Deep-Learning-Based Real-Time and Automatic Target-to-Background Ratio Calculation in Fluorescence Endoscopy for Cancer Detection and Localization
Abstract
:1. Introduction
1.1. Detection and Classification
Localization and Segmentation
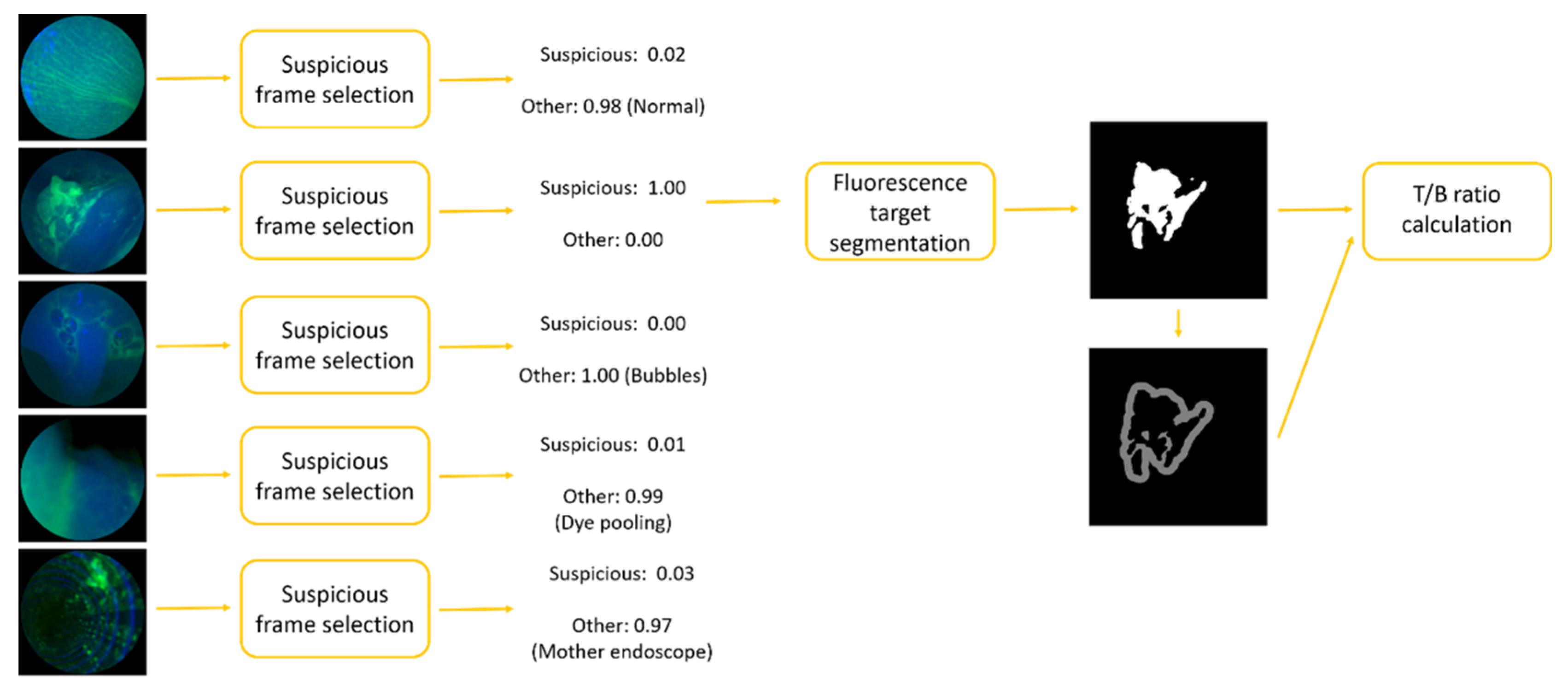
2. Materials and Methods
2.1. Suspicious Frame Selection Model
2.2. Fluorescence Target Segmentation Model
2.3. Regularization
3. Experiments
3.1. Datasets and Preprocessing
- The first set contained a total of 15 videos from an in vivo Multiplex imaging study (NCT03589443) performed in patients with Barrett’s neoplasia. The fluorescence-labeled peptides of QRH*-Cy5 and KSP*-IRDye800 targeting EGFR and ErbB2, respectively, were topically applied in the lower esophagus. Videos were recorded using SFE with a resolution of 720 720 at 30 Hz. Separate channels were used to record fluorescence images from QRH*-Cy5 (red) and KSP*-IRDye800 (green), as well as reflectance (blue). The combined duration of 15 videos was approximately 96 min (~173,000 frames) [7].
