Automatic Segmentation and Quantitative Assessment of Stroke Lesions on MR Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Manual Tracing
2.3. Preprocessing and Dataset Partitioning
2.4. Lesion Characteristics
2.5. Model Architecture
2.6. Model Training
2.7. Model Performance Evaluation
2.8. Statistical Analysis
3. Results
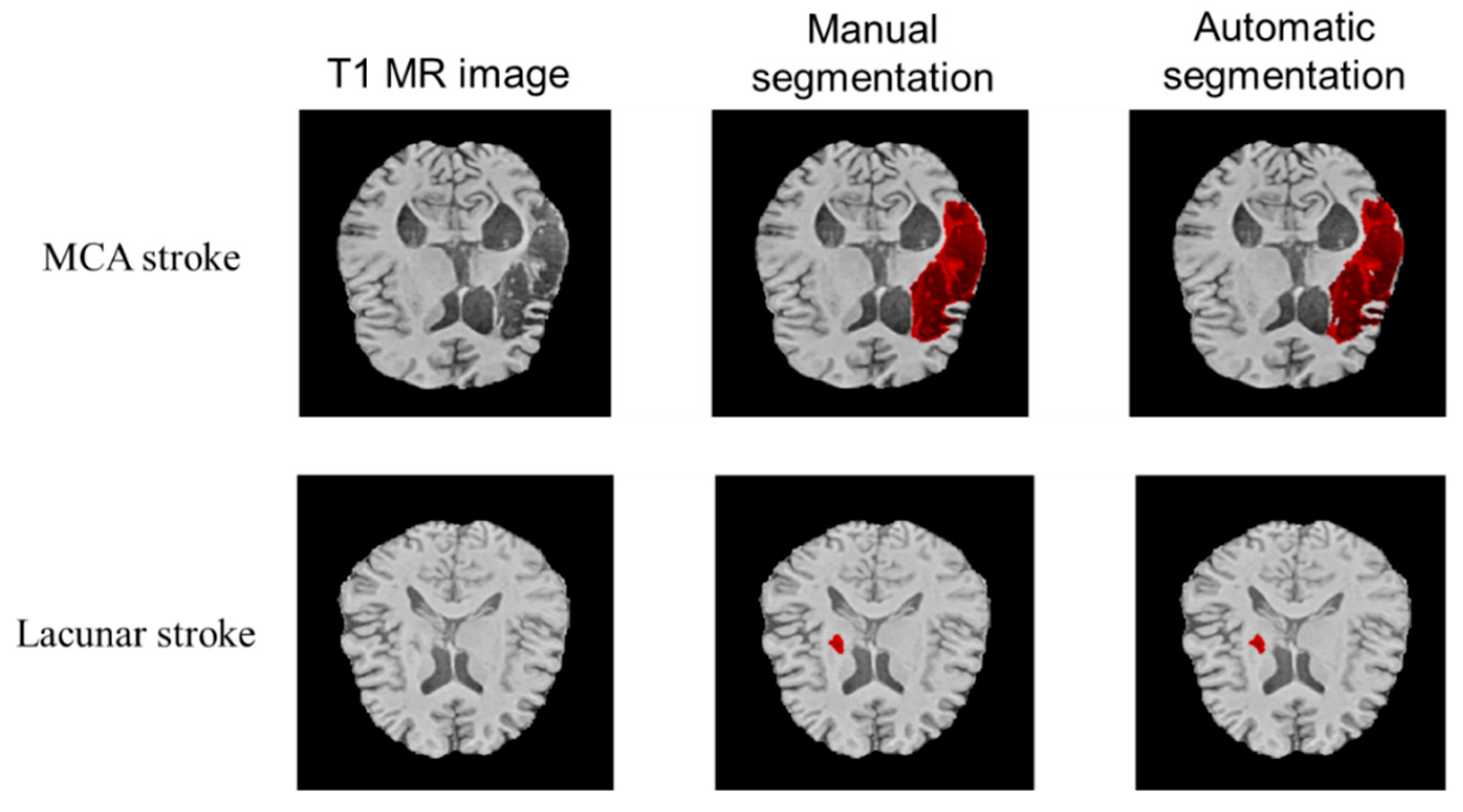
3.1. Qualitative Performance
3.2. Quantitative Performance
4. Discussion
4.1. Regularization Methods to Overcome Overfitting
4.2. A Combination Approach for Quantitative Analysis
4.3. Limitations
4.4. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adolphs, R. Human Lesion Studies in the 21st Century. Neuron 2016, 90, 1151–1153. [Google Scholar] [CrossRef] [PubMed]
- Liew, S.-L.; Lo, B.P.; Donnelly, M.R.; Zavaliangos-Petropulu, A.; Jeong, J.N.; Barisano, G.; Hutton, A.; Simon, J.P.; Juliano, J.M.; Suri, A.; et al. A Large, Curated, Open-Source Stroke Neuroimaging Dataset to Improve Lesion Segmentation Algorithms. Sci. Data 2022, 9, 320. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shapiro, L.G. Image Segmentation Techniques. Comput. Vis. Graph. Image Process. 1985, 29, 100–132. [Google Scholar] [CrossRef]
- Nazari-Farsani, S.; Nyman, M.; Karjalainen, T.; Bucci, M.; Isojärvi, J.; Nummenmaa, L. Automated Segmentation of Acute Stroke Lesions Using a Data-Driven Anomaly Detection on Diffusion Weighted MRI. J. Neurosci. Methods 2020, 333, 108575. [Google Scholar] [CrossRef]
- Chen, L.; Bentley, P.; Rueckert, D. Fully Automatic Acute Ischemic Lesion Segmentation in DWI Using Convolutional Neural Networks. NeuroImage Clin. 2017, 15, 633–643. [Google Scholar] [CrossRef]
- Clèrigues, A.; Valverde, S.; Bernal, J.; Freixenet, J.; Oliver, A.; Lladó, X. Acute Ischemic Stroke Lesion Core Segmentation in CT Perfusion Images Using Fully Convolutional Neural Networks. Comput. Biol. Med. 2019, 115, 103487. [Google Scholar] [CrossRef]
- Tomita, N.; Jiang, S.; Maeder, M.E.; Hassanpour, S. Automatic Post-Stroke Lesion Segmentation on MR Images Using 3D Residual Convolutional Neural Network. NeuroImage Clin. 2020, 27, 102276. [Google Scholar] [CrossRef]
- Schellinger, P.D.; Fiebach, J.B.; Hacke, W. Imaging-Based Decision Making in Thrombolytic Therapy for Ischemic Stroke. Stroke 2003, 34, 575–583. [Google Scholar] [CrossRef]
- Liew, S.L.; Anglin, J.M.; Banks, N.W.; Sondag, M.; Ito, K.L.; Kim, H.; Chan, J.; Ito, J.; Jung, C.; Khoshab, N.; et al. A Large, Open Source Dataset of Stroke Anatomical Brain Images and Manual Lesion Segmentations. Sci. Data 2018, 5, 180011. [Google Scholar] [CrossRef]
