, 189, 105124. [Google Scholar] [CrossRef]
- Saudi Vision 2030. Available online: https://www.vision2030.gov.sa/thekingdom/explore/energy/ (accessed on 8 June 2022).
- Khan, D.A.; Razak, S.; Raj, B.; Singh, R. Human behaviour recognition using wifi channel state information. In Proceedings of the ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 7625–7629. [Google Scholar]
- Yan, L.C.; Yoshua, B.; Geoffrey, H. Deep learning. Nature 2015, 521, 436–444. [Google Scholar]
- Wu, J. Introduction to Convolutional Neural Networks; National Key Lab for Novel Software Technology/Nanjing University: Nanjing, China, 2017; Volume 5, p. 495. [Google Scholar]
- Delahoz, Y.S.; Labrador, M.A. Survey on fall detection and fall prevention using wearable and external sensors. Sensors 2014, 14, 19806–19842. [Google Scholar] [CrossRef]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar]
- Ibrahim, D.M.; Elshennawy, N.M.; Sarhan, A.M. Deep-chest: Multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Comput. Biol. Med. 2021, 132, 104348. [Google Scholar] [CrossRef]
- Guo, L.; Wei, D.; Zhang, X.; Wu, Y.; Li, Q.; Zhou, M.; Qu, J. Clinical features predicting mortality risk in patients with viral pneumonia: The mulbsta score. Front. Microbiol. 2019, 10, 2752. [Google Scholar] [CrossRef]
- Das, A.K.; Mishra, S.; Gopalan, S.S. Predicting COVID-19 community mortality risk using machine learning and development of an online prognostic tool. PeerJ 2020, 8, e10083. [Google Scholar] [CrossRef]
- Zhu, J.S.; Ge, P.; Jiang, C.; Zhang, Y.; Li, X.; Zhao, Z.; Zhang, L.; Duong, T.Q. Deep-learning artificial intelligence analysis of clinical variables predicts mortality in COVID-19 patients. J. Am. Coll. Emerg. Physicians Open 2020, 1, 1364–1373. [Google Scholar] [CrossRef]
- O’Mahony, C.; Jichi, F.; Pavlou, M.; Monserrat, L.; Anastasakis, A.; Rapezzi, C.; Biagini, E.; Gimeno, J.R.; Limongelli, G.; McKenna, W.J. A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (hcm risk-scd). Eur. Heart J. 2014, 35, 2010–2020. [Google Scholar] [CrossRef]
- Killock, D. Ai outperforms radiologists in mammographic screening. Nat. Rev. Clin. Oncol. 2020, 17, 134. [Google Scholar] [CrossRef]
- Shankar, V.; Kumar, V.; Devagade, U.; Karanth, V.; Rohitaksha, K. Heart disease prediction using cnn algorithm. SN Comput. Sci. 2020, 1, 170. [Google Scholar] [CrossRef]
- Barik, S.; Mohanty, S.; Mohanty, S.; Singh, D. Analysis of prediction accuracy of diabetes using classifier and hybrid machine learning techniques. In Intelligent and Cloud Computing; Springer: Berlin/Heidelberg, Germany, 2021; pp. 399–409. [Google Scholar]
- Bergamaschi, L.; D’Angelo, E.C.; Paolisso, P.; Toniolo, S.; Fabrizio, M.; Angeli, F.; Donati, F.; Magnani, I.; Rinaldi, A.; Bartoli, L.; et al. The value of ECG changes in risk stratification of COVID-19 patients. Ann. Noninvasive Electrocardiol. 2021, 26, e12815. [Google Scholar] [CrossRef] [PubMed]
- Bikku, T. Multi-layered deep learning perceptron approach for health risk prediction. J. Big Data 2020, 7, 50. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, H.-T.; Goncalves, J.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Zhang, M. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020, 2, 283–288. [Google Scholar] [CrossRef]