- A separate set contained a total of 20 videos from an in vivo Dimer imaging study (NCT03852576) performed in patients with Barrett’s neoplasia. Only IRDye800 was used to label dimer peptide QRH*-KSP*-E3-IRDye800, targeting both EGFR and ErbB2, and was again topically applied. The NIR fluorescence projected to the green channel was recorded with reflectance as the blue channel. The combined duration of 20 videos was approximately 88 min (~158,000 frames) [8].
3.2. Model Development
3.2.1. Frame Selection Model
3.2.2. Fluorescence Target Segmentation Model
- Training and validation: 1006 images from 11 videos from the Multiplex study were randomly separated into training (910) and validation (96) groups;
- Testing A: 100 images were extracted from 4 different videos from the Multiplex study;
- Testing B: 100 images were extracted from the Dimer study.
4. Results
4.1. Suspicious Frame Selection
4.2. Fluorescence Target Segmentation
- (1)
- Networks with geometric augmentation during training surpassed those without geometric augmentation by a significant margin (one-tailed paired t-test, p-value), which was consistent with our findings for the frame selection model;
- (2)
- Neither (a) adjusting the brightness and contrast of the images nor (b) adding noise or blur to the images during training showed an improvement in performance;
- (3)
- UNet outperformed BiSeNet in segmentation accuracy (one-tailed paired t-test, p-value);
- (4)
- Using pretrained ImageNet weights as initialization did not improve the performance;
- (5)
- BiSeNet models using the Xception backbone outperformed BiSeNet models using the MobileNetV2 backbone regarding to accuracy and mIOU (one-tailed paired t-test, p-value).
4.3. Speed
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Torre, and Ahmedin Jemal. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melina, A.; Rutherford, M.J.; Bardot, A.; Ferlay, J.; Andersson, T.M.L.; Myklebust, T.Å.; Tervonen, H.; Grad, V.T.; Ransom, D.; Shack, L.; et al. Progress in cancer survival, mortality, and incidence in seven high-income countries 1995–2014 ICBP SURVMARK-2: A population-based study. Lancet Oncol. 2019, 20, 1493–1505. [Google Scholar]
- Vingeliene, S.; Chan, D.S.M.; Vieira, A.R.; Polemiti, E.; Stevens, C.; Abar, L.; Rosenblatt, D.N.; Greenwood, D.C.; Norat, T. An update of the WCRF/AICR systematic literature review and meta-analysis on dietary and anthropometric factors and esophageal cancer risk. Ann. Oncol. 2017, 28, 2409–2419. [Google Scholar] [CrossRef]
- Bhat, S.; Coleman, H.G.; Yousef, F.; Johnston, B.T.; McManus, D.T.; Gavin, A.T.; Murray, L.J. Risk of malignant progression in Barrett’s esophagus patients: Results from a large population-based study. J. Natl. Cancer Inst. 2011, 103, 1049–1057. [Google Scholar] [CrossRef] [Green Version]
- Shaheen, N.J.; Falk, G.W.; Iyer, P.G.; Gerson, L.B. ACG clinical guideline: Diagnosis and management of Barrett’s esophagus. Off. J. Am. Coll. Gastroenterol. ACG 2016, 111, 30–50. [Google Scholar] [CrossRef]
