A Mini-Review Regarding the Clinical Outcomes of In Vitro Fertilization (IVF) Following Pre-Implantation Genetic Testing (PGT)-Next Generation Sequencing (NGS) Approach
Abstract
:1. Introduction
2. Methodology
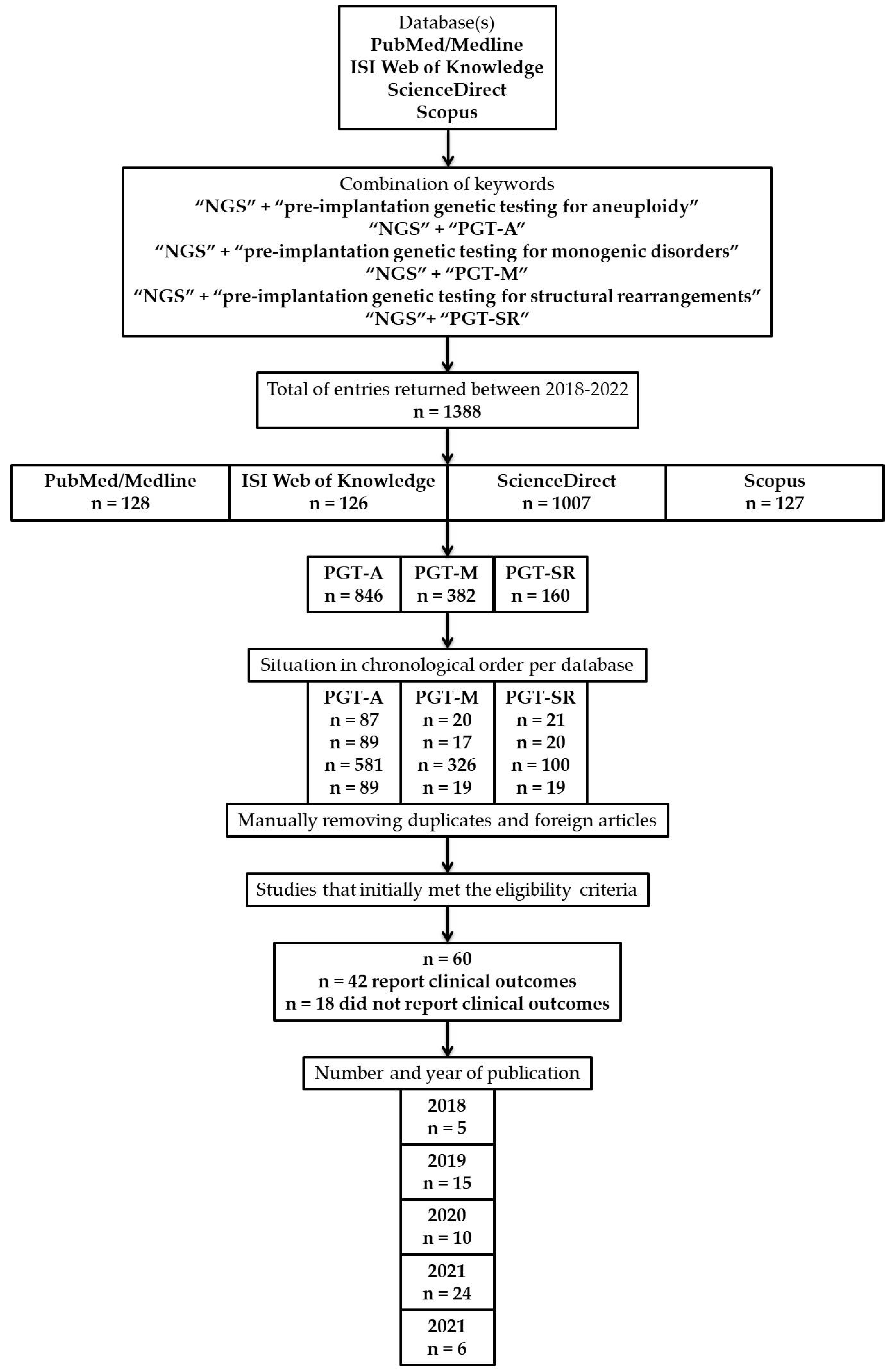
2.1. Database Search Strategy
2.2. Inclusion Criteria
2.3. Exclusion Criteria
2.4. Study Selection
2.5. Limitations of the Study
3. Results
- (1)
- Illumina
- (2)
- Ion Torrent [42]
- (1)
- (2)
- While age exerts a detrimental effect, it is mitigated through SNP-based PGT-A [39], and the embryos’ morphology possesses a significant threat with greater impact [11,37,47], but contradicted on several occasions [21,29,38]; Embryo morphokinetic [11,34,37] and inner cell mass (ICM) morphology constitute an optimal predictor of sustained implantation [48];
- (3)
- Mitochondrial DNA (mtDNA) copy numbers are higher in day 5 blastocysts of older women than day 6 blastocysts, further associated with a low chance of ongoing pregnancies [14,24]; The content of mtDNA is unable to predict the miscarriage risk [12] and additionally refuted when comparing the outcome differences between them [19] despite the cryo-storage [49];
- (4)
- (5)
- Mosaic embryos have poor reproductive potential but retain the ability to result in live births [13,33], further sustaining that TE biopsy did not add detectable adverse effects [42] but as a supplement for the management of recurrent implantation failure (RIF) [45]; However, zona pellucida opening combined with TE biopsy increases the risk of mosaicism [32], TE mosaicism deriving after TE and ICM differentiation [30], while re-biopsy may rescue those with developmental potential [31];
- (6)
- (7)
- (8)
- Public coverage of ART should be strongly encouraged [20].