- Liew, S.-L.; Zavaliangos-Petropulu, A.; Jahanshad, N.; Lang, C.E.; Hayward, K.S.; Lohse, K.R.; Juliano, J.M.; Assogna, F.; Baugh, L.A.; Bhattacharya, A.K.; et al. The ENIGMA Stroke Recovery Working Group: Big Data Neuroimaging to Study Brain–Behavior Relationships after Stroke. Hum. Brain Mapp. 2022, 43, 129–148. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional Neural Networks: An Overview and Application in Radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Paing, M.P.; Tungjitkusolmun, S.; Bui, T.H.; Visitsattapongse, S.; Pintavirooj, C. Automated Segmentation of Infarct Lesions in T1-Weighted MRI Scans Using Variational Mode Decomposition and Deep Learning. Sensors 2021, 21, 1952. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Farhat, F.G.; Boukrina, O.; Barrett, A.M.; Binder, J.R.; Roshan, U.W.; Graves, W.W. A Multi-Path 2.5 Dimensional Convolutional Neural Network System for Segmenting Stroke Lesions in Brain MRI Images. NeuroImage Clin. 2020, 25, 102118. [Google Scholar] [CrossRef]
- Chen, X.; You, S.; Tezcan, K.C.; Konukoglu, E. Unsupervised Lesion Detection via Image Restoration with a Normative Prior. Med. Image Anal. 2020, 64, 101713. [Google Scholar] [CrossRef]
- Hui, H.; Zhang, X.; Li, F.; Mei, X.; Guo, Y. A Partitioning-Stacking Prediction Fusion Network Based on an Improved Attention U-Net for Stroke Lesion Segmentation. IEEE Access 2020, 8, 47419–47432. [Google Scholar] [CrossRef]
- Liu, X.; Yang, H.; Qi, K.; Dong, P.; Liu, Q.; Liu, X.; Wang, R.; Wang, S. MSDF-Net: Multi-Scale Deep Fusion Network for Stroke Lesion Segmentation. IEEE Access 2019, 7, 178486–178495. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Liu, Y.; Chen, Y.; Wu, E.X.; Tang, X. MI-UNet: Multi-Inputs UNet Incorporating Brain Parcellation for Stroke Lesion Segmentation from T1-Weighted Magnetic Resonance Images. IEEE J. Biomed. Health Inform. 2021, 25, 526–535. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. Lect. Notes Comput. Sci. 2016, 9901, 424–432. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, W.; Dong, P.; Xia, Y.; Wang, S. D-UNet: A Dimension-Fusion U Shape Network for Chronic Stroke Lesion Segmentation. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 940–950. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Gerig, G. ITK-SNAP: An Intractive Medical Image Segmentation Tool to Meet the Need for Expert-Guided Segmentation of Complex Medical Images. IEEE Pulse 2017, 8, 54–57. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Schell, M.; Pflueger, I.; Brugnara, G.; Bonekamp, D.; Neuberger, U.; Wick, A.; Schlemmer, H.-P.; Heiland, S.; Wick, W.; et al. Automated Brain Extraction of Multisequence MRI Using Artificial Neural Networks. Hum. Brain Mapp. 2019, 40, 4952–4964. [Google Scholar] [CrossRef] [PubMed]