- Goetz, M.; Wang, T.D. Molecular imaging in gastrointestinal endoscopy. Gastroenterology 2010, 138, 828–833. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Jiang, Y.; Chang, T.-S.; Joshi, B.; Zhou, J.; Rubenstein, J.H.; Wamsteker, E.J.; Kwon, R.S.; Appelman, H.; Beer, D.G.; et al. Multiplexed endoscopic imaging of Barrett’s neoplasia using targeted fluorescent heptapeptides in a phase 1 proof-of-concept study. Gut 2021, 70, 1010–1013. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, Y.; Chang, T.-S.; Rubenstein, J.H.; Kwon, R.S.; Wamsteker, E.-J.; Prabhu, A.; Zhao, L.; Appelman, H.D.; Owens, S.R.; et al. Detection of Barrett’s neoplasia with a near-infrared fluorescent heterodimeric peptide. Endoscopy 2022. [Google Scholar] [CrossRef]
- Jiang, Y.; Gong, Y.; Rubenstein, J.H.; Wang, T.D.; Seibel, E.J. Toward real-time quantification of fluorescence molecular probes using target/background ratio for guiding biopsy and endoscopic therapy of esophageal neoplasia. J. Med. Imaging 2017, 4, 024502. [Google Scholar] [CrossRef] [Green Version]
- Christian, S.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Joseph, R.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Ilya, S.; Vinyals, O.; Le, Q.V. Sequence to sequence learning with neural networks. In Proceedings of the 27th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014. [Google Scholar]
- Kyunghyun, C.; van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing EMNLP, Doha, Qatar, 25–29 October 2014. [Google Scholar]
- Silver, D.; Huang, A.; Maddison, C.J.; Guez, A.; Sifre, L.; van den Driessche, G.; Schrittwieser, J.; Antonoglou, I.; Panneershelvam, V.; Lanctot, M.; et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016, 529, 484–489. [Google Scholar] [CrossRef]
- Seonwoo, M.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2017, 18, 851–869. [Google Scholar]
- Milletari, F.; Ahmadi, S.-A.; Kroll, C.; Plate, A.; Rozanski, V.; Maiostre, J.; Levin, J.; Dietrich, O.; Ertl-Wagner, B.; Bötzel, K.; et al. Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput. Vis. Image Underst. 2017, 164, 92–102. [Google Scholar] [CrossRef] [Green Version]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-Van de Kaa, C.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Kooi, T.; Litjens, G.; van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Trebeschi, S.; Van Griethuysen, J.J.M.; Lambregts, D.; Lahaye, M.J.; Parmar, C.; Bakers, F.C.H.; Peters, N.H.G.M.; Beets-Tan, R.G.H.; Aerts, H.J.W.L. Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci. Rep. 2017, 7, 5301. [Google Scholar] [CrossRef]
- Olaf, R.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.; Heng, P.-A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [Green Version]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.-M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Xiao, X.; Brown, J.R.G.; Berzin, T.M.; Tu, M.; Xiong, F.; Hu, X.; Liu, P.; Song, Y.; Zhang, D.; et al. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat. Biomed. Eng. 2018, 2, 741–748. [Google Scholar] [CrossRef]