3.1. Pre-Implantation Genetic Testing for Monogenic Diseases
3.2. Pre-Implantation Genetic Testing for Structural Rearrangements
3.3. Pre-Implantation Genetic Testing for Aneuploidy
4. Discussions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macklon, N.S.; Geraedts, J.P.M.; Fauser, B.C.J.M. Conception to ongoing pregnancy: The ‘black box’ of early pregnancy loss. Hum. Reprod. Update 2002, 8, 333–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rycke, M.; Berckmoes, V. Preimplantation Genetic Testing for Monogenic Disorders. Genes 2020, 11, 871. [Google Scholar] [CrossRef] [PubMed]
- Handyside, A.H.; Kontogianni, E.H.; Hardy, K.; Winston, R.M.L. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature 1990, 344, 768–770. [Google Scholar] [CrossRef]
- Griffin, D.K.; Ogur, C. Chromosomal analysis in IVF: Just how useful is it? Reproduction 2018, 156, F29–F50. [Google Scholar] [CrossRef]
- Zegers-Hochschild, F.; Adamson, G.D.; Dyer, S.; Racowsky, C.; de Mouzon, J.; Sokol, R.; Rienzi, L.; Sunde, A.; Schmidt, L.; Cooke, I.D.; et al. The International Glossary on Infertility and Fertility Care, 2017 †,‡,§. Hum. Reprod. 2017, 32, 1786–1801. [Google Scholar] [CrossRef] [Green Version]
- ESHRE PGT Consortium Steering Committee; Carvalho, F.; Coonen, E.; Goossens, V.; Kokkali, G.; Rubio, C.; Meijer-Hoogeveen, M.; Moutou, C.; Vermeulen, N.; De Rycke, M. ESHRE PGT Consortium good practice recommendations for the organisation of PGT †. Hum. Reprod. Open 2020, 2020, hoaa021. [Google Scholar] [CrossRef]
- ESHRE PGT Consortium and SIG-Embryology Biopsy Working Group; Kokkali, G.; Coticchio, G.; Bronet, F.; Celebi, C.; Cimadomo, D.; Goossens, V.; Liss, J.; Nunes, S.; Sfontouris, I.; et al. ESHRE PGT Consortium and SIG Embryology good practice recommendations for polar body and embryo biopsy for PGT †. Hum. Reprod. Open 2020, 2020, hoaa020. [Google Scholar] [CrossRef] [PubMed]
- ESHRE PGT-M Working Group; Carvalho, F.; Moutou, C.; Dimitriadou, E.; Dreesen, J.; Giménez, C.; Goossens, V.; Kakourou, G.; Vermeulen, N.; Zuccarello, D.; et al. ESHRE PGT Consortium good practice recommendations for the detection of monogenic disorders †. Hum. Reprod. Open 2020, 2020, hoaa018. [Google Scholar] [CrossRef]
- ESHRE PGT-SR/PGT-A Working Group; Coonen, E.; Rubio, C.; Christopikou, D.; Dimitriadou, E.; Gontar, J.; Goossens, V.; Maurer, M.; Spinella, F.; Vermeulen, N.; et al. ESHRE PGT Consortium good practice recommendations for the detection of structural and numerical chromosomal aberrations. Hum. Reprod. Open 2020, 2020, hoaa017. [Google Scholar] [CrossRef]
- Green, B.N.; Johnson, C.D.; Adams, A. Writing narrative literature reviews for peer-reviewed journals: Secrets of the trade. J. Chiropr. Med. 2006, 5, 101–117. [Google Scholar] [CrossRef] [Green Version]
- Gazzo, E.; Peña, F.; Valdéz, F.; Chung, A.; Velit, M.; Ascenzo, M.; Escudero, E. Blastocyst contractions are strongly related with aneuploidy, lower implantation rates, and slow-cleaving embryos: A time lapse study. JBRA Assist. Reprod. 2020, 24, 77–81. [Google Scholar] [CrossRef] [PubMed]
- El-Damen, A.; Elkhatib, I.; Bayram, A.; Arnanz, A.; Abdala, A.; Samir, S.; Lawrenz, B.; De Munck, N.; Fatemi, H.M. Does blastocyst mitochondrial DNA content affect miscarriage rate in patients undergoing single euploid frozen embryo transfer? J. Assist. Reprod. Genet. 2021, 38, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Zore, T.; Kroener, L.L.; Wang, C.; Liu, L.; Buyalos, R.; Hubert, G.; Shamonki, M. Transfer of embryos with segmental mosaicism is associated with a significant reduction in live-birth rate. Fertil. Steril. 2019, 111, 69–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.S.-Y.; Weng, S.-P.; Shen, M.-S.; Ma, P.-C.; Wu, P.-K.; Lee, N.-C. Suboptimal trophectoderm mitochondrial DNA level is associated with delayed blastocyst development. J. Assist. Reprod. Genet. 2021, 38, 587–594. [Google Scholar] [CrossRef]
- Walters-Sen, L.; Neitzel, D.; Bristow, S.L.; Mitchell, A.; Alouf, C.A.; Aradhya, S.; Faulkner, N. Experience analysing over 190,000 embryo trophectoderm biopsies using a novel FAST-SeqS preimplantation genetic testing assay. Reprod. Biomed. Online 2022, 44, 228–238. [Google Scholar] [CrossRef]