- Verma, K.; Kumar, S. Automatic Segmentation of Stroke Lesions on MR Images for Voxel-Based Lesion-Symptom Mapping. In Proceedings of the Annual Meeting of the Society for Neuroscience, Society for Neuroscience, Virtual, 8 November 2021. [Google Scholar]
- DiCarlo, J.J.; Zoccolan, D.; Rust, N.C. How Does the Brain Solve Visual Object Recognition? Neuron 2012, 73, 415–434. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Yeghiazaryan, V.; Voiculescu, I. Family of Boundary Overlap Metrics for the Evaluation of Medical Image Segmentation. J. Med. Imaging 2018, 5, 15006. [Google Scholar] [CrossRef]
- Zhang, Y.J. A Survey on Evaluation Methods for Image Segmentation. Pattern Recognit. 1996, 29, 1335–1346. [Google Scholar] [CrossRef]
- Rorden, C.; Karnath, H.O. Using Human Brain Lesions to Infer Function: A Relic from a Past Era in the FMRI Age? Nat. Rev. Neurosci. 2004, 5, 812–819. [Google Scholar] [CrossRef]
- Kimberg, D.Y.; Coslett, H.B.; Schwartz, M.F. Power in Voxel-Based Lesion-Symptom Mapping. J. Cogn. Neurosci. 2007, 19, 1067–1080. [Google Scholar] [CrossRef]
- Poldrack, R.A.; Huckins, G.; Varoquaux, G. Establishment of Best Practices for Evidence for Prediction: A Review. JAMA Psychiatry 2020, 77, 534–540. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Crum, W.R.; Camara, O.; Hill, D.L.G. Generalized Overlap Measures for Evaluation and Validation in Medical Image Analysis. IEEE Trans. Med. Imaging 2006, 25, 1451–1461. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.L.; Kim, H.; Liew, S.-L. A Comparison of Automated Lesion Segmentation Approaches for Chronic Stroke T1-Weighted MRI Data. Hum. Brain Mapp. 2019, 40, 4669–4685. [Google Scholar] [CrossRef] [PubMed]
- Mah, Y.-H.; Nachev, P.; MacKinnon, A.D. Quantifying the Impact of Chronic Ischemic Injury on Clinical Outcomes in Acute Stroke with Machine Learning. Front. Neurol. 2020, 11, 15. [Google Scholar] [CrossRef]
- Han, T.S.; Fry, C.H.; Gulli, G.; Affley, B.; Robin, J.; Irvin-Sellers, M.; Fluck, D.; Kakar, P.; Sharma, S.; Sharma, P. Prestroke Disability Predicts Adverse Poststroke Outcome. Stroke 2020, 51, 594–600. [Google Scholar] [CrossRef]
- Chao, L.L.; Knight, R.T. Contribution of Human Prefrontal Cortex to Delay Performance. J. Cogn. Neurosci. 1998, 10, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Bates, E.; Wilson, S.M.; Saygin, A.P.; Dick, F.; Sereno, M.I.; Knight, R.T.; Dronkers, N.F. Voxel-Based Lesion–Symptom Mapping. Nat. Neurosci. 2003, 6, 448–450. [Google Scholar] [CrossRef]