- Urban, G.; Tripathi, P.; Alkayali, T.; Mittal, M.; Jalali, F.; Karnes, W.; Baldi, P. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 2018, 155, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Zhou, F.; Braden, B.; Bailey, A.; Yang, S.; Cheng, G.; Zhang, P.; Li, X.; Kayser, M.; Soberanis-Mukul, R.D.; et al. An objective comparison of detection and segmentation algorithms for artefacts in clinical endoscopy. Sci. Rep. 2020, 10, 2748. [Google Scholar] [CrossRef] [PubMed]
- Izadyyazdanabadi, M.; Belykh, E.; Mooney, M.A.; Eschbacher, J.M.; Nakaji, P.; Yang, Y.; Preul, M.C. Prospects for theranostics in neurosurgical imaging: Empowering confocal laser endomicroscopy diagnostics via deep learning. Front. Oncol. 2018, 8, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caicedo, J.C.; Roth, J.; Goodman, A.; Becker, T.; Karhohs, K.W.; Broisin, M.; Molnar, C.; McQuin, C.; Singh, S.; Theis, F.J.; et al. Evaluation of deep learning strategies for nucleus segmentation in fluorescence images. Cytom. Part A 2019, 95, 952–965. [Google Scholar] [CrossRef] [Green Version]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Van Valen, D. Deep learning for cellular image analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef]
- van der Sommen, F.; de Groof, J.; Struyvenberg, M.; van der Putten, J.; Boers, T.; Fockens, K.; Schoon, E.J.; Curvers, W.; De With, P.; Mori, Y.; et al. Machine learning in GI endoscopy: Practical guidance in how to interpret a novel field. Gut 2020, 69, 2035–2045. [Google Scholar] [CrossRef]
- Mark, S.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- François, C. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Yu, C.; Wang, J.; Peng, C.; Gao, C.; Yu, G.; Sang, N. Bisenet: Bilateral segmentation network for real-time semantic segmentation. In Proceedings of the European Conference on Computer Vision ECCV, Munich, Germany, 8–14 September 2018; pp. 325–341. [Google Scholar]
- Lutz, P. Early stopping-but when? In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 1998; pp. 55–69. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- P, K.D.; Ba, J.L. Adam: A method for stochastic gradient descent. In Proceedings of the ICLR: International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized intersection over union: A metric and a loss for bounding box regression. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 658–666. [Google Scholar]
- Martín, A.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. {TensorFlow}: A system for {Large-Scale} machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation OSDI 16, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Jasper, V.; de Wit, J.G.; jan Voskuil, F.; Witjes, M.J.H. Improving oral cavity cancer diagnosis and treatment with fluorescence molecular imaging. Oral Dis. 2021, 27, 21–26. [Google Scholar]
- Pan, Y.; Volkmer, J.-P.; Mach, K.E.; Rouse, R.V.; Liu, J.-J.; Sahoo, D.; Chang, T.C.; Metzner, T.J.; Kang, L.; van de Rijn, M.; et al. Endoscopic molecular imaging of human bladder cancer using a CD47 antibody. Sci. Transl. Med. 2014, 6, ra148–ra260. [Google Scholar] [CrossRef]
