Phantom Study on the Robustness of MR Radiomics Features: Comparing the Applicability of 3D Printed and Biological Phantoms
Abstract
:1. Introduction
2. Methods
2.1. Biological Phantoms
2.2. 3D Printed Phantoms
2.3. MR Scanning
2.4. Image Visualization and Segmentation
2.5. Normalization and Discretization
2.6. Texture Calculation
3. Statistical Analysis
3.1. Coefficient of Variation
3.2. Relative Parameter Differences
3.3. Interclass Correlation Coefficient
3.4. QR Code Readability Test
4. Results
4.1. Visual Comparison
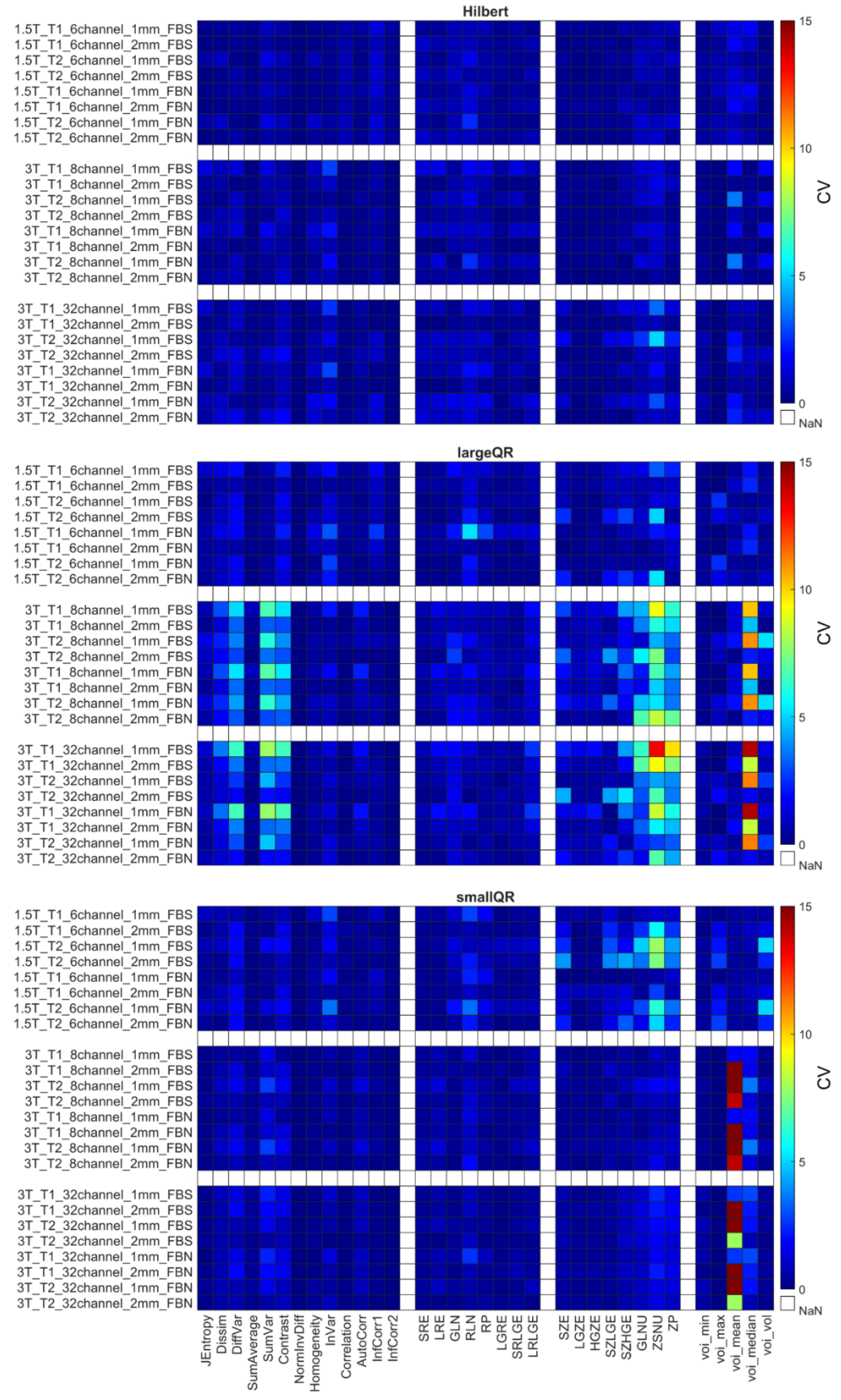
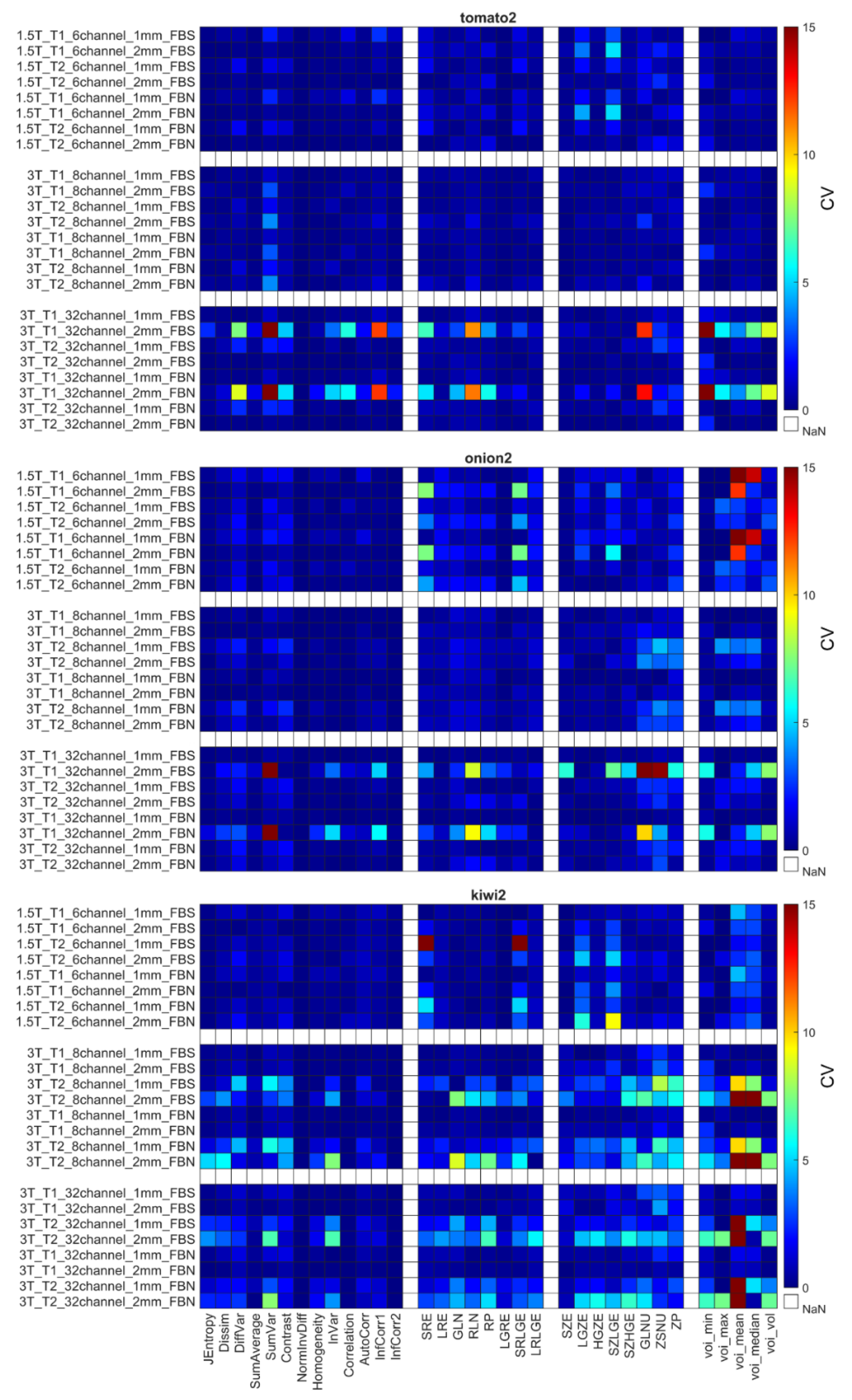
4.2. Coefficient of Variation
4.3. Relative Difference
4.4. Interclass Correlation Coefficient
4.5. QR Code Readability Test
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Schick, U.; Lucia, F.; Bourbonne, V.; Dissaux, G.; Pradier, O.; Jaouen, V.; Tixier, F.; Visvikis, D.; Hatt, M. Use of radiomics in the radiation oncology setting: Where do we stand and what do we need? Cancer/Radiotherapie 2020, 24, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Bibault, J.E.; Xing, L.; Giraud, P.; El Ayachy, R.; Giraud, N.; Decazes, P.; Burgun, A. Radiomics: A primer for the radiation oncologist. Cancer/Radiotherapie 2020, 24, 403–410. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; De Jong, E.E.C.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Veres, G.; Vas, N.F.; Lassen, M.L.; Béresová, M.; Krizsan, A.K.; Forgács, A.; Berényi, E.; Balkay, L. Effect of grey-level discretization on texture feature on different weighted MRI images of diverse disease groups. PLoS ONE 2021, 16, e0253419. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Szomolanyi, P.; Jirak, D.; Materka, A.; Trattnig, S. Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: An application-oriented study. Med. Phys. 2009, 36, 1236–1243. [Google Scholar] [CrossRef]
- Mi, H.; Yuan, M.; Suo, S.; Cheng, J.; Li, S.; Duan, S.; Lu, Q. Impact of different scanners and acquisition parameters on robustness of MR radiomics features based on women’s cervix. Sci. Rep. 2020, 10, 20407. [Google Scholar] [CrossRef] [PubMed]
- Ammari, S.; Pitre-Champagnat, S.; Dercle, L.; Chouzenoux, E.; Moalla, S.; Reuze, S.; Talbot, H.; Mokoyoko, T.; Hadchiti, J.; Diffetocq, S.; et al. Influence of Magnetic Field Strength on Magnetic Resonance Imaging Radiomics Features in Brain Imaging, an In Vitro and In Vivo Study. Front. Oncol. 2021, 10, 541663. [Google Scholar] [CrossRef] [PubMed]
- Forgács, A.; Béresová, M.; Garai, I.; Lassen, M.L.; Beyer, T.; Difranco, M.D.; Berényi, E.; Balkay, L. Impact of intensity discretization on textural indices of [18F]FDG-PET tumour heterogeneity in lung cancer patients. Phys. Med. Biol. 2019, 64, 125016. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Abdalah, M.A.; Apte, A.; Ashrafinia, S.; Beukinga, J.; Bogowicz, M.; Dinh, C.V.; Götz, M.; Hatt, M.; Leijenaar, R.T.H.; et al. PO-0981: Results from the Image Biomarker Standardisation Initiative. Radiother. Oncol. 2018, 127, 258–264. [Google Scholar] [CrossRef]
- Bernatz, S.; Zhdanovich, Y.; Ackermann, J.; Koch, I.; Wild, P.J.; dos Santos, D.P.; Vogl, T.J.; Kaltenbach, B.; Rosbach, N. Impact of rescanning and repositioning on radiomic features employing a multi-object phantom in magnetic resonance imaging. Sci. Rep. 2021, 11, 14248. [Google Scholar] [CrossRef] [PubMed]
- Dreher, C.; Kuder, T.A.; König, F.; Mlynarska-Bujny, A.; Tenconi, C.; Paech, D.; Schlemmer, H.P.; Ladd, M.E.; Bickelhaupt, S. Radiomics in diffusion data: A test–retest, inter- and intra-reader DWI phantom study. Clin. Radiol. 2020, 75, 798.e13–798.e22. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.; Lim, J.; Lee, J.G.; McCarthy, M.J.; Kim, S.M. Investigation of the maturity changes of cherry tomato using magnetic resonance imaging. Appl. Sci. 2020, 10, 5188. [Google Scholar] [CrossRef]