- Cai, Y.; Ding, M.; Lin, F.; Diao, Z.; Zhang, N.; Sun, H.; Zhou, J. Evaluation of preimplantation genetic testing based on next-generation sequencing for balanced reciprocal translocation carriers. Reprod. Biomed. Online 2019, 38, 669–675. [Google Scholar] [CrossRef]
- Giles, J.; Meseguer, M.; Mercader, A.; Rubio, C.; Alegre, L.; Vidal, C.; Trabalon, M.; Bosch, E. Preimplantation genetic testing for aneuploidy in patients with partial X monosomy using their own oocytes: Is this a suitable indication? Fertil. Steril. 2020, 114, 346–353. [Google Scholar] [CrossRef]
- Riestenberg, C.; Kroener, L.; Quinn, M.; Ching, K.; Ambartsumyan, G. Routine endometrial receptivity array in first embryo transfer cycles does not improve live birth rate. Fertil. Steril. 2021, 115, 1001–1006. [Google Scholar] [CrossRef]
- Stankewicz, T.; Ruiz-Alonso, M.; Soler-Ibañez, M.; Simón, C.; Valbuena, D. Do clinical outcomes differ for day-5 versus day-6 single embryo transfers controlled for endometrial factor? Reprod. Biomed. Online 2022, 44, 478–485. [Google Scholar] [CrossRef]
- Shao, Y.-H.; Zhang, X.Y.; Buckett, W.; Ao, A. Impact of in vitro fertilization-preimplantation genetic testing (IVF-PGT) funding policy on clinical outcome: An issue that stems beyond effectiveness of treatment. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 235, 1–5. [Google Scholar] [CrossRef]
- Hou, W.; Xu, Y.; Li, R.; Song, J.; Wang, J.; Zeng, Y.; Pan, J.; Zhou, C.; Xu, Y. Role of aneuploidy screening in preimplantation genetic testing for monogenic diseases in young women. Fertil. Steril. 2019, 111, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Friedenthal, J.; Alkon-Meadows, T.; Hernandez-Nieto, C.; Gounko, D.; Lee, J.A.; Copperman, A.; Buyuk, E. The association between prior cesarean delivery and subsequent in vitro fertilization outcomes in women undergoing autologous, frozen-thawed single euploid embryo transfer. Am. J. Obstet. Gynecol. 2021, 225, 287.e1–287.e8. [Google Scholar] [CrossRef] [PubMed]
- Jaswa, E.G.; McCulloch, C.E.; Simbulan, R.; Cedars, M.I.; Rosen, M.P. Diminished ovarian reserve is associated with reduced euploid rates via preimplantation genetic testing for aneuploidy independently from age: Evidence for concomitant reduction in oocyte quality with quantity. Fertil. Steril. 2021, 115, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Lledo, B.; Ortiz, J.A.; Morales, R.; García-Hernández, E.; Ten, J.; Bernabeu, A.; Llácer, J.; Bernabeu, R. Comprehensive mitochondrial DNA analysis and IVF outcome. Hum. Reprod. Open 2018, 2018, hoy023. [Google Scholar] [CrossRef] [Green Version]
- Xie, P.; Hu, L.; Tan, Y.; Gong, F.; Zhang, S.; Xiong, B.; Peng, Y.; Lu, G.X.; Lin, G. Retrospective analysis of meiotic segregation pattern and interchromosomal effects in blastocysts from inversion preimplantation genetic testing cycles. Fertil. Steril. 2019, 112, 336–342.e3. [Google Scholar] [CrossRef]
- Yuan, P.; Zheng, L.; Ou, S.; Zhao, H.; Li, R.; Luo, H.; Tan, X.; Zhang, Q.; Wang, W. Evaluation of chromosomal abnormalities from preimplantation genetic testing to the reproductive outcomes: A comparison between three different structural rearrangements based on next-generation sequencing. J. Assist. Reprod. Genet. 2021, 38, 709–718. [Google Scholar] [CrossRef]
- Li, S.; Shen, Y.; Zhu, Y.; Li, H.; Jiang, W.; Yan, J.; Chen, Z.-J. The interaction effect between advanced paternal age and paternal obesity is associated with the low implantation rate in couples with unexplained recurrent pregnancy loss. Gynecol. Obstet. Clin. Med. 2021, 1, 197–204. [Google Scholar] [CrossRef]
- Niu, W.; Wang, L.; Xu, J.; Li, Y.; Shi, H.; Li, G.; Jin, H.; Song, W.; Wang, F.; Sun, Y. Improved clinical outcomes of preimplantation genetic testing for aneuploidy using MALBAC-NGS compared with MDA-SNP array. BMC Pregnancy Childbirth 2020, 20, 388. [Google Scholar] [CrossRef]
- Viñals Gonzalez, X.; Odia, R.; Naja, R.; Serhal, P.; Saab, W.; Seshadri, S.; Ben-Nagi, J. Euploid blastocysts implant irrespective of their morphology after NGS-(PGT-A) testing in advanced maternal age patients. J. Assist. Reprod. Genet. 2019, 36, 1623–1629. [Google Scholar] [CrossRef]