- Kelly, K.; Alencar, H.; Funovics, M.; Mahmood, U.; Weissleder, R. Detection of invasive colon cancer using a novel, targeted, library-derived fluorescent peptide. Cancer Res. 2004, 64, 6247–6251. [Google Scholar] [CrossRef] [Green Version]
- Hsiung, P.-L.; Hardy, J.; Friedland, S.; Soetikno, R.; Du, C.B.; Wu, A.; Sahbaie, P.; Crawford, J.M.; Lowe, A.W.; Contag, C.; et al. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat. Med. 2008, 14, 454–458. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Daniel, B.; Zhou, C.; Xiao, F.; Lee, Y.J. Yolact: Real-time instance segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 9157–9166. [Google Scholar]
- Vorontsov, E.; Cerny, M.; Régnier, P.; Di Jorio, L.; Pal, C.J.; Lapointe, R.; Vandenbroucke-Menu, F.; Turcotte, S.; Kadoury, S.; Tang, A. Deep learning for automated segmentation of liver lesions at ct in patients with colorectal cancer liver metastases. Radiol. Artif. Intell. 2019, 1, 180014. [Google Scholar] [CrossRef] [PubMed]
- Karimi, D.; Dou, H.; Warfield, S.K.; Gholipour, A. Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis. Med. Image Anal. 2020, 65, 101759. [Google Scholar] [CrossRef] [PubMed]
- Thekumparampil, K.K.; Khetan, A.; Lin, Z.; Oh, S. Robustness of conditional gans to noisy labels. In Proceedings of the 31th International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Bekker, A.J.; Goldberger, J. Training deep neural-networks based on unreliable labels. In Proceedings of the 2016 IEEE International Conference on Acoustics, Speech and Signal Processing ICASSP, Shanghai, China, 20–25 March 2016; pp. 2682–2686. [Google Scholar]
- Ghosh, A.; Kumar, H.; Sastry, P.S. Robust loss functions under label noise for deep neural networks. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31. [Google Scholar] [CrossRef]
- Giorgio, P.; Rozza, A.; Menon, A.K.; Nock, R.; Qu, L. Making deep neural networks robust to label noise: A loss correction approach. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1944–1952. [Google Scholar]
- Shen, Y.; Sanghavi, S. Learning with bad training data via iterative trimmed loss minimization. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 5739–5748. [Google Scholar]
- Lee, K.; Yun, S.; Lee, K.; Lee, H.; Li, B.; Shin, J. Robust inference via generative classifiers for handling noisy labels. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 3763–3772. [Google Scholar]
- Gong, C.; Bin, K.; Seibel, E.J.; Wang, X.; Yin, Y.; Song, Q. Synergistic Network Learning and Label Correction for Noise-Robust Image Classification. In Proceedings of the ICASSP 2022-2022 IEEE International Conference on Acoustics, Speech and Signal Processing ICASSP, Singapore, 23–27 May 2022; pp. 4253–4257. [Google Scholar]