- Kalne, A.; Kotwaliwale, N.; Singh, K.; Singh, V.K. Non-Destructive Inspection of Mango Fruit Using Digital Radiography, Computed Tomography and Magnetic Resonance Imaging. J. Agric. Eng. 2012, 49, 33–41. [Google Scholar]
- Mayerhoefer, M.E.; Szomolanyi, P.; Jirak, D.; Berg, A.; Materka, A.; Dirisamer, A.; Trattnig, S. Effects of magnetic resonance image interpolation on the results of texture-based pattern classification a phantom study. Investig. Radiol. 2009, 44, 405–411. [Google Scholar] [CrossRef]
- Wong, O.L.; Yuan, J.; Zhou, Y.; Yu, S.K.; Cheung, K.Y. Longitudinal acquisition repeatability of MRI radiomics features: An ACR MRI phantom study on two MRI scanners using a 3D T1W TSE sequence. Med. Phys. 2021, 48, 1239–1249. [Google Scholar] [CrossRef]
- Jirák, D.; Dezortová, M.; Hájek, M. Phantoms for texture analysis of MR images. Long-term and multi-center study. Med. Phys. 2004, 31, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Valladares, A.; Beyer, T.; Rausch, I. Physical imaging phantoms for simulation of tumor heterogeneity in PET, CT, and MRI: An overview of existing designs. Med. Phys. 2020, 47, 2023–2037. [Google Scholar] [CrossRef]
- Rai, R.; Holloway, L.C.; Brink, C.; Field, M.; Christiansen, R.L.; Sun, Y.; Barton, M.B.; Liney, G.P. Multicenter evaluation of MRI-based radiomic features: A phantom study. Med. Phys. 2020, 47, 3054–3063. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, L.; Botta, F.; Origgi, D.; Rizzo, S.; Mariani, M.; Summers, P.; García-Polo, P.; Cremonesi, M.; Lascialfari, A. PETER PHAN: An MRI phantom for the optimisation of radiomic studies of the female pelvis. Phys. Medica 2020, 71, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, L.; Santinha, J.; Loução, N.; Figueiredo, M.; Botta, F.; Origgi, D.; Cremonesi, M.; Cassano, E.; Papanikolaou, N.; Lascialfari, A. A multicenter study on radiomic features from T2-weighted images of a customized MR pelvic phantom setting the basis for robust radiomic models in clinics. Magn. Reson. Med. 2020, 85, 1713–1726. [Google Scholar] [CrossRef] [PubMed]
- Orlhac, F.; Lecler, A.; Savatovski, J.; Goya-Outi, J.; Nioche, C.; Charbonneau, F.; Ayache, N.; Frouin, F.; Duron, L.; Buvat, I. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure. Eur. Radiol. 2021, 31, 2272–2280. [Google Scholar] [CrossRef]
- Baeßler, B.; Weiss, K.; Santos, D.P. Dos Robustness and Reproducibility of Radiomics in Magnetic Resonance Imaging: A Phantom Study. Investig. Radiol. 2019, 54, 221–228. [Google Scholar] [CrossRef]
- Cattell, R.; Chen, S.; Huang, C. Robustness of radiomic features in magnetic resonance imaging: Review and a phantom study. Vis. Comput. Ind. Biomed. Art 2019, 2, 19. [Google Scholar] [CrossRef]
- Rai, R.; Wang, Y.F.; Manton, D.; Dong, B.; Deshpande, S.; Liney, G.P. Development of multi-purpose 3D printed phantoms for MRI. Phys. Med. Biol. 2019, 64, 075010. [Google Scholar] [CrossRef]
- Filippou, V.; Tsoumpas, C. Recent advances on the development of phantoms using 3D printing for imaging with CT, MRI, PET, SPECT, and ultrasound. Med. Phys. 2018, 45, e740–e760. [Google Scholar] [CrossRef]
- Jin, Z.; Li, Y.; Yu, K.; Liu, L.; Fu, J.; Yao, X.; Zhang, A.; He, Y. 3D Printing of Physical Organ Models: Recent Developments and Challenges. Adv. Sci. 2021, 8, 2101394. [Google Scholar] [CrossRef]
- Aimar, A.; Palermo, A.; Innocenti, B. The Role of 3D Printing in Medical Applications: A State of the Art. J. Healthc. Eng. 2019, 2019, 5340616. [Google Scholar] [CrossRef]
- Bieniosek, M.F.; Lee, B.J.; Levin, C.S. Technical Note: Characterization of custom 3D printed multimodality imaging phantoms. Med. Phys. 2015, 42, 5913–5918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagan, H. A three-dimensional hilbert curve. Int. J. Math. Educ. Sci. Technol. 1993, 24, 541–545. [Google Scholar] [CrossRef]