- Capalbo, A.; Poli, M.; Rienzi, L.; Girardi, L.; Patassini, C.; Fabiani, M.; Cimadomo, D.; Benini, F.; Farcomeni, A.; Cuzzi, J.; et al. Mosaic human preimplantation embryos and their developmental potential in a prospective, non-selection clinical trial. Am. J. Hum. Genet. 2021, 108, 2238–2247. [Google Scholar] [CrossRef]
- Zhou, S.; Xie, P.; Zhang, S.; Hu, L.; Luo, K.; Gong, F.; Lu, G.; Lin, G. Complex mosaic blastocysts after preimplantation genetic testing: Prevalence and outcomes after re-biopsy and re-vitrification. Reprod. Biomed. Online 2021, 43, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Liu, W.; Wang, J.; Liu, J.; Gao, Y.; Wu, L.; Zhu, J.; Hao, X.; Li, J.; Liu, D.; et al. Trophectoderm biopsy protocols may impact the rate of mosaic blastocysts in cycles with pre-implantation genetic testing for aneuploidy. J. Assist. Reprod. Genet. 2021, 38, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.-Y.; Lee, C.-I.; Cheng, E.-H.; Huang, C.-C.; Lee, T.-H.; Shih, H.-H.; Pai, Y.-P.; Chen, Y.-C.; Lee, M.-S. Clinical Outcomes of Single Mosaic Embryo Transfer: High-Level or Low-Level Mosaic Embryo, Does it Matter? J. Clin. Med. 2020, 9, 1695. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-I.; Chen, C.-H.; Huang, C.-C.; Cheng, E.-H.; Chen, H.-H.; Ho, S.-T.; Lin, P.; Lee, M.-S.; Lee, T.-H. Embryo morphokinetics is potentially associated with clinical outcomes of single-embryo transfers in preimplantation genetic testing for aneuploidy cycles. Reprod. Biomed. Online 2019, 39, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Gorodeckaja, J.; Neumann, S.; McCollin, A.; Ottolini, C.S.; Wang, J.; Ahuja, K.; Handyside, A.; Summers, M. High implantation and clinical pregnancy rates with single vitrified-warmed blastocyst transfer and optional aneuploidy testing for all patients. Hum. Fertil. 2020, 23, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Kort, J.; Westphal, L. Miscarriage history association with euploid embryo transfer outcomes. Reprod. Biomed. Online 2019, 39, 617–623. [Google Scholar] [CrossRef]
- Ozbek, I.Y.; Mumusoglu, S.; Polat, M.; Bozdag, G.; Sokmensuer, L.K.; Yarali, H. Comparison of single euploid blastocyst transfer cycle outcome derived from embryos with normal or abnormal cleavage patterns. Reprod. Biomed. Online 2021, 42, 892–900. [Google Scholar] [CrossRef]
- Munné, S.; Kaplan, B.; Frattarelli, J.L.; Child, T.; Nakhuda, G.; Shamma, F.N.; Silverberg, K.; Kalista, T.; Handyside, A.H.; Katz-Jaffe, M.; et al. Preimplantation genetic testing for aneuploidy versus morphology as selection criteria for single frozen-thawed embryo transfer in good-prognosis patients: A multicenter randomized clinical trial. Fertil. Steril. 2019, 112, 1071–1079.e7. [Google Scholar] [CrossRef]
- Simon, A.L.; Kiehl, M.; Fischer, E.; Proctor, J.G.; Bush, M.R.; Givens, C.; Rabinowitz, M.; Demko, Z.P. Pregnancy outcomes from more than 1800 in vitro fertilization cycles with the use of 24-chromosome single-nucleotide polymorphism-based preimplantation genetic testing for aneuploidy. Fertil. Steril. 2018, 110, 113–121. [Google Scholar] [CrossRef]
- Boynukalin, F.K.; Abalı, R.; Gultomruk, M.; Yarkiner, Z.; Mutlu, A.; Bahceci, M. Which factors affect the likelihood of miscarriage after single euploid blastocyst transfer? Reprod. Biomed. Online 2021, 42, 1187–1195. [Google Scholar] [CrossRef]
- Xiao, M.; Lei, C.-X.; Xi, Y.-P.; Lu, Y.-L.; Wu, J.-P.; Li, X.-Y.; Zhang, S.; Zhu, S.-J.; Zhou, J.; Li, X.; et al. Next-Generation Sequencing Is More Efficient at Detecting Mosaic Embryos and Improving Pregnancy Outcomes than Single-Nucleotide Polymorphism Array Analysis. J. Mol. Diagn. 2021, 23, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Tiegs, A.W.; Tao, X.; Zhan, Y.; Whitehead, C.; Kim, J.; Hanson, B.; Osman, E.; Kim, T.J.; Patounakis, G.; Gutmann, J.; et al. A multicenter, prospective, blinded, nonselection study evaluating the predictive value of an aneuploid diagnosis using a targeted next-generation sequencing-based preimplantation genetic testing for aneuploidy assay and impact of biopsy. Fertil. Steril. 