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Al-Madeed, S.; Zughaier, S.M.; Doi, S.A.; Hassen, H.; Islam, M.T. An early warning tool for predicting mortality risk of COVID-19 patients using machine learning. Cogn. Comput. 2021, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, A.; Garg, A.; Vinod, P.; Priyakumar, U.D. Machine learning based clinical decision support system for early COVID-19 mortality prediction. Front. Public Health 2021, 9, 6697. [Google Scholar] [CrossRef] [PubMed]
- Kar, S.; Chawla, R.; Haranath, S.P.; Ramasubban, S.; Ramakrishnan, N.; Vaishya, R.; Sibal, A.; Reddy, S. Multivariable mortality risk prediction using machine learning for COVID-19 patients at admission (aiCOVID). Sci. Rep. 2021, 11, 12801. [Google Scholar] [CrossRef]
- Aktar, S.; Talukder, A.; Ahamad, M.; Kamal, A.; Khan, J.R.; Protikuzzaman, M.; Hossain, N.; Azad, A.; Quinn, J.M.; Summers, M.A. Machine learning approaches to identify patient comorbidities and symptoms that increased risk of mortality in COVID-19. Diagnostics 2021, 11, 1383. [Google Scholar] [CrossRef] [PubMed]
- Tezza, F.; Lorenzoni, G.; Azzolina, D.; Barbar, S.; Leone, L.A.C.; Gregori, D. Predicting in-hospital mortality of patients with COVID-19 using machine learning techniques. J. Pers. Med. 2021, 11, 343. [Google Scholar] [CrossRef] [PubMed]
- Khozeimeh, F.; Sharifrazi, D.; Izadi, N.H.; Joloudari, J.H.; Shoeibi, A.; Alizadehsani, R.; Gorriz, J.M.; Hussain, S.; Sani, Z.A.; Moosaei, H. Combining a convolutional neural network with autoencoders to predict the survival chance of COVID-19 patients. Sci. Rep. 2021, 11, 15343. [Google Scholar] [CrossRef]
- Paolisso, P.; Bergamaschi, L.; D’Angelo, E.C.; Donatim, F.; Giannella, M.; Tedeschi, S.; Pascale, R.; Bartoletti, M.; Tesini, G.; Biffi, M.; et al. Preliminary Experience with Low Molecular Weight Heparin Strategy in COVID-19 Patients. Front. Pharmacol. 2020, 11, 1124. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhang, H.-T.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Li, S.; Zhang, M. Prediction of criticality in patients with severe COVID-19 infection using three clinical features: A machine learning-based prognostic model with clinical data in wuhan. MedRxiv 2020. [Google Scholar] [CrossRef]
- Wang, K.; Zuo, P.; Liu, Y.; Zhang, M.; Zhao, X.; Xie, S.; Zhang, H.; Chen, X.; Liu, C. Clinical and laboratory predictors of in-hospital mortality in patients with coronavirus disease-2019: A cohort study in wuhan, china. Clin. Infect. Dis. 2020, 71, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Rechtman, E.; Curtin, P.; Navarro, E.; Nirenberg, S.; Horton, M.K. Vital signs assessed in initial clinical encounters predict COVID-19 mortality in an nyc hospital system. Sci. Rep. 2020, 10, 21545. [Google Scholar] [CrossRef]