- Denso Wave. QRcode.com. 2014. Available online: https://www.QRCode.com. (accessed on 1 July 2022).
- Tiwari, S. An introduction to QR code technology. In Proceedings of the 2016 15th International Conference on Information Technology, Bhubaneswar, India, 22–24 December 2016. [Google Scholar]
- Teramoto, D.; Ushioda, Y.; Sasaki, A.; Sakurai, Y.; Nagahama, H.; Nakamura, M.; Sugimori, H.; Sakata, M. [Can fruits and vegetables be used as substitute phantoms for normal human brain tissues in magnetic resonance imaging?]. Nihon Hoshasen Gijutsu Gakkai Zasshi 2013, 69, 1146–1152. [Google Scholar] [CrossRef]
- Werz, K.; Braun, H.; Vitha, D.; Bruno, G.; Martirosian, P.; Steidle, G.; Schick, F. Relaxation times T1, T2, and T2 * of apples, pears, citrus fruits, and potatoes with a comparison to human tissues. Z. Med. Phys. 2011, 21, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, R.; Pastor-Juan, M.D.R.; Canales-Vázquez, J.; Castro-García, M.; Villas, M.V.; Legorburo, F.M.; Sabater, S. Radiomics of CT features may be nonreproducible and redundant: Influence of CT acquisition parameters. Radiology 2018, 288, 407–415. [Google Scholar] [CrossRef]
- Mueller-Lisse, U.G.; Murer, S.; Mueller-Lisse, U.L.; Kuhn, M.; Scheidler, J.; Scherr, M. Everyman’s prostate phantom: Kiwi-fruit substitute for human prostates at magnetic resonance imaging, diffusion-weighted imaging and magnetic resonance spectroscopy. Eur. Radiol. 2017, 27, 3362–3371. [Google Scholar] [CrossRef] [PubMed]
- Stupic, K.F.; Ainslie, M.; Boss, M.A.; Charles, C.; Dienstfrey, A.M.; Evelhoch, J.L.; Finn, P.; Gimbutas, Z.; Gunter, J.L.; Hill, D.L.G.; et al. A standard system phantom for magnetic resonance imaging. Magn. Reson. Med. 2021, 86, 1194–1211. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Peng, H.; Liu, L.; Lu, L. 3D printed perforated QR codes. Comput. Graph. 2019, 81, 117–124. [Google Scholar] [CrossRef]
- Papp, G.; Hoffmann, M.; Papp, I. Improved Embedding of QR Codes onto Surfaces to be 3D Printed. CAD Comput. Aided Des. 2021, 131, 102961. [Google Scholar] [CrossRef]
- Alber, J.; Niedermeier, R. On multidimensional curves with hilbert property. Theory Comput. Syst. 2000, 33, 295–312. [Google Scholar] [CrossRef]
- Garcia, J.; Yang, Z.L.; Mongrain, R.; Leask, R.L.; Lachapelle, K. 3D printing materials and their use in medical education: A review of current technology and trends for the future. BMJ Simul. Technol. Enhanc. Learn. 2017, 4, 27–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mugler, J.P. Optimized three-dimensional fast-spin-echo MRI. J. Magn. Reson. Imaging 2014, 39, 745–767. [Google Scholar] [CrossRef] [PubMed]
- Bapst, B.; Amegnizin, J.L.; Vignaud, A.; Kauv, P.; Maraval, A.; Kalsoum, E.; Tuilier, T.; Benaissa, A.; Brugières, P.; Leclerc, X.; et al. Post-contrast 3D T1-weighted TSE MR sequences (SPACE, CUBE, VISTA/BRAINVIEW, isoFSE, 3D MVOX): Technical aspects and clinical applications. J. Neuroradiol. 2020, 47, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Nair, G.; Absinta, M.; Reich, D.S. Optimized T1-MPRAGE sequence for better visualization of spinal cord multiple sclerosis lesions at 3T. Am. J. Neuroradiol. 2013, 34, 2215–2222. [Google Scholar] [CrossRef] [PubMed]
- Pinter, C.; Lasso, A.; Fichtinger, G. Polymorph segmentation representation for medical image computing. Comput. Methods Programs Biomed. 2019, 171, 19–26. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Parmar, C.; Velazquez, E.R.; Leijenaar, R.; Jermoumi, M.; Carvalho, S.; Mak, R.H.; Mitra, S.; Shankar, B.U.; Kikinis, R.; Haibe-Kains, B.; et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS ONE 2014, 9, e102107. [Google Scholar] [CrossRef]
- NIfTI: Neuroimaging Informatics Technology Initiative. 2012. Available online: https://nifti.nimh.nih.gov. (accessed on 1 July 2022).