- Yu, X.; Han, B.; Yao, J.; Niu, G.; Tsang, I.; Sugiyama, M. How does disagreement help generalization against label corruption? In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 7164–7173. [Google Scholar]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Pérez, M.L.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019, 68, 94–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernal, J.; Tajkbaksh, N.; Sanchez, F.J.; Matuszewski, B.J.; Chen, H.; Yu, L.; Angermann, Q.; Romain, O.; Rustad, B.; Balasingham, I.; et al. Comparative validation of polyp detection methods in video colonoscopy: Results from the MICCAI 2015 endoscopic vision challenge. IEEE Trans. Med. Imaging 2017, 36, 1231–1249. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.M.; Engelbrecht, C.J.; Soper, T.D.; Helmchen, F.; Seibel, E.J. Scanning fiber endoscopy with highly flexible, 1 mm catheterscopes for wide-field, full-color imaging. J. Biophotonics 2010, 3, 385–407. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.J.; Lee, C.M.; Joshi, B.P.; Gaustad, A.; Seibel, E.J.; Wang, T.D. Special section on endomicroscopy technologies and biomedical applications: Targeted detection of murine colonic dysplasia in vivo with flexible multispectral scanning fiber endoscopy. J. Biomed. Opt. 2012, 17, 021103. [Google Scholar] [CrossRef]
- Deepak, S.; van der Putten, P.; Plaat, A. On the impact of data set size in transfer learning using deep neural networks. In Proceedings of the International Symposium on Intelligent Data Analysis, Stockholm, Sweden, 13–15 October 2016; Springer: Cham, Switzerland, 2016; pp. 50–60. [Google Scholar]
- de La Comble, A.; Prepin, K. Efficient transfer learning for multi-channel convolutional neural networks. In Proceedings of the 2021 17th International Conference on Machine Vision and Applications MVA, Aichi, Japan, 25–27 July 2021; pp. 1–6. [Google Scholar]
- Ophoff, T.; Van Beeck, K.; Goedemé, T. Exploring RGB+ Depth fusion for real-time object detection. Sensors 2019, 19, 866. [Google Scholar] [CrossRef] [Green Version]
- Choe, G.; Kim, S.-H.; Im, S.; Lee, J.-Y.; Narasimhan, S.G.; Kweon, I.S. RANUS: RGB and NIR urban scene dataset for deep scene parsing. IEEE Robot. Autom. Lett. 2018, 3, 1808–1815. [Google Scholar] [CrossRef]
- Zhou, T.; Ruan, S.; Canu, S. A review: Deep learning for medical image segmentation using multi-modality fusion. Array 2019, 3, 100004. [Google Scholar] [CrossRef]
- Guo, Z.; Li, X.; Huang, H.; Guo, N.; Li, Q. Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans. Radiat. Plasma Med. Sci. 2019, 3, 162–169. [Google Scholar] [CrossRef]
- Nabil, I.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar]
- Jose, D.; Gopinath, K.; Yuan, J.; Lombaert, H.; Desrosiers, C.; Ayed, I.B. HyperDense-Net: A hyper-densely connected CNN for multi-modal image segmentation. IEEE Trans. Med. Imaging 2018, 38, 1116–1126. [Google Scholar]
- Jose, D.; Desrosiers, C.; Ayed, I.B. IVD-Net: Intervertebral disc localization and segmentation in MRI with a multi-modal UNet. In Proceedings of the International Workshop and Challenge on Computational Methods and Clinical Applications for Spine Imaging, Granada, Spain, 16 September 2018; Springer: Cham, Switzerland, 2018; pp. 130–143. [Google Scholar]