- Bertsimas, D.; Lukin, G.; Mingardi, L.; Nohadani, O.; Orfanoudaki, A.; Stellato, B.; Wiberg, H.; Gonzalez-Garcia, S.; Parra-Calderon, C.L.; Robinson, K. COVID-19 mortality risk assessment: An international multi-center study. PLoS ONE 2020, 15, e0243262. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Zhang, B.; Fu, M.; Li, M.; Yuan, X.; Zhu, Y.; Peng, J.; Guo, H.; Lu, Y. Clinical and inflammatory features based machine learning model for fatal risk prediction of hospitalized COVID-19 patients: Results from a retrospective cohort study. Ann. Med. 2021, 53, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Booth, A.L.; Abels, E.; McCaffrey, P. Development of a prognostic model for mortality in COVID-19 infection using machine learning. Mod. Pathol. 2021, 34, 522–531. [Google Scholar] [CrossRef]
- Sun, L.; Song, F.; Shi, N.; Liu, F.; Li, S.; Li, P.; Zhang, W.; Jiang, X.; Zhang, Y.; Sun, L. Combination of four clinical indicators predicts the severe/critical symptom of patients infected COVID-19. J. Clin. Virol. 2020, 128, 104431. [Google Scholar] [CrossRef]
- Yao, H.; Zhang, N.; Zhang, R.; Duan, M.; Xie, T.; Pan, J.; Peng, E.; Huang, J.; Zhang, Y.; Xu, X. Severity detection for the coronavirus disease 2019 (COVID-19) patients using a machine learning model based on the blood and urine tests. Front. Cell Dev. Biol. 2020, 8, 683. [Google Scholar] [CrossRef]
- Zhao, C.; Bai, Y.; Wang, C.; Zhong, Y.; Lu, N.; Tian, L.; Cai, F.; Jin, R. Risk factors related to the severity of COVID-19 in Wuhan. Int. J. Med. Sci. 2021, 18, 120. [Google Scholar] [CrossRef]
- Hu, C.; Liu, Z.; Jiang, Y.; Shi, O.; Zhang, X.; Xu, K.; Suo, C.; Wang, Q.; Song, Y.; Yu, K. Early prediction of mortality risk among patients with severe COVID-19, using machine learning. Int. J. Epidemiol. 2020, 49, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chen, A.; Hou, W.; Graham, J.M.; Li, H.; Richman, P.S.; Thode, H.C.; Singer, A.J.; Duong, T.Q. Prediction model and risk scores of icu admission and mortality in COVID-19. PLoS ONE 2020, 15, e0236618. [Google Scholar] [CrossRef]
- Huang, H.; Cai, S.; Li, Y.; Li, Y.; Fan, Y.; Li, L.; Lei, C.; Tang, X.; Hu, F.; Li, F. Prognostic factors for COVID-19 pneumonia progression to severe symptoms based on earlier clinical features: A retrospective analysis. Front. Med. 2020, 7, 643. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, Z.; Guo, Y.; Geng, S.; Gao, S.; Ye, S.; Hu, Y.; Wang, Y. A new predictor of disease severity in patients with COVID-19 in Wuhan, China. MedRxiv 2020, 1, 1–19. [Google Scholar] [CrossRef]
- Zhu, Z.; Cai, T.; Fan, L.; Lou, K.; Hua, X.; Huang, Z.; Gao, G. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int. J. Infect. Dis. 2020, 95, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Wang, S.; Zha, Y.; Li, W.; Wu, Q.; Li, X.; Niu, M.; Wang, M.; Qiu, X.; Li, H.; et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020, 56, 2000775. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Chen, Y.; Wu, Q.; Chen, G.; Shen, X. A tool to early predict severe corona virus disease 2019 (COVID-19): A multicenter study using the risk nomogram in Wuhan and Guangdong, China. Cancer 2020, 46, ciaa443. [Google Scholar] [CrossRef]
- Liu, Q.; Song, N.; Zheng, Z.; Li, J.; Li, S. Laboratory findings and a combined multifactorial approach to predict death in critically ill patients with COVID-19: A retrospective study. Epidemiol. Infect. 2020, 148, e129. [Google Scholar] [CrossRef]
- Luo, M.; Liu, J.; Jiang, W.; Yue, S.; Liu, H.; Wei, S. Il-6 and CD8+ T cell counts combined are an early predictor of in-hospital mortality of patients with COVID-19. JCI Insight 2020, 5, e139024. [Google Scholar] [CrossRef]