- Samuel, S.; Moore, M.; Sheridan, H.; Sorensen, C.; Patterson, B. Touring a Data Curation Network Primer: A Focus on Neuroimaging Data. J. eScience Libr. 2021, 10, 5. [Google Scholar] [CrossRef]
- Collewet, G.; Strzelecki, M.; Mariette, F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn. Reson. Imaging 2004, 22, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Hatt, M.; Vallieres, M.; Visvikis, D.; Zwanenburg, A. IBSI: An international community radiomics standardization initiative. J. Nucl. Med. 2018, 59, 287. [Google Scholar]
- Depeursinge, A.; Andrearczyk, V.; Whybra, P.; van Griethuysen, J.; Müller, H.; Schaer, R.; Vallières, M.; Zwanenburg, A. Standardised convolutional filtering for radiomics: Image biomarker standardisation initiative (IBSI). arXiv 2020, arXiv:2006.05470v5. [Google Scholar]
- Lacroix, M.; Frouin, F.; Dirand, A.S.; Nioche, C.; Orlhac, F.; Bernaudin, J.F.; Brillet, P.Y.; Buvat, I. Correction for Magnetic Field Inhomogeneities and Normalization of Voxel Values Are Needed to Better Reveal the Potential of MR Radiomic Features in Lung Cancer. Front. Oncol. 2020, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhuang, Q.; Xiao, Y.; Wu, G.; Shi, Z.; Chen, L.; Wang, Y.; Yu, J. MIL normalization —— prerequisites for accurate MRI radiomics analysis. Comput. Biol. Med. 2021, 133, 104403. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Leger, S.; Vallières, M.; Löck, S. Radiomics Digital Phantom. CancerData 2016, 41, 366–373. [Google Scholar]
- Orlhac, F.; Soussan, M.; Maisonobe, J.-A.; Garcia, C.A.; Vanderlinden, B.; Buvat, I. Tumor Texture Analysis in 18F-FDG PET: Relationships Between Texture Parameters, Histogram Indices, Standardized Uptake Values, Metabolic Volumes, and Total Lesion Glycolysis. J. Nucl. Med. 2014, 55, 414–422. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Schwier, M.; van Griethuysen, J.; Vangel, M.G.; Pieper, S.; Peled, S.; Tempany, C.; Aerts, H.J.W.L.; Kikinis, R.; Fennessy, F.M.; Fedorov, A. Repeatability of Multiparametric Prostate MRI Radiomics Features. Sci. Rep. 2019, 9, 9441. [Google Scholar] [CrossRef]
- Carbonell, G.; Kennedy, P.; Bane, O.; Kirmani, A.; El Homsi, M.; Stocker, D.; Said, D.; Mukherjee, P.; Gevaert, O.; Lewis, S.; et al. Precision of MRI radiomics features in the liver and hepatocellular carcinoma. Eur. Radiol. 2021, 32, 2030–2040. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Scalco, E.; Belfatto, A.; Mastropietro, A.; Rancati, T.; Avuzzi, B.; Messina, A.; Valdagni, R.; Rizzo, G. T2w-MRI signal normalization affects radiomics features reproducibility. Med. Phys. 2020, 47, 1680–1691. [Google Scholar] [CrossRef]
- Ta, D.; Khan, M.; Ishaque, A.; Seres, P.; Eurich, D.; Yang, Y.H.; Kalra, S. Reliability of 3D texture analysis: A multicenter MRI study of the brain. J. Magn. Reson. Imaging 2020, 51, 1200–1209. [Google Scholar] [CrossRef] [PubMed]
- Moradmand, H.; Aghamiri, S.M.R.; Ghaderi, R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J. Appl. Clin. Med. Phys. 2020, 21, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Fiset, S.; Welch, M.L.; Weiss, J.; Pintilie, M.; Conway, J.L.; Milosevic, M.; Fyles, A.; Traverso, A.; Jaffray, D.; Metser, U.; et al. Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother. Oncol. 2019, 135, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Xing, B.; Zhang, B.; Yang, H. Optimizing QR code readability for curved agro-food packages using response surface methodology to improve mobile phone-based traceability. Food Packag. Shelf Life 2021, 28, 100638. [Google Scholar] [CrossRef]