- Chen, L.; Wu, Y.; Souza, A.M.D.; Abidin, A.Z.; Wismüller, A.; Xu, C. MRI tumor segmentation with densely connected 3D CNN. In Proceedings of the Medical Imaging 2018: Image Processing, Houston, TX, USA, 11–13 February 2018; Volume 10574, pp. 357–364. [Google Scholar]
- Kamnitsas, K.; Bai, W.; Ferrante, E.; McDonagh, S.; Sinclair, M.; Pawlowski, N.; Rajchl, M.; Lee, M.; Kainz, B.; Rueckert, D.; et al. Ensembles of multiple models and architectures for robust brain tumour segmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Quebec City, QC, Canada, 14 September 2017; Springer: Cham, Switerland, 2017; pp. 450–462. [Google Scholar]
- Rokach, L. Ensemble-based classifiers. Artif. Intell. Rev. 2010, 33, 1–39. [Google Scholar] [CrossRef]
- Aygün, M.; Şahin, Y.H.; Ünal, G. Multi modal convolutional neural networks for brain tumor segmentation. arXiv 2018, arXiv:1809.06191. [Google Scholar]
- Zhe, G.; Li, X.; Huang, H.; Guo, N.; Li, Q. Medical image segmentation based on multi-modal convolutional neural network: Study on image fusion schemes. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging ISBI 2018, Washington, DC, USA, 4–7 April 2018; pp. 903–907. [Google Scholar]
- Gianfrancesco, M.A.; Tamang, S.; Yazdany, J.; Schmajuk, G. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern. Med. 2018, 178, 1544–1547. [Google Scholar] [CrossRef]






| Backbone | ImageNet | Rotation, Shift, Scale, Flip | Brightness, Contrast | Blur, Gaussian Noise | AUC |
|---|---|---|---|---|---|
| MobileNetV2 | ✔ | ✔ | ✔ | 0.926 | |
| ✔ | ✔ | ✔ | ✔ | 0.976 | |
| ✔ | ✔ | ✔ | 0.973 | ||
| ✔ | ✔ | ✔ | 0.97 | ||
| ✔ | ✔ | 0.969 | |||
| ✔ | ✔ | ✔ | 0.887 | ||
| ✔ | ✔ | 0.953 | |||
| ✔ | ✔ | 0.926 | |||
| ✔ | 0.915 | ||||
| Xception | ✔ | ✔ | ✔ | ✔ | 0.989 |
| ✔ | ✔ | ✔ | 0.989 | ||
| ✔ | ✔ | ✔ | 0.986 | ||
| ✔ | ✔ | 0.981 | |||
| ✔ | ✔ | ✔ | 0.95 | ||
| ✔ | ✔ | 0.948 | |||
| ✔ | ✔ | 0.966 | |||
| ✔ | 0.928 |
| Model + Backbone | ImageNet | Rotation, Shift, Scale, Flip | Brightness, Contrast | Blur, Gaussian Noise | Multiplex | Dimer | ||
|---|---|---|---|---|---|---|---|---|
| Accuracy | mIOU | Accuracy | mIOU | |||||
| BiSeNet + MobileNetV2 | ✔ | ✔ | ✔ | ✔ | 0.977 | 0.868 | 0.959 | 0.844 |
| ✔ | ✔ | ✔ | 0.977 | 0.869 | 0.959 | 0.849 | ||
| ✔ | ✔ | ✔ | 0.976 | 0.864 | 0.962 | 0.858 | ||
| ✔ | ✔ | 0.973 | 0.848 | 0.958 | 0.846 | |||
| ✔ | ✔ | ✔ | 0.972 | 0.839 | 0.954 | 0.824 | ||
| ✔ | ✔ | 0.97 | 0.828 | 0.952 | 0.824 | |||
| ✔ | ✔ | 0.976 | 0.864 | 0.956 | 0.832 | |||
| ✔ | 0.967 | 0.818 | 0.952 | 0.818 | ||||
| ✔ | ✔ | ✔ | 0.977 | 0.871 | 0.957 | 0.841 | ||
| ✔ | ✔ | 0.977 | 0.87 | 0.959 | 0.846 | |||
| ✔ | ✔ | 0.978 | 0.874 | 0.964 | 0.865 | |||
| ✔ | 0.977 | 0.872 | 0.962 | 0.858 | ||||
| ✔ | ✔ | 0.968 | 0.819 | 0.948 | 0.805 | |||
| ✔ | 0.968 | 0.826 | 0.955 | 0.831 | ||||
| ✔ | 0.971 | 0.836 | 0.952 | 0.817 | ||||
| 0.969 | 0.828 | 0.954 | 0.829 | |||||
| BiSeNet + Xception | ✔ | ✔ | ✔ | ✔ | 0.978 | 0.873 | 0.964 | 0.864 |
| ✔ | ✔ | ✔ | 0.978 | 0.874 | 0.962 | 0.859 | ||
| ✔ | ✔ | ✔ | 0.978 | 0.878 | 0.966 | 0.873 | ||
| ✔ | ✔ | 0.98 | 0.886 | 0.965 | 0.868 | |||
| ✔ | ✔ | ✔ | 0.973 | 0.841 | 0.953 | 0.821 | ||
| ✔ | ✔ | 0.974 | 0.85 | 0.954 | 0.829 | |||