- Li, Y.; Horowitz, M.A.; Liu, J.; Chew, A.; Lan, H.; Liu, Q.; Sha, D.; Yang, C. Individual-level fatality prediction of COVID-19 patients using ai methods. Front. Publ. Health 2020, 8, 566. [Google Scholar] [CrossRef]
- de Terwangne, C.; Laouni, J.; Jouffe, L.; Lechien, J.R.; Bouillon, V.; Place, S.; Capulzini, L.; Machayekhi, S.; Ceccarelli, A.; Saussez, S. Predictive accuracy of COVID-19 world health organization (who) severity classification and comparison with a bayesian-method-based severity score (epi-score). Pathogens 2020, 9, 880. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Gutierrez, B.; Mekaru, S.; Sewalk, K.; Goodwin, L.; Loskill, A.; Cohn, E.L.; Hswen, Y.; Hill, S.C.; Cobo, M.M. Epidemiological data from the COVID-19 outbreak, real-time case information. Sci. Data 2020, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Schwab, P.; Schütte, A.; Dietz, B.; Bauer, S. PreCOVID-19: A systematic study of clinical predictive models for coronavirus disease 2019. arXiv 2020, arXiv:2005.08302. [Google Scholar]
- Tekerek, A. A novel architecture for web-based attack detection using convolutional neural network. Comput. Secur. 2021, 100, 102096. [Google Scholar] [CrossRef]
- Bisong, E. Building Machine Learning and Deep Learning Models on Google Cloud Platform; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- GitHub, Deep-Risk. Available online: https://github.com/Dr-Dina-M-Ibrahim/Deep_RMP (accessed on 8 June 2022).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
















| Study | Method | ML/DL | Performance |
|---|---|---|---|
| Pourhomayoun et al. [1] | SVM, NN, and RF | Machine learning | 89.98% (Accuracy) |
| Yan et al. [19] | XGBoost | Machine learning | Accuracy 90% |
| Yan et al. [27] | XGBoost | Machine learning | 93% (Accuracy) |
| Wang et al. [28] | XGBoost | Machine learning | 83% (AUC for clinical model) 88% (AUC for laboratory model) |
| Rechtman et al. [29] | XGBoost | Machine learning | 86% (AUC) |
| Bertsimas et al. [30] | XGBoost | Machine learning | 81%, 87%, and 92% (AUCs using three validation cohorts) |
| Guan et al. [31] | XGBoost | Machine learning | Precision >90%, Sensitivity >85% F1 scores 0.9 |
| Booth et al. [32] | SVM | Machine learning | 93% (AUC), 91% (Specificity), 91 % (Sensitivity) |
| Sun et al. [33] | SVM | Machine learning | 97.57% (AUC) |
| Yao et al. [34] | SVM | Machine learning | 81.48% (Accuracy) |
| Zhao et al. [35] | SVM | Machine learning | 91.38% (Accuracy), 94% (Specificity) 90 % (Sensitivity) |
| Chowdhury et al. [20] | XGBoost | Machine learning | 96.1% (AUC) |
| Karthikeyan et al. [21] | NN, SVM, LR, random forests, XGBoost, and DT | Machine learning | NN model performance 96.53% (Accuracy), 98.9% (AUC), 96.9 % (F1 scores) |
| Kar et al. [22] | XGBoost | Machine learning | 78.2% (AUC), 93% (Accuracy score), 77% (Precision) |
| Hu et al. [36] | partial least squares regression, elastic net model, RF, bagged FDA, and LR | Machine learning | LR model performance 88.1% (AUC), 79.4% (Specificity), 83.9% (Sensitivity) |
| Zhao et al. [37] | LR | Machine learning | 83% (AUC for mortality prediction), 74% (AUC for ICU admission prediction) |