2021, 115, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Ding, Y.; Wang, Y.; He, Y.; Sun, Y.; Lu, Y.; Yao, N. Comparison of preimplantation genetic testing for aneuploidy versus intracytoplasmic sperm injection in severe male infertility. Andrologia 2021, 53, e14065. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Y.; Fan, Q.; Wang, J.; Li, R.; Xu, Y.; Guo, J.; Wang, Y.-Z.; Zeng, Y.-H.; Ding, C.-H.; Cai, B.; et al. Higher chromosomal abnormality rate in blastocysts from young patients with idiopathic recurrent pregnancy loss. Fertil. Steril. 2020, 113, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Niu, Y.; Wan, A.; Zhang, T. Next-Generation Sequencing (NGS)-Based Preimplantation Genetic Testing for Aneuploidy (PGT-A) of Trophectoderm Biopsy for Recurrent Implantation Failure (RIF) Patients: A Retrospective Study. Reprod. Sci. 2021, 28, 1923–1929. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, M.; García-Velasco, J.A.; Meseguer, M.; Pellicer, A.; Bellver, J. Female obesity increases the risk of miscarriage of euploid embryos. Fertil. Steril. 2021, 115, 1495–1502. [Google Scholar] [CrossRef]
- Awadalla, M.S.; Vestal, N.L.; McGinnis, L.K.; Ahmady, A.; Paulson, R.J. Effect of age and morphology on sustained implantation rate after euploid blastocyst transfer. Reprod. Biomed. Online 2021, 43, 395–403. [Google Scholar] [CrossRef]
- Nazem, T.G.; Sekhon, L.; Lee, J.A.; Overbey, J.; Pan, S.; Duke, M.; Briton-Jones, C.; Whitehouse, M.; Copperman, A.B.; Stein, D.E. The correlation between morphology and implantation of euploid human blastocysts. Reprod. Biomed. Online 2019, 38, 169–176. [Google Scholar] [CrossRef]
- Cimadomo, D.; Fabozzi, G.; Dovere, L.; Maggiulli, R.; Albricci, L.; Innocenti, F.; Soscia, D.; Giancani, A.; Vaiarelli, A.; Guido, M.; et al. Clinical, obstetric and perinatal outcomes after vitrified-warmed euploid blastocyst transfer are independent of cryo-storage duration. Reprod. Biomed. Online 2022, 44, 221–227. [Google Scholar] [CrossRef]
- Friedenthal, J.; Maxwell, S.M.; Tiegs, A.W.; Besser, A.G.; McCaffrey, C.; Munné, S.; Noyes, N.; Grifo, J.A. Clinical error rates of next generation sequencing and array comparative genomic hybridization with single thawed euploid embryo transfer. Eur. J. Med. Genet. 2020, 63, 103852. [Google Scholar] [CrossRef]
- Sekhon, L.; Feuerstein, J.; Pan, S.; Overbey, J.; Lee, J.A.; Briton-Jones, C.; Flisser, E.; Stein, D.E.; Mukherjee, T.; Grunfeld, L.; et al. Endometrial preparation before the transfer of single, vitrified-warmed, euploid blastocysts: Does the duration of estradiol treatment influence clinical outcome? Fertil. Steril. 2019, 111, 1177–1185.e3. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, X.; Hao, Y.; Ji, D.; Zhang, Z.; Wei, Z.; Cao, Y.; Zhou, P. Comprehensive assessment of a clinic’s experience of preimplantation genetic testing by a cumulative rate. Taiwan. J. Obstet. Gynecol. 2021, 60, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Niu, Y.; Wan, A.; Zhang, T. Effect of parental origin and predictors for obtaining a euploid embryo in balanced translocation carriers. Reprod. Biomed. Online 2022, 44, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Cheng, D.; Ouyang, Q.; Xie, P.; Lu, C.; Gong, F.; Hu, L.; Tan, Y.; Lu, G.; Lin, G. Prevalence and authenticity of de-novo segmental aneuploidy (>16 Mb) in human blastocysts as detected by next-generation sequencing. Reprod. Biomed. Online 2018, 37, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Nair, J.; Shetty, S.; Kasi, C.I.; Thondehalmath, N.; Ganesh, D.; Bhat, V.R.; Mannadia, S.; Ranganath, A.; Nayak, R.; Gunasheela, D.; et al. Preimplantation genetic testing for aneuploidy (PGT-A)—A single-center experience. J. Assist. Reprod. Genet. 2022, 39, 729–738. [Google Scholar] [CrossRef]
- Liu, Y.-L.; Yu, T.-N.; Wang, P.-H.; Tzeng, C.-R.; Chen, C.-H.; Chen, C.-H. Could PGT-A pick up true abnormalities that have clinical relevance? Retrospective analysis of 1043 embryos. Taiwan. J. Obstet. Gynecol. 2020, 59, 496–501. [Google Scholar] [CrossRef]