| ✔ | ✔ | 0.975 | 0.856 | 0.957 | 0.837 | |||
| ✔ | 0.975 | 0.859 | 0.955 | 0.831 | ||||
| ✔ | ✔ | ✔ | 0.978 | 0.873 | 0.963 | 0.861 | ||
| ✔ | ✔ | 0.978 | 0.875 | 0.962 | 0.859 | |||
| ✔ | ✔ | 0.98 | 0.885 | 0.964 | 0.865 | |||
| ✔ | 0.979 | 0.88 | 0.965 | 0.87 | ||||
| ✔ | ✔ | 0.97 | 0.827 | 0.956 | 0.83 | |||
| ✔ | 0.971 | 0.833 | 0.951 | 0.815 | ||||
| ✔ | 0.972 | 0.84 | 0.956 | 0.834 | ||||
| 0.972 | 0.845 | 0.956 | 0.833 | |||||
| Model + Backbone | ImageNet | Rotation, Shift, Scale, Flip | Brightness, Contrast | Blur, Gaussian Noise | Multiplex | Dimer | ||
|---|---|---|---|---|---|---|---|---|
| Accuracy | mIOU | Accuracy | mIOU | |||||
| UNet + MobileNetV2 | ✔ | ✔ | ✔ | ✔ | 0.98 | 0.889 | 0.962 | 0.857 |
| ✔ | ✔ | ✔ | 0.982 | 0.897 | 0.962 | 0.862 | ||
| ✔ | ✔ | ✔ | 0.981 | 0.891 | 0.968 | 0.88 | ||
| ✔ | ✔ | 0.979 | 0.881 | 0.963 | 0.863 | |||
| ✔ | ✔ | ✔ | 0.974 | 0.847 | 0.955 | 0.829 | ||
| ✔ | ✔ | 0.973 | 0.845 | 0.955 | 0.83 | |||
| ✔ | ✔ | 0.976 | 0.865 | 0.959 | 0.845 | |||
| ✔ | 0.97 | 0.829 | 0.952 | 0.816 | ||||
| ✔ | ✔ | ✔ | 0.979 | 0.88 | 0.963 | 0.865 | ||
| ✔ | ✔ | 0.979 | 0.877 | 0.963 | 0.86 | |||
| ✔ | ✔ | 0.979 | 0.882 | 0.967 | 0.877 | |||
| ✔ | 0.979 | 0.881 | 0.964 | 0.865 | ||||
| ✔ | ✔ | 0.972 | 0.839 | 0.949 | 0.81 | |||
| ✔ | 0.97 | 0.834 | 0.962 | 0.857 | ||||
| ✔ | 0.972 | 0.84 | 0.958 | 0.844 | ||||
| 0.96 | 0.797 | 0.957 | 0.839 | |||||
| UNet + Xception | ✔ | ✔ | ✔ | ✔ | 0.982 | 0.897 | 0.966 | 0.872 |
| ✔ | ✔ | ✔ | 0.979 | 0.883 | 0.965 | 0.87 | ||
| ✔ | ✔ | ✔ | 0.981 | 0.892 | 0.966 | 0.875 | ||
| ✔ | ✔ | 0.978 | 0.877 | 0.964 | 0.867 | |||
| ✔ | ✔ | ✔ | 0.967 | 0.823 | 0.954 | 0.83 | ||
| ✔ | ✔ | 0.972 | 0.84 | 0.957 | 0.838 | |||
| ✔ | ✔ | 0.976 | 0.862 | 0.957 | 0.838 | |||
| ✔ | 0.972 | 0.845 | 0.956 | 0.835 | ||||
| ✔ | ✔ | ✔ | 0.981 | 0.892 | 0.964 | 0.866 | ||
| ✔ | ✔ | 0.981 | 0.893 | 0.966 | 0.875 | |||
| ✔ | ✔ | 0.981 | 0.895 | 0.966 | 0.875 | |||
| ✔ | 0.981 | 0.891 | 0.966 | 0.875 | ||||
| ✔ | ✔ | 0.969 | 0.831 | 0.96 | 0.852 | |||
| ✔ | 0.971 | 0.841 | 0.958 | 0.844 | ||||
| ✔ | 0.973 | 0.851 | 0.96 | 0.85 | ||||
| 0.972 | 0.842 | 0.957 | 0.837 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Chen, J.; Gong, C.; Wang, T.D.; Seibel, E.J. Deep-Learning-Based Real-Time and Automatic Target-to-Background Ratio Calculation in Fluorescence Endoscopy for Cancer Detection and Localization. Diagnostics 2022, 12, 2031. https://doi.org/10.3390/diagnostics12092031
Jiang Y, Chen J, Gong C, Wang TD, Seibel EJ. Deep-Learning-Based Real-Time and Automatic Target-to-Background Ratio Calculation in Fluorescence Endoscopy for Cancer Detection and Localization. Diagnostics. 2022; 12(9):2031. https://doi.org/10.3390/diagnostics12092031
Chicago/Turabian StyleJiang, Yang, Jing Chen, Chen Gong, Thomas D. Wang, and Eric J. Seibel. 2022. "Deep-Learning-Based Real-Time and Automatic Target-to-Background Ratio Calculation in Fluorescence Endoscopy for Cancer Detection and Localization" Diagnostics 12, no. 9: 2031. https://doi.org/10.3390/diagnostics12092031