| Huang et al. [38] | LR | Machine learning | 94.4% (AUC), 90.2% (Specificity), 94.1% (Sensitivity) |
| Zhou et al. [39] | LR | Machine learning | 87.9% (AUC), 73.7% (Specificity), 88.6% (Sensitivity) |
| Zhu et al. [40] | LR | Machine learning | 90% (AUC) |
| Gong et al. [41] | LASSO regression, DT, RF, SVM, and LR | Machine learning | LR model performance 85.3% (AUC), 78.4% (Specificity) 77.5% (Sensitivity) |
| Liu et al [43] | Multivariable LR | Machine learning | 99.4% (AUC), 97.2% (Specificity) 100% (Sensitivity) |
| Li et al. [45] | Autoencoder, LR, RF, SVM, one-class SVM, isolation forest, and local outlier factor | Machine learning | Autoencoder model performance 97% (Accuracy) and 73% (AUC) |
| Terwangne et al. [46] | Bayesian network analysis | Machine learning | 83.8% (ROC for WHO classification model) and 91% (ROC for EPI-SCORE model), |
| Aktar et al. [23] | Random Forest, DT, GBM, XGBoost, SVM, and LGBM | Machine learning | 88% (Accuracy of comorbidity and mortality for LGBM model), 90% (Accuracy of symptoms for GBM and LGBM models) |
| Tezza et al. [24] | RPART, SVM, GBM, and Random Forest | Machine learning | Random Forest model performance 84% (ROC) |
| Khozeimeh et al. [25] | CNN and autoencoders | Deep learning | 96.05% (Average Accuracy) |
| Feature Type | Feature Name | ||
|---|---|---|---|
| Symptoms | anorexia | fever | shortness of breath |
| chest pain | gasp | somnolence | |
| chills | headache | sore throat | |
| conjunctivitis | kidney failure | sputum | |
| cough | lesions on chest radiographs | septic shock | |
| diarrhea | hypertension | Heart attack | |
| dizziness | Myalgia | old | |
| dyspnea | Obnubilation | cardiac disease | |
| emesis | pneumonia | hypoxia | |
| expectoration | myelofibrosis | fatigue | |
| eye irritation | respiratory distress | rhinorrhea | |
| Pre-existing Conditions | diabetes | COPD | coronary heart disease |
| hypertension | Parkinson’s disease | prostate hypertrophy | |
| chronic kidney disease | asthma | Tuberculosis | |
| hypothyroidism | cancer | hepatitis B | |
| cerebral infarction | HIV positive | chronic bronchitis | |
| cardiac disease | dyslipidemia | any chronic disease | |
| Demographics | age | country | province |
| gender | city | travel history | |
| Layer (Type) | Output Shape | Parameters |
|---|---|---|
| conv1d_5 (Conv1D) | (None, 52, 256) | 1024 |
| conv1d_6 (Conv1D) | (None, 50, 256) | 196,864 |
| conv1d_7 (Conv1D) | (None, 48, 256) | 196,864 |
| max_pooling1d_1 (MaxPooling1D) | (None, 47, 256) | 0 |
| flatten_2 (Flatten) | (None, 12,032) | 0 |
| dense_6 (Dense) | (None, 64) | 770,112 |
| batch_normalization_4 (BatchNormalization) | (None, 64) | 256 |
| dropout_4 (Dropout) | (None, 64) | 0 |
| dense_7 (Dense) | (None, 32) | 2080 |
| batch_normalization_5 (BatchNormalization) | (None, 32) | 128 |
| dropout_5 (Dropout) | (None, 32) | 0 |
| dense_8 (Dense) | (None, 1) | 33 |
| Total parameters: 1,167,361 | ||
| Trainable parameters: 1,167,169 | ||
| Non-trainable parameters: 192 | ||
| Layer (Type) | Output Shape | Parameters |
|---|---|---|
| batch_normalization_2 (BatchNormalization) | (None, 54, 1) | 4 |
| Reshape (Reshape) | (None, 9, 6, 1) | 0 |