- Thorne, J.; Loza, A.; Kaye, L.; Nulsen, J.; Benadiva, C.; Grow, D.; Engmann, L. Euploidy rates between cycles triggered with gonadotropin-releasing hormone agonist and human chorionic gonadotropin. Fertil. Steril. 2019, 112, 258–265. [Google Scholar] [CrossRef]
- Stovezky, Y.R.; Romanski, P.A.; Bortoletto, P.; Spandorfer, S.D. Body mass index is not associated with embryo ploidy in patients undergoing in vitro fertilization with preimplantation genetic testing. Fertil. Steril. 2021, 116, 388–395. [Google Scholar] [CrossRef]
- de Los Santos, M.J.; Diez Juan, A.; Mifsud, A.; Mercader, A.; Meseguer, M.; Rubio, C.; Pellicer, A. Variables associated with mitochondrial copy number in human blastocysts: What can we learn from trophectoderm biopsies? Fertil. Steril. 2018, 109, 110–117. [Google Scholar] [CrossRef]
- Hanson, B.M.; Tao, X.; Zhan, Y.; Kim, J.G.; Klimczak, A.M.; Herlihy, N.S.; Scott, R.T., Jr.; Seli, E. Shorter telomere length of white blood cells is associated with higher rates of aneuploidy among infertile women undergoing in vitro fertilization. Fertil. Steril. 2021, 115, 957–965. [Google Scholar] [CrossRef]
- Cascales, A.; Lledó, B.; Ortiz, J.A.; Morales, R.; Ten, J.; Llácer, J.; Bernabeu, R. Effect of ovarian stimulation on embryo aneuploidy and mosaicism rate. Syst. Biol. Reprod. Med. 2021, 67, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Dviri, M.; Madjunkova, S.; Koziarz, A.; Antes, R.; Abramov, R.; Mashiach, J.; Moskovtsev, S.; Kuznyetsova, I.; Librach, C. Is there a correlation between paternal age and aneuploidy rate? An analysis of 3118 embryos derived from young egg donors. Fertil. Steril. 2020, 114, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Zeyad, A.; Al-Abdulkareem, B.; Hammadeh, M.E. The relationship between preimplantation morphokinetics of human embryos and sex chromosome pattern. Reprod. Biol. 2018, 18, 385–389. [Google Scholar] [CrossRef]
- Rubio, C.; Navarro-Sánchez, L.; García-Pascual, C.M.; Ocali, O.; Cimadomo, D.; Venier, W.; Barroso, G.; Kopcow, L.; Bahçeci, M.; Kulmann, M.I.R.; et al. Multicenter prospective study of concordance between embryonic cell-free DNA and trophectoderm biopsies from 1301 human blastocysts. Am. J. Obstet. Gynecol. 2020, 223, 751.e1–751.e13. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Ding, M.; Zhang, Y.; Sun, Y.; Lin, F.; Diao, Z.; Zhou, J. A mathematical model for predicting the number of transferable blastocysts in next-generation sequencing-based preimplantation genetic testing. Arch. Gynecol. Obstet. 2022, 305, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K. Pre-implantation genetic testing: Past, present, future. Reprod. Med. Biol. 2020, 20, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Hoyos, L.R.; Cheng, C.Y.; Brennan, K.; Hubert, G.; Wang, B.; Buyalos, R.P.; Quinn, M.; Shamonki, M. Euploid rates among oocyte donors: Is there an optimal age for donation? J. Assist. Reprod. Genet. 2020, 37, 589–594. [Google Scholar] [CrossRef]
- Roos Kulmann, M.I.; Lumertz Martello, C.; Bos-Mikich, A.; Frantz, N. Pronuclear and blastocyst morphology are associated age-dependently with embryo ploidy in in vitro fertilization cycles. Hum. Fertil. 2020, 369–379. [Google Scholar] [CrossRef]
- Karlıkaya, G.; Boynukalin, F.K.; Gultomruk, M.; Kavrut, M.; Abalı, R.; Demir, B.; Ecemis, S.; Yarkiner, Z.; Bahceci, M. Euploidy rates of embryos in young patients with good and low prognosis according to the POSEIDON criteria. Reprod. Biomed. Online 2021, 42, 733–741. [Google Scholar] [CrossRef]
- Clua, E.; Rodríguez, I.; Arroyo, G.; Racca, A.; Martínez, F.; Polyzos, N.P. Blastocyst transfer increases cumulative-live-birth-rates and reduces time and cost to livebirth compared with cleavage stage in recipients of donated oocytes. A randomized controlled trial. Reprod. Biomed. Online 2022, 44, 995–1004. [Google Scholar] [CrossRef]