- Tarjan, L.; Šenk, I.; Tegeltija, S.; Stankovski, S.; Ostojic, G. A readability analysis for QR code application in a traceability system. Comput. Electron. Agric. 2014, 109, 1–11. [Google Scholar] [CrossRef]
- Chen, R.; Zheng, Z.; Zhao, H.; Ren, J.; Tan, H. Fast Blind Restoration of QR Code Images Based on Blurred Imaging Mechanism. Guangzi Xuebao/Acta Photonica Sin. 2021, 50, 91. [Google Scholar] [CrossRef]
- Kim, Y.; Cho, K.-O.; Han, J.; Cho, M. Three-dimensional QR Code Using Integral Imaging. J. Korea Inst. Inf. Commun. Eng. 2016, 20, 2363–2369. [Google Scholar] [CrossRef]
- Calabrese, A.; Santucci, D.; Landi, R.; Beomonte Zobel, B.; Faiella, E.; de Felice, C. Radiomics MRI for lymph node status prediction in breast cancer patients: The state of art. J. Cancer Res. Clin. Oncol. 2021, 147, 1587–1597. [Google Scholar] [CrossRef]
- Hassani, C.; Saremi, F.; Varghese, B.A.; Duddalwar, V. Myocardial radiomics in cardiac MRI. Am. J. Roentgenol. 2020, 214, 536–545. [Google Scholar] [CrossRef]
- Lohmann, P.; Kocher, M.; Ruge, M.I.; Visser-Vandewalle, V.; Shah, N.J.; Fink, G.R.; Langen, K.J.; Galldiks, N. PET/MRI Radiomics in Patients With Brain Metastases. Front. Neurol. 2020, 11, 1. [Google Scholar] [CrossRef]
- Sun, Y.; Reynolds, H.M.; Parameswaran, B.; Wraith, D.; Finnegan, M.E.; Williams, S.; Haworth, A. Multiparametric MRI and radiomics in prostate cancer: A review. Australas. Phys. Eng. Sci. Med. 2019, 42, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Cutaia, G.; la Tona, G.; Comelli, A.; Vernuccio, F.; Agnello, F.; Gagliardo, C.; Salvaggio, L.; Quartuccio, N.; Sturiale, L.; Stefano, A.; et al. Radiomics and prostate MRI: Current role and future applications. J. Imaging 2021, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.M.; Wang, H.T.; Yu, T. The application of radiomics in breast MRI: A review. Technol. Cancer Res. Treat. 2020, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Schick, U.; Lucia, F.; Dissaux, G.; Visvikis, D.; Badic, B.; Masson, I.; Pradier, O.; Bourbonne, V.; Hatt, M. MRI-derived radiomics: Methodology and clinical applications in the field of pelvic oncology. Br. J. Radiol. 2019, 92, 20190105. [Google Scholar] [CrossRef] [PubMed]
- Caruso, D.; Polici, M.; Zerunian, M.; Pucciarelli, F.; Guido, G.; Polidori, T.; Landolfi, F.; Nicolai, M.; Lucertini, E.; Tarallo, M.; et al. Radiomics in oncology, part 1: Technical principles and gastrointestinal application in ct and mri. Cancers 2021, 13, 2522. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, M.; Barat, M.; Dohan, A.; Gaujoux, S.; Coriat, R.; Hoeffel, C.; Cassinotto, C.; Chassagnon, G.; Soyer, P. CT and MRI of pancreatic tumors: An update in the era of radiomics. Jpn. J. Radiol. 2020, 38, 1111–1124. [Google Scholar] [CrossRef]
- Feng, Q.; Ding, Z. MRI Radiomics Classification and Prediction in Alzheimer’s Disease and Mild Cognitive Impairment: A Review. Curr. Alzheimer Res. 2020, 17, 297–309. [Google Scholar] [CrossRef]
- Cusumano, D.; Meijer, G.; Lenkowicz, J.; Chiloiro, G.; Boldrini, L.; Masciocchi, C.; Dinapoli, N.; Gatta, R.; Casà, C.; Damiani, A.; et al. A field strength independent MR radiomics model to predict pathological complete response in locally advanced rectal cancer. Radiol. Medica 2021, 126, 421–429. [Google Scholar] [CrossRef]