| Time_distribution (TimeDistributed) | (None, 9, 6, 256) | 264,192 |
| dropout_2 (Dropout) | (None, 9, 6, 256) | 0 |
| batch_normalization_3 (BatchNormalization) | (None, 9, 6, 256) | 1024 |
| Time_distribution_1 (TimeDistributed) | (None, 9, 6, 256) | 26,2400 |
| conv1d_4 (Conv1D) | (None, 9, 4, 256) | 196,864 |
| average_pooling2d_1 (AveragePooling2D) | (None, 4, 2, 256) | 0 |
| flatten_1 (Flatten) | (None, 2048) | 0 |
| dropout_3 (Dropout) | (None, 2048) | 0 |
| dense_3 (Dense) | (None, 128) | 262,272 |
| dense_4 (Dense) | (None, 64) | 8256 |
| dense_5 (Dense) | (None, 1) | 65 |
| Total parameters: 995,077 | ||
| Trainable parameters: 994,563 | ||
| Non-trainable parameters: 514 | ||
| Layer (Type) | Output Shape | Parameters |
|---|---|---|
| Conv2d (Conv2D) | (None, 224, 224, 256) | 7168 |
| Activation (Activation) | (None, 224, 224, 256) | 0 |
| batch_normalization (BatchNormalization) | (None, 224, 224, 256) | 1024 |
| Conv2d_1 (Conv2D) | (None, 224, 224, 128) | 295,040 |
| Activation_1 (Activation) | (None, 224, 224, 128) | 0 |
| max_pooling2d (MaxPooling2D) | (None, 74, 74, 128) | 0 |
| dropout (Dropout) | (None, 74, 74, 128) | 0 |
| Conv2d_2 (Conv2D) | (None, 72, 72, 64) | 73,792 |
| Activation_2 (Activation) | (None, 72, 72, 64) | 0 |
| batch_normalization_1 (BatchNormalization) | (None, 72, 72, 64) | 256 |
| flatten (Flatten) | (None, 331,776) | 0 |
| dense (Dense) | (None, 512) | 169,869,824 |
| dropout_1 (Dropout) | (None, 512) | 0 |
| dense_1 (Dense) | (None, 64) | 32,832 |
| dense_2 (Dense) | (None, 1) | 65 |
| Total parameters: 170,280,001 | ||
| Trainable parameters: 170,279,361 | ||
| Non-trainable parameters: 640 | ||
| Fold | Precision Recovered | Died | Macro | Weight | Recall Recovered | Died | Macro | Weight | F1-Score Recovered | Died | Macro | Weight | Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.79 | 0.93 | 0.86 | 0.86 | 0.94 | 0.75 | 0.85 | 0.85 | 0.86 | 0.83 | 0.85 | 0.85 | 0.85 |
| 2 | 0.80 | 0.93 | 0.86 | 0.86 | 0.94 | 0.76 | 0.85 | 0.85 | 0.86 | 0.83 | 0.85 | 0.85 | 0.85 |
| 3 | 0.53 | 0.95 | 0.74 | 0.74 | 1.00 | 0.09 | 0.54 | 0.55 | 0.69 | 0.17 | 0.43 | 0.43 | 0.55 |
| 4 | 0.59 | 0.95 | 0.77 | 0.77 | 0.98 | 0.30 | 0.64 | 0.64 | 0.74 | 0.46 | 0.60 | 0.60 | 0.64 |
| 5 | 0.69 | 0.93 | 0.81 | 0.81 | 0.96 | 0.57 | 0.77 | 0.77 | 0.80 | 0.71 | 0.76 | 0.76 | 0.77 |
| 6 | 0.74 | 0.93 | 0.84 | 0.83 | 0.95 | 0.66 | 0.80 | 0.81 | 0.83 | 0.77 | 0.80 | 0.80 | 0.81 |
| 7 | 0.81 | 0.93 | 0.87 | 0.87 | 0.94 | 0.78 | 0.86 | 0.86 | 0.87 | 0.85 | 0.86 | 0.86 | 0.86 |
| 8 | 0.50 | 0.00 | 0.25 | 0.25 | 1.00 | 0.00 | 0.50 | 0.50 | 0.67 | 0.00 | 0.33 | 0.34 | 0.50 |
| 9 | 0.81 | 0.92 | 0.87 | 0.87 | 0.94 | 0.77 | 0.86 | 0.86 | 0.87 | 0.84 | 0.86 | 0.86 | 0.86 |
| 10 | 0.65 | 0.92 | 0.78 | 0.78 | 0.96 | 0.47 | 0.71 | 0.72 | 0.77 | 0.62 | 070 | 0.70 | 0.72 |
| Fold | Precision Recovered | Died | Macro | Weight | Recall Recovered | Died | Macro | Weight | F1-Score Recovered | Died | Macro | Weight | Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.81 | 0.93 | 0.87 | 0.87 | 0.94 | 0.77 | 0.86 | 0.86 | 0.87 | 0.84 | 0.85 | 0.85 | 0.86 |
| 2 | 0.83 | 0.63 | 0.73 | 0.73 | 0.48 | 0.90 | 0.69 | 0.69 | 0.61 | 0.74 | 0.68 | 0.67 | 0.69 |