- He, H.; Jing, S.; Lu, C.F.; Tan, Y.Q.; Luo, K.L.; Zhang, S.P.; Gong, F.; Lu, G.X.; Lin, G. Neonatal outcomes of live births after blastocyst biopsy in preimplantation genetic testing cycles: A follow-up of 1721 children. Fertil. Steril. 2019, 112, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Frese, K.S.; Katus, H.A.; Meder, B. Next-generation sequencing: From understanding biology to personalized medicine. Biology 2013, 2, 378–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of Next-Generation Sequencing Systems. J. Biomed. Biotechnol. 2012, 2012, 251364. [Google Scholar] [CrossRef]
- Mardis, E.R. Next-Generation Sequencing Platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [Google Scholar] [CrossRef] [Green Version]
- Mardis, E.R. Next-Generation DNA Sequencing Methods. Annu. Rev. Genomics Hum. Genet. 2008, 9, 387–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Gupta, U.D. Next generation sequencing and its applications. In Animal Biotechnology; Academic Press: Cambridge, MA, USA, 2020; pp. 395–421. ISBN 9780128117101. [Google Scholar]
- Bras, J.; Guerreiro, R.; Hardy, J. Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nat. Rev. Neurosci. 2012, 13, 453–464. [Google Scholar] [CrossRef]
- Papageorgiou, E.A.; Patsalis, P.C. Non-invasive prenatal diagnosis of aneuploidies: New technologies and clinical applications. Genome Med. 2012, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- Norwitz, E.R.; Levy, B. Noninvasive prenatal testing: The future is now. Rev. Obstet. Gynecol. 2013, 6, 48–62. [Google Scholar]
- Cogulu, O. Next Generation Sequencing as a Tool for Noninvasive Prenatal Tests. In Clinical Applications for Next-Generation Sequencing; Demkow, U., Płoski, R., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 171–188. ISBN 978-0-12-801739-5. [Google Scholar]
- Di Resta, C.; Galbiati, S.; Carrera, P.; Ferrari, M. Next-generation sequencing approach for the diagnosis of human diseases: Open challenges and new opportunities. Electron. J. IFCC (eJIFCC) 2018, 29, 4–14. [Google Scholar]
- Płoski, R. Next Generation Sequencing—General Information about the Technology, Possibilities, and Limitations. In Clinical Applications for Next-Generation Sequencing; Demkow, U., Płoski, R., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 1–18. ISBN 978-0-12-801739-5. [Google Scholar]

| No. of Patients or Couples | Reproductive Outcomes | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Implantation | Pregnancy | Clinical Pregnancy | Ongoing Pregnancy | Miscarriage/ Early Pregnancy Loss | Spontaneous Pregnancy Loss | Late Pregnancy Loss | Biochemical Pregnancy | Ectopic Pregnancy | ||
| NO ALLOCATION PER GROUPS | ||||||||||
| 270 patients | 63.10% (n = 53) vs. 46.67% (n = 30) | [11] | ||||||||
| 314 patients | 66.2% (n = 235) | 52.4% (n = 186) | 5.6% (n = 20) | 2.3% (n = 8) | 7.6% (n = 27) | 0.6% (n = 2) | [12] | |||
| 330 patients | 60% (n = 215) vs. 40% (n = 8) | 18% (n = 65) vs. 40% (n = 8) | [13] | |||||||
| 460 patients | 69.7% (n = 159) vs. 57.2 (n = 63) | [14] | ||||||||
| 31649 patients | 62% | [15] | ||||||||
| 100 couples | 65.38% (n = 34) | n = 8 | 2.94% (n = 1) | 69.23% (n = 36) | [16] | |||||
| WITH ALLOCATION PER GROUPS | ||||||||||
| 166 patients | 29.0% (n = 9) vs. 45.7% (n = 64) vs. 24.0% (n = 6) | 29.0% (n = 9) vs. 40.7% (n = 57) vs. 16% (n = 4) | 30.9% (n = 4) vs. 21.9% (n = 16) vs. 42.8% (n = 3) | 0% (n = 0) vs. 0% (n = 0) vs. 0% (n = 0) | [17] | |||||
| 228 patients | 65.4% (n = 53) vs. 67.4% (n = 99) | 13.2% (n = 7) vs. 15.2% (n = 15) | 14.8% (n = 9) vs. 15.4% (n = 18) | [18] | ||||||
| 260 patients | 67.8% (n = 124) vs. 63.6% (n = 49) | 75.4% (n = 138) vs. 70.1% (n = 54) | 57.9% (n = 106) vs. 58.4% (n = 45) | [19] | ||||||
| 275 patients | * 31.5% vs. 28.8% ** 16.40% vs. 20.50% | [20] | ||||||||
| 364 patients | *** 64.29% (n = 63) vs. 50.38% (n = 134) **** 64.12% (n = 84) vs. 51.60% (n = 226) | *** 3.17% (n = 2) vs. 11.94% (n = 16) **** 4.76% (n = 4) vs. 12.39% (n = 28) | *** 6.12% (n = 6) vs. 11.26% (n = 17) **** 8.70% (n = 8) vs. 9.96% (n = 25) | [21] | ||||||
| 525 patients | 68.0% (n = 221) vs. 55.5% (n = 111) | 13.1% (n = 29) vs. 11.7% (n = 13) | 16.9% (n = 45) vs. 20.1% (n = 28) | [22] | ||||||
| 1152 patients | 3.2% vs. 6.8% | 4.2% vs. 3.9% | 1.1% vs. 0.4% | [23] | ||||||