- Doran, S.J.; Kumar, S.; Orton, M.; d’Arcy, J.; Kwaks, F.; O’Flynn, E.; Ahmed, Z.; Downey, K.; Dowsett, M.; Turner, N.; et al. “Real-world” radiomics from multi-vendor MRI: An original retrospective study on the prediction of nodal status and disease survival in breast cancer, as an exemplar to promote discussion of the wider issues. Cancer Imaging 2021, 21. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; He, X.; Li, Y.; Pang, P.; Shu, Z.; Gong, X. The Nomogram of MRI-based Radiomics with Complementary Visual Features by Machine Learning Improves Stratification of Glioblastoma Patients: A Multicenter Study. J. Magn. Reson. Imaging 2021, 54, 37. [Google Scholar] [CrossRef]
- Alvarez-Jimenez, C.; Antunes, J.T.; Talasila, N.; Bera, K.; Brady, J.T.; Gollamudi, J.; Marderstein, E.; Kalady, M.F.; Purysko, A.; Willis, J.E.; et al. Radiomic texture and shape descriptors of the rectal environment on post-chemoradiation T2-weighted MRI are associated with pathologic tumor stage regression in rectal cancers: A retrospective, multi-institution study. Cancers 2020, 12, 2027. [Google Scholar] [CrossRef]
- Masokano, I.B.; Liu, W.; Xie, S.; Marcellin, D.F.H.; Pei, Y.; Li, W. The application of texture quantification in hepatocellular carcinoma using CT and MRI: A review of perspectives and challenges. Cancer Imaging 2020, 20, 67. [Google Scholar] [CrossRef] [PubMed]
- Leandrou, S.; Lamnisos, D.; Kyriacou, P.A.; Constanti, S.; Pattichis, C.S. Comparison of 1.5 T and 3 T MRI hippocampus texture features in the assessment of Alzheimer’s disease. Biomed. Signal Process. Control 2020, 62, 102098. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Talluri, S.; Beebi, S.K.; Rajesh Kumar, B. Magnetic Resonance Imaging for Quality Evaluation of Fruits: A Review. Food Anal. Methods 2018, 11, 2943–2960. [Google Scholar] [CrossRef]
- Lee, J.; Steinmann, A.; Ding, Y.; Lee, H.; Owens, C.; Wang, J.; Yang, J.; Followill, D.; Ger, R.; MacKin, D.; et al. Radiomics feature robustness as measured using an MRI phantom. Sci. Rep. 2021, 11, 3973. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef]
- Haralick, R.M.; Dinstein, I.; Shanmugam, K. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Tian, Z.; Chen, C.; Fan, Y.; Ou, X.; Wang, J.; Ma, X.; Xu, J. Glioblastoma and Anaplastic Astrocytoma: Differentiation Using MRI Texture Analysis. Front. Oncol. 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Zhang, B.; Tian, J.; Dong, D.; Gu, D.; Dong, Y.; Zhang, L.; Lian, Z.; Liu, J.; Luo, X.; Pei, S.; et al. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin. Cancer Res. 2017, 23, 4259–4269. [Google Scholar] [CrossRef] [PubMed]
- Loizou, C.P.; Pantziaris, M.; Seimenis, I.; Pattichis, C.S. Brain MR image normalization in texture analysis of multiple sclerosis. In Proceedings of the the Final Program and Abstract Book—9th International Conference on Information Technology and Applications in Biomedicine, Larnaka, Cyprus, 4–7 November 2009. [Google Scholar]
- Sun, X.; Shi, L.; Luo, Y.; Yang, W.; Li, H.; Liang, P.; Li, K.; Mok, V.C.T.; Chu, W.C.W.; Wang, D. Histogram-based normalization technique on human brain magnetic resonance images from different acquisitions. Biomed. Eng. Online 2015, 14, 73. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, R.T.; Sweeney, E.M.; Goldsmith, J.; Shiee, N.; Mateen, F.J.; Calabresi, P.A.; Jarso, S.; Pham, D.L.; Reich, D.S.; Crainiceanu, C.M. Statistical normalization techniques for magnetic resonance imaging. NeuroImage Clin. 2014, 6, 9–19. [Google Scholar] [CrossRef]
- Cernadas, E.; Fernández-Delgado, M.; González-Rufino, E.; Carrión, P. Influence of normalization and color space to color texture classification. Pattern Recognit. 2017, 61, 120–138. [Google Scholar] [CrossRef]