| 3 | 0.81 | 0.92 | 0.86 | 0.86 | 0.93 | 0.78 | 0.85 | 0.85 | 0.87 | 0.84 | 0.85 | 0.85 | 0.85 |
| 4 | 0.76 | 0.61 | 0.68 | 0.69 | 0.47 | 0.85 | 0.66 | 0.66 | 0.58 | 0.71 | 0.65 | 0.65 | 0.66 |
| 5 | 0.81 | 0.93 | 0.87 | 0.87 | 0.94 | 0.77 | 0.86 | 0.86 | 0.87 | 0.84 | 0.86 | 0.86 | 0.86 |
| 6 | 0.81 | 0.92 | 0.86 | 0.86 | 0.93 | 0.77 | 0.85 | 0.85 | 0.86 | 0.84 | 0.85 | 0.85 | 0.85 |
| 7 | 0.63 | 0.50 | 0.56 | 0.56 | 0.01 | 0.99 | 0.50 | 0.50 | 0.03 | 0.66 | 0.35 | 0.34 | 0.50 |
| 8 | 0.96 | 0.50 | 0.73 | 0.73 | 0.02 | 1.00 | 0.51 | 0.51 | 0.04 | 0.67 | 0.35 | 0.35 | 0.51 |
| 9 | 0.79 | 0.92 | 0.86 | 0.86 | 0.94 | 0.75 | 0.84 | 0.84 | 0.86 | 0.83 | 0.84 | 0.84 | 0.84 |
| 10 | 0.10 | 0.49 | 0.30 | 0.30 | 0.00 | 0.98 | 0.49 | 0.49 | 0.00 | 0.66 | 0.33 | 0.33 | 0.49 |
| Performance Metric | Value | Performance Metric | Value | Performance Metric | Value | Performance Metric | Value |
|---|---|---|---|---|---|---|---|
| Tp | 124 | val_Tp | 305 | accuracy | 0.85 | val_accuracy | 0.94 |
| Fp | 2 | val_Fp | 0 | precision | 0.98 | val_precision | 1 |
| Tn | 94 | val_Tn | 177 | recall | 0.77 | val_recall | 0.91 |
| Fn | 36 | val_Fn | 30 | AUC | 0.94 | val_AUC | 0.936 |
| loss | 0.27 | val_loss | 0.22 |
| Models | Precision Recovered | Died | Avg. | Recall Recovered | Died | Avg. | F1-Score Recovered | Died | Avg. | Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|
| CV-LSTM + CNN | 81% | 92% | 86.5% | 93% | 77% | 85% | 86% | 84% | 85% | 85.27% |
| CV-CNN | 81% | 93% | 87% | 94% | 78% | 86% | 87% | 85% | 86% | 86.06% |
| IMG-CNN | 83% | 100% | 91.5% | 100% | 84% | 92% | 91% | 91% | 91% | 94.14% |
| Models | Rank | Accuracy (%) | AUC (%) |
|---|---|---|---|
| NN [1] | 2 | 89.98 | 93 |
| KNN [1] | 3 | 89.83 | 90 |
| SVM [1] | 4 | 89.02 | 88 |
| RF [1] | 5 | 87.93 | 94 |
| LR [1] | 6 | 87.91 | 92 |
| DT [1] | 7 | 86.87 | 93 |
| IMG-CNN (proposed) | 1 | 94.14 | 93.6 |
| Models | Rank | Accuracy (%) | Precision (%) | Recall (%) | Specificity (%) | F1-score (%) | AUC (%) | Loss |
|---|---|---|---|---|---|---|---|---|
| ABC-CNN [25] | 3 | 92.32 | 94.7 | 97.4 | 89.65 | 96.0 | 53.3 | 0.25 |
| ACO-CNN [25] | 2 | 93.10 | 95.6 | 97.3 | 90.57 | 96.4 | 62.5 | 0.26 |
| BOA-CNN [25] | 7 | 91.37 | 94.1 | 97.0 | 89.08 | 95.1 | 53.5 | 0.28 |
| EHO-CNN [25] | 5 | 91.86 | 94.1 | 98.0 | 85.69 | 95.9 | 53.2 | 0.23 |
| GA-CNN [25] | 4 | 92.18 | 94.8 | 97.8 | 88.32 | 96.1 | 57.5 | 0.29 |
| PSO-CNN [25] | 6 | 91.85 | 95.0 | 96.4 | 88.17 | 95.5 | 61.5 | 0.28 |
| IMG-CNN (proposed) | 1 | 94.14 | 100 | 91.0 | 100 | 95.3 | 93.6 | 0.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elshennawy, N.M.; Ibrahim, D.M.; Sarhan, A.M.; Arafa, M. Deep-Risk: Deep Learning-Based Mortality Risk Predictive Models for COVID-19. Diagnostics 2022, 12, 1847. https://doi.org/10.3390/diagnostics12081847
Elshennawy NM, Ibrahim DM, Sarhan AM, Arafa M. Deep-Risk: Deep Learning-Based Mortality Risk Predictive Models for COVID-19. Diagnostics. 2022; 12(8):1847. https://doi.org/10.3390/diagnostics12081847
Chicago/Turabian StyleElshennawy, Nada M., Dina M. Ibrahim, Amany M. Sarhan, and Mohamed Arafa. 2022. "Deep-Risk: Deep Learning-Based Mortality Risk Predictive Models for COVID-19" Diagnostics 12, no. 8: 1847. https://doi.org/10.3390/diagnostics12081847