| 142 couples | 45.77% (n = 65) vs. 29.41% (n = 5) | 59.15% (n = 84) vs. 47.10% (n = 8) | 42.96% (n = 61) vs. 17.65% (n = 3) | 6.15% (n = 4) vs. 40.0% (n = 2) | 13.38% (n = 19) vs. 17.65% (n = 3) | [24] | ||||
| 150 couples | 3.4% (n = 1) vs. 14.7% (n = 8) | 6.9% (n = 2) vs. 1.8% (n = 1) | [25] | |||||||
| 180 couples | n = 9 vs. n = 2 vs. n = 2 | [26] | ||||||||
| 779 couples | ***** 67.9% (n = 106) vs. 69.6% (n = 117) vs. 75.6% (n = 68) ****** 86.8% (n = 33) vs. 78.4% (n = 29) vs. 46.4% (n = 13) | ***** 51.2% (n = 66) vs. 47.4% (n = 65) vs. 62.2% (n = 51) ****** 44.7% (n = 17) vs. 64.7% (n = 22) vs. 69.2% (n = 18) | [27] | |||||||
| 1418 couples | 50.5% (n = 341) vs. 41.7% (n = 228) | 15.5% (n = 54) vs. 22.8% (n = 52) | [28] | |||||||
| No. of Patients or Couples | Reproductive Outcomes | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Implantation | Pregnancy | Clinical Pregnancy | Ongoing Pregnancy | Miscarriage/ Early Pregnancy Loss | Spontaneous Pregnancy Loss | Late Pregnancy Loss | Biochemical Pregnancy | Ectopic Pregnancy | ||
| NO ALLOCATION PER GROUPS | ||||||||||
| 296 patients | 85.7% (n = 12) vs. 84.0% (n = 84) vs. 80.0% (n = 44) vs. 80.0% (n = 8) | 7.1% (n = 1) vs. 6% (n = 6) vs. 5.4% (n = 3) vs. 10% (n = 1) | [29] | |||||||
| 783 patients | 12.0% (n = 29) vs. 11.0% (n = 15) vs. 12.7% (n = 8) | 10.7% (n = 29) vs. 12.3% (n = 19) vs. 13.7% (n = 10) | [30] | |||||||
| 1531 patients | 44.4% (n = 8) | 38.9% (n = 7) | [31] | |||||||
| WITH ALLOCATION PER GROUPS | ||||||||||
| 206 patients | 64.71% (n = 22) vs. 65.71% (n = 23) | n = 1 vs. n = 3 | [32] | |||||||
| 108 patients | 51.8% (n = 43) vs. 52% (n = 13) | 47% (n = 39) vs. 52% (n = 13) | 47% (n = 39) vs. 36% (n = 9) | 5.1% (n = 2) vs. 30.7% (n = 4) | [33] | |||||
| 108 patients | 79.4% (n = 50) vs. 66.7% (n = 16) vs. 25.0% (n = 5) | 76.2% (n = 48) vs. 62.5% (n = 15) vs. 25.0% (n = 5) | 68.3% (n = 43) vs. 62.5% (n = 15) vs. 10.0% (n = 2) | [34] | ||||||
| 155 patients | 80% (n = 68) | 66% (n = 56) | [35] | |||||||
| 283 patients | 52.3% (n = 148) | [36] | ||||||||
| 554 patients | 63.9% (n = 186) | 19.3% (n = 36) | [37] | |||||||
| 661 patients | 50.0% (n = 137) vs. 45.7% (n = 143) | 9.9% (n = 27) vs. 9.6% (n = 30) | 10.6% (n = 29) vs. 8.3% (n = 26) | [38] | ||||||
| 974 patients | 69.9% vs. 64.9% | n = 472 vs. n = 94 | n = 21 vs. n = 6 | [39] | ||||||
| 1051 patients | 14.5% (n = 100) | n = 68 | n = 11 | [40] | ||||||
| 1513 patients | 51.34% (n = 306) vs. 49.56% (n = 227) | 10.07% (n = 60) vs. 6.33% (n = 29) | 9.56% (n = 57) vs. 10.48% (n = 48) | [41] | ||||||
| 648 couples | 64.7% (n = 202) vs. 0% (n = 0) vs. 68.8% (n = 11) vs. 30.8% (n = 12) | 73.1% vs. 23.5% | 7.4% (n = 23) vs. 23.5% (n = 24 vs. 12.5% (n = 2) vs. 5.1% (n = 2) | 9% (n = 28) vs. 16.7% (n = 17) vs. 12.5% (n = 2) vs. 12.8% (n = 5) | 1.0% (n = 3) vs. 0% (n = 0) vs. 0% (n = 0) vs. 0% (n = 0) | [42] | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doroftei, B.; Ilie, O.-D.; Anton, N.; Armeanu, T.; Ilea, C. A Mini-Review Regarding the Clinical Outcomes of In Vitro Fertilization (IVF) Following Pre-Implantation Genetic Testing (PGT)-Next Generation Sequencing (NGS) Approach. Diagnostics 2022, 12, 1911. https://doi.org/10.3390/diagnostics12081911
Doroftei B, Ilie O-D, Anton N, Armeanu T, Ilea C. A Mini-Review Regarding the Clinical Outcomes of In Vitro Fertilization (IVF) Following Pre-Implantation Genetic Testing (PGT)-Next Generation Sequencing (NGS) Approach. Diagnostics. 2022; 12(8):1911. https://doi.org/10.3390/diagnostics12081911
Chicago/Turabian StyleDoroftei, Bogdan, Ovidiu-Dumitru Ilie, Nicoleta Anton, Theodora Armeanu, and Ciprian Ilea. 2022. "A Mini-Review Regarding the Clinical Outcomes of In Vitro Fertilization (IVF) Following Pre-Implantation Genetic Testing (PGT)-Next Generation Sequencing (NGS) Approach" Diagnostics 12, no. 8: 1911. https://doi.org/10.3390/diagnostics12081911
APA StyleDoroftei, B., Ilie, O.-D., Anton, N., Armeanu, T., & Ilea, C. (2022). A Mini-Review Regarding the Clinical Outcomes of In Vitro Fertilization (IVF) Following Pre-Implantation Genetic Testing (PGT)-Next Generation Sequencing (NGS) Approach. Diagnostics, 12(8), 1911. https://doi.org/10.3390/diagnostics12081911