- Sollini, M.; Cozzi, L.; Antunovic, L.; Chiti, A.; Kirienko, M. PET Radiomics in NSCLC: State of the art and a proposal for harmonization of methodology. Sci. Rep. 2017, 7, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Yi, Y.; Shen, J.; Shan, F.; Zhang, Z.; Yang, G.; Shi, Y. Preliminary study of 3 T-MRI native T1-mapping radiomics in differential diagnosis of non-calcified solid pulmonary nodules/masses. Cancer Cell Int. 2021, 20, 346. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Kwon, H.; Yang, J.J.; Park, M.; Cha, Y.J.; Suh, S.H.; Lee, J.M. Contrast-enhanced T1-weighted image radiomics of brain metastases may predict EGFR mutation status in primary lung cancer. Sci. Rep. 2020, 10, 8905. [Google Scholar] [CrossRef]
- Carré, A.; Klausner, G.; Edjlali, M.; Lerousseau, M.; Briend-Diop, J.; Sun, R.; Ammari, S.; Reuzé, S.; Andres, E.A.; Estienne, T.; et al. Standardization of brain MR images across machines and protocols: Bridging the gap for MRI-based radiomics. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Xiao, D.D.; Yan, P.F.; Wang, Y.X.; Osman, M.S.; Zhao, H.Y. Glioblastoma and primary central nervous system lymphoma: Preoperative differentiation by using MRI-based 3D texture analysis. Clin. Neurol. Neurosurg. 2018, 173, 1–26. [Google Scholar] [CrossRef]
- Forgacs, A.; Kallos-Balogh, P.; Nagy, F.; Krizsan, A.K.; Garai, I.; Tron, L.; Dahlbom, M.; Balkay, L. Activity painting: PET images of freely defined activity distributions applying a novel phantom technique. PLoS ONE 2019, 14, e0207658. [Google Scholar] [CrossRef]
- Jha, A.K.; Mithun, S.; Jaiswar, V.; Sherkhane, U.B.; Purandare, N.C.; Prabhash, K.; Rangarajan, V.; Dekker, A.; Wee, L.; Traverso, A. Repeatability and reproducibility study of radiomic features on a phantom and human cohort. Sci. Rep. 2021, 11, 2055. [Google Scholar] [CrossRef]
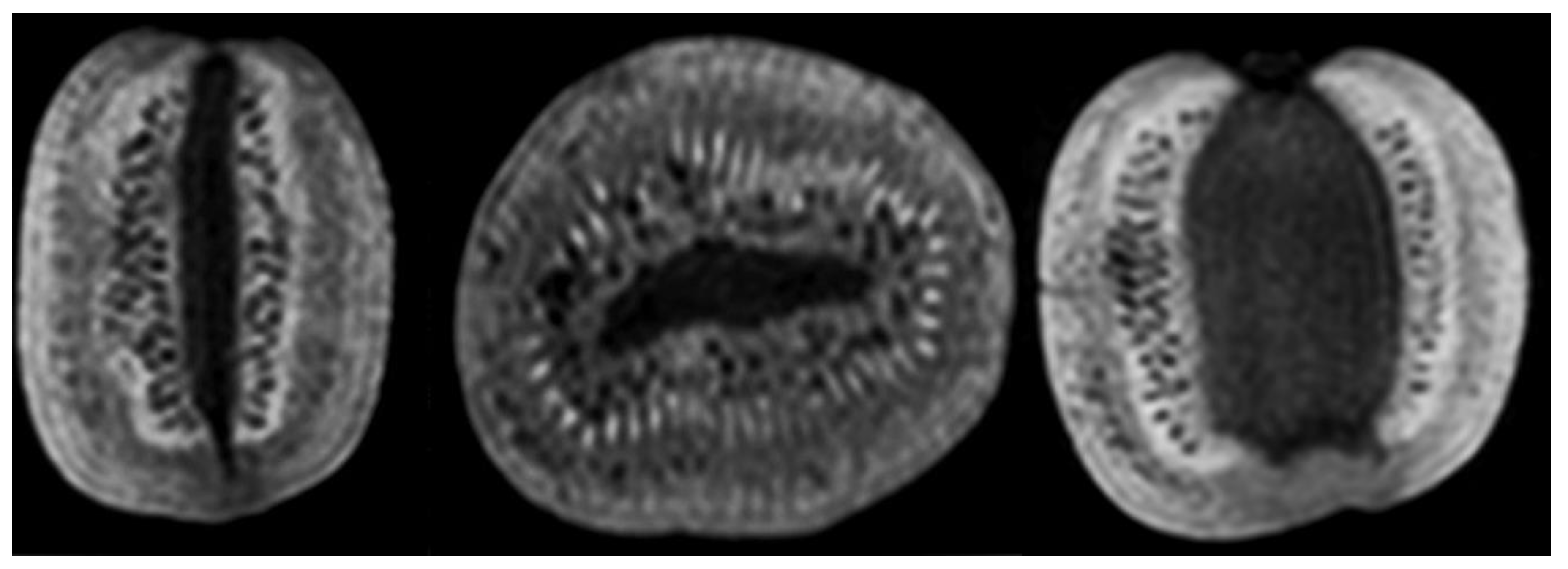






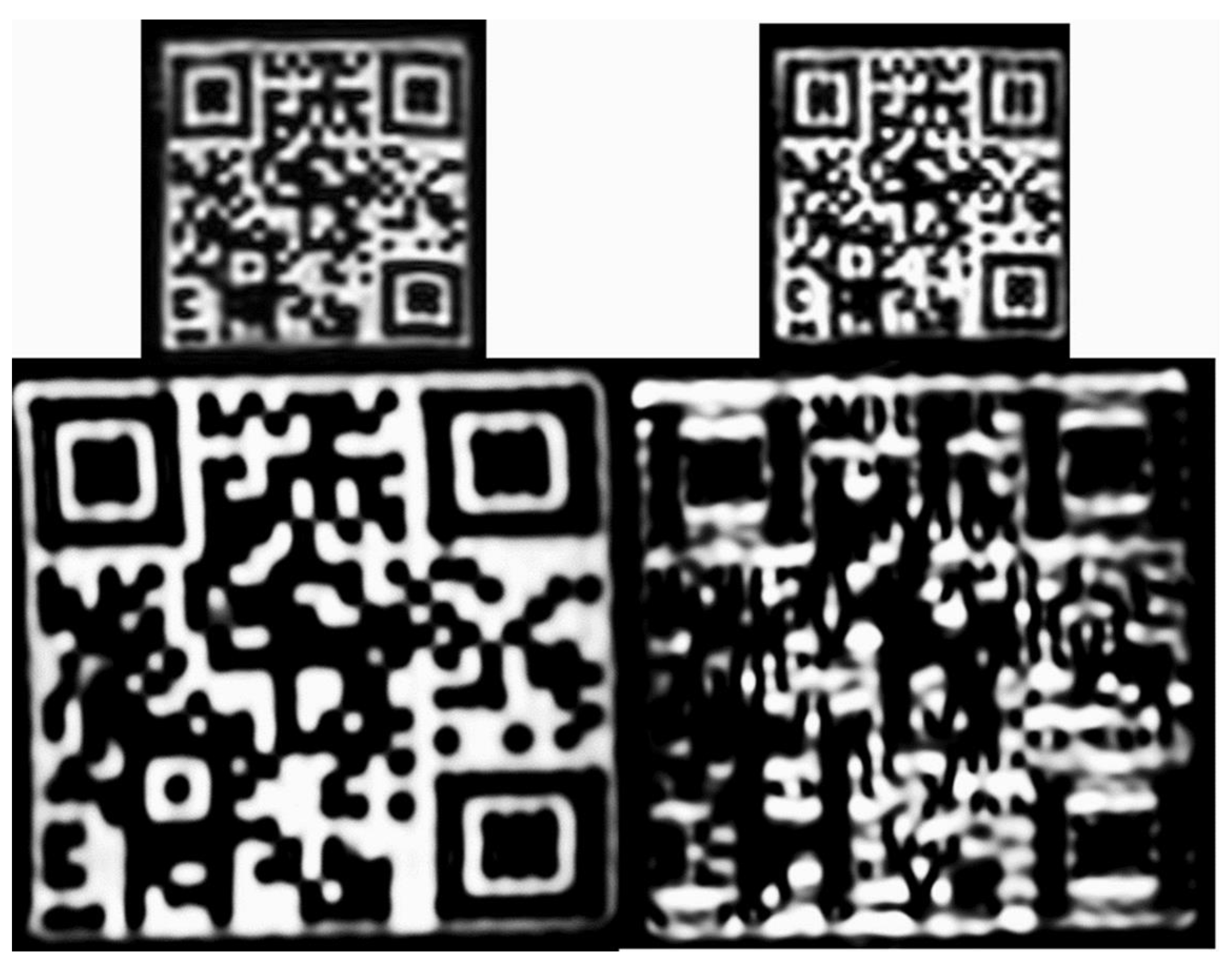
| MR | Sequences | TR (ms) | TE (ms) | NSA | Voxel Size (iso mm) |
|---|---|---|---|---|---|
| 3 Tesla | 3D T2 BrainVIEW | 2500 | 233 | 3 | 1, 2 |
| 3D T1 MPRAGE | 600 | 28.3 | 2 | 1, 2 | |
| 1.5 Tesla | 3D T2 SPACE | 1200 | 97 | 2 | 1, 2 |
| 3D T1 MPRAGE | 1040 | 4 | 2 | 1, 2 |
| # | Abbreviation | Field Strength | Weighting | Number of Channels | Voxel Size (mm3) | Discretization |
|---|---|---|---|---|---|---|
| 1 | 1.5T_T1_6ch_1mm_FBS | 1.5 T | T1 | 6 | 1 × 1 × 1 | FBS |
| 2 | 1.5T_T1_6ch_1mm_FBN | 1.5 T | T1 | 6 | 1 × 1 × 1 | FBN |
| 3 | 1.5T_T1_6ch_2mm_FBS | 1.5 T | T1 | 6 | 2 × 2 × 2 | FBS |
| 4 | 1.5T_T1_6ch_2mm_FBN | 1.5 T | T1 | 6 | 2 × 2 × 2 | FBN |
| 5 | 3T_T1_8ch_1mm_FBS | 3 T | T1 | 8 | 1 × 1 × 1 | FBS |
| 6 | 3T_T1_8ch_1mm_FBN | 3 T | T1 | 8 | 1 × 1 × 1 | FBN |
| 7 | 3T_T1_8ch_2mm_FBS | 3 T | T1 | 8 | 2 × 2 × 2 | FBS |
| 8 | 3T_T1_8ch_2mm_FBN | 3 T | T1 | 8 | 2 × 2 × 2 | FBN |
| 9 | 3T_T1_32ch_1mm_FBS | 3 T | T1 | 32 | 1 × 1 × 1 | FBS |
| 10 | 3T_T1_32ch_1mm_FBN | 3 T | T1 | 32 | 1 × 1 × 1 | FBN |
| 11 | 3T_T1_32ch_2m1m_FBS | 3 T | T1 | 32 | 2 × 2 × 2 | FBS |
| 12 | 3T_T1_32ch_2mm_FBN | 3 T | T1 | 32 | 2 × 2 × 2 | FBN |
| 13 | 1.5T_T2_6ch_1mm_FBS | 1.5 T | T2 | 6 | 1 × 1 × 1 | FBS |
| 14 | 1.5T_T2_6ch_1mm_FBN | 1.5 T | T2 | 6 | 1 × 1 × 1 | FBN |
| 15 | 1.5T_T2_6ch_2mm_FBS | 1.5 T | T2 | 6 | 2 × 2 × 2 | FBS |
| 16 | 1.5T_T2_6ch_2mm_FBN | 1.5 T | T2 | 6 | 2 × 2 × 2 | FBN |
| 17 | 3T_T2_8ch_1mm_FBS | 3 T | T2 | 8 | 1 × 1 × 1 | FBS |
| 18 | 3T_T2_8ch_1mm_FBN | 3 T | T2 | 8 | 1 × 1 × 1 | FBN |
| 19 | 3T_T2_8ch_2mm_FBS | 3 T | T2 | 8 | 2 × 2 × 2 | FBS |
| 20 | 3T_T2_8ch_2mm_FBN | 3 T | T2 | 8 | 2 × 2 × 2 | FBN |
| 21 | 3T_T2_32ch_1mm_FBS | 3 T | T2 | 32 | 1 × 1 × 1 | FBS |
| 22 | 3T_T2_32ch_1mm_FBN | 3 T | T2 | 32 | 1 × 1 × 1 | FBN |
| 23 | 3T_T2_32ch_2mm_FBS | 3 T | T2 | 32 | 2 × 2 × 2 | FBS |