- Munsch, F.; Sagnier, S.; Asselineau, J.; Bigourdan, A.; Guttmann, C.R.; Debruxelles, S.; Poli, M.; Renou, P.; Perez, P.; Dousset, V. Stroke Location Is an Independent Predictor of Cognitive Outcome. Stroke 2016, 47, 66–73. [Google Scholar] [CrossRef]
- Biesbroek, J.M.; van Zandvoort, M.J.E.; Kappelle, L.J.; Velthuis, B.K.; Biessels, G.J.; Postma, A. Shared and Distinct Anatomical Correlates of Semantic and Phonemic Fluency Revealed by Lesion-Symptom Mapping in Patients with Ischemic Stroke. Brain Struct. Funct. 2016, 221, 2123–2134. [Google Scholar] [CrossRef]
- Biesbroek, J.M.; van Zandvoort, M.J.E.; Kappelle, L.J.; Schoo, L.; Kuijf, H.J.; Velthuis, B.K.; Biessels, G.J.; Postma, A.; Group, U.V.C.I. study Distinct Anatomical Correlates of Discriminability and Criterion Setting in Verbal Recognition Memory Revealed by Lesion-symptom Mapping. Hum. Brain Mapp. 2015, 36, 1292–1303. [Google Scholar] [CrossRef]
- Kalénine, S.; Shapiro, A.D.; Buxbaum, L.J. Dissociations of Action Means and Outcome Processing in Left-Hemisphere Stroke. Neuropsychologia 2013, 51, 1224–1233. [Google Scholar] [CrossRef] [Green Version]
- Jwa, A.S.; Poldrack, R.A. The Spectrum of Data Sharing Policies in Neuroimaging Data Repositories. Hum. Brain Mapp. 2022, 43, 2707–2721. [Google Scholar] [CrossRef] [PubMed]
- Alter, G.; Gonzalez, R. Responsible Practices for Data Sharing. Am. Psychol. 2018, 73, 146–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Subjects (n = 655) | Location |
|---|---|
| Cortical lesions | Left 12%; Right 13.5% |
| Subcortical lesions | Left 30.2%; Right 29.4% |
| Other lesions | 14.8% |
| Performance Metric | Mean (Bootstrap 95% CI) |
|---|---|
| Sørensen–Dice coefficient | 0.65 (0.61–0.67), median: 0.73 |
| Average symmetric surface distance (ASSD) on MNI152 template (in mm) | 12.04 (8.44–19.53), median: 2.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, K.; Kumar, S.; Paydarfar, D. Automatic Segmentation and Quantitative Assessment of Stroke Lesions on MR Images. Diagnostics 2022, 12, 2055. https://doi.org/10.3390/diagnostics12092055
Verma K, Kumar S, Paydarfar D. Automatic Segmentation and Quantitative Assessment of Stroke Lesions on MR Images. Diagnostics. 2022; 12(9):2055. https://doi.org/10.3390/diagnostics12092055
Chicago/Turabian StyleVerma, Khushboo, Satwant Kumar, and David Paydarfar. 2022. "Automatic Segmentation and Quantitative Assessment of Stroke Lesions on MR Images" Diagnostics 12, no. 9: 2055. https://doi.org/10.3390/diagnostics12092055
APA StyleVerma, K., Kumar, S., & Paydarfar, D. (2022). Automatic Segmentation and Quantitative Assessment of Stroke Lesions on MR Images. Diagnostics, 12(9), 2055. https://doi.org/10.3390/diagnostics12092055