| 24 | 3T_T2_32ch_2mm_FBN | 3 T | T2 | 32 | 2 × 2 × 2 | FBN |
| Abbreviation | Compared Setups | |
|---|---|---|
| Setup 1 (Row # of Table 2) | Setup 2 (Row # of Table 2) | |
| 1.5T_T1_6ch_1mm-2mm | 1 | 3 |
| 1.5T_T1_6ch-3T_T1_8ch_1mm | 1 | 5 |
| 1.5T_T1_6ch-3T_T1_8ch_2mm | 3 | 7 |
| 3T_T1_8ch_1mm-2mm | 5 | 7 |
| 3T_T1_8ch-32ch_1mm | 5 | 9 |
| 3T_T1_8ch-32ch_2mm | 7 | 11 |
| 3T_T1_32ch_1mm-2mm | 9 | 11 |
| 1.5T_T2_6ch_1mm-2mm | 13 | 15 |
| 1.5T_T2_6ch-3T_T2_8ch_1mm | 13 | 17 |
| 1.5T_T2_6ch-3T_T2_8ch_2mm | 15 | 19 |
| 3T_T2_8ch_1mm-2mm | 17 | 19 |
| 3T_T2_8ch-32ch_1mm | 17 | 21 |
| 3T_T2_8ch-32ch_2mm | 19 | 23 |
| 3T_T2_32ch_1mm-2mm | 21 | 23 |
| Object | without Normalization | with Normalization |
|---|---|---|
| Hilbert | 1.55 | 1.22 |
| smallQR | 2.75 | 2.08 |
| largeQR | 1.90 | 1.86 |
| tomato1 | 2.91 | 1.59 |
| tomato2 | 2.75 | 1.21 |
| tomato3 | 2.28 | 1.39 |
| onion1 | 2.90 | 1.03 |
| onion2 | 2.73 | 1.05 |
| onion3 | 2.77 | 1.15 |
| kiwirot | 4.82 | 4.40 |
| kiwi1 | 2.98 | 3.87 |
| kiwi2 | 3.60 | 0.46 |
| kiwi3 | 1.73 | 2.71 |
| Radiomics Parameter Group | without Normalization | with Normalization |
|---|---|---|
| GLCM | 3.08 | 2.72 |
| GLRLM | 2.90 | 2.04 |
| GLSZM | 3.87 | 2.26 |
| Histogram based | 3.09 | 6.11 |
| Acquisition Setup | Texture Parameter | Discretization Method | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Object | 1.5 T | 3 T | 1 mm | 2 mm | T1 | T2 | GLCM | GLRLM | GLSZM | Histogram Base | FBS | FBN |
| Hilbert | 0.60 | 0.75 | 0.85 | 0.55 | 0.71 | 0.69 | 0.55 | 0.64 | 1.09 | 0.64 | 0.67 | 0.73 |
| largeQR | 0.89 | 2.08 | 1.98 | 1.38 | 1.86 | 1.50 | 1.57 | 0.79 | 2.69 | 1.82 | 1.71 | 1.65 |
| smallQR | 1.08 | 2.04 | 1.21 | 2.23 | 2.17 | 1.27 | 0.66 | 0.55 | 1.30 | 7.46 | 1.76 | 1.68 |
| onion1 | 0.73 | 2.04 | 1.48 | 1.73 | 1.60 | 1.61 | 1.49 | 1.05 | 1.51 | 2.98 | 1.61 | 1.60 |
| onion2 | 1.50 | 1.38 | 1.03 | 1.80 | 1.80 | 1.03 | 1.26 | 1.17 | 1.46 | 2.22 | 1.46 | 1.38 |
| onion3 | 1.23 | 2.03 | 1.43 | 2.09 | 1.78 | 1.75 | 1.37 | 1.26 | 1.60 | 4.00 | 1.74 | 1.78 |
| tomato1 | 0.96 | 1.53 | 0.96 | 1.73 | 1.86 | 0.83 | 1.64 | 1.06 | 1.29 | 0.98 | 1.33 | 1.36 |
| tomato2 | 0.72 | 1.61 | 0.65 | 1.98 | 1.99 | 0.65 | 1.75 | 0.78 | 1.11 | 1.23 | 1.32 | 1.32 |
| tomato3 | 1.09 | 1.46 | 0.91 | 1.77 | 1.97 | 0.70 | 1.66 | 0.81 | 1.07 | 1.65 | 1.34 | 1.33 |
| kiwi1 | 3.52 | 3.66 | 2.86 | 4.37 | 1.83 | 5.40 | 1.63 | 2.81 | 3.78 | 10.59 | 3.67 | 3.55 |
| kiwi2 | 1.06 | 4.17 | 2.70 | 3.57 | 0.66 | 5.60 | 1.19 | 1.51 | 2.23 | 12.97 | 3.16 | 3.10 |
| kiwi3 | 10.72 | 2.38 | 1.43 | 8.88 | 6.99 | 3.33 | 1.10 | 1.42 | 2.06 | 28.27 | 5.15 | 5.16 |
| kiwirot | 1.15 | 2.28 | 1.43 | 2.38 | 0.69 | 3.12 | 1.23 | 1.31 | 2.04 | 4.64 | 1.91 | 1.90 |
| All included | 1.94 | 2.11 | 1.46 | 2.65 | 1.99 | 2.11 | 1.32 | 1.17 | 1.79 | 6.11 | 2.06 | 2.04 |
| # of Successful Read-Outs | |||||
|---|---|---|---|---|---|
| Read-Out Ratio | Phone 1 | Phone 2 | Phone 3 | Phone 4 | Phone 5 |
| 3T_T1_32channel_1mm_smallQR | 2/3 | - | - | - | - |
| 3T_T1_32channel_1mm_largeQR | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 |
| 3T_T2_32channel_1mm_smallQR | - | - | - | - | - |
| 3T_T2_32channel_1mm_largeQR | - | - | - | - | - |
| 3T_T1_8channel_1mm_smallQR | 2/3 | - | - | - | - |
| 3T_T1_8channel_1mm_largeQR | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 |
| 3T_T2_8channel_1mm_smallQR | - | - | - | - | - |
| 3T_T2_8channel_1mm_largeQR | - | - | - | - | - |
| 1.5T_T1_6channel_1mm_smallQR | - | - | - | - | - |
| 1.5T_T1_6channel_1mm_largeQR | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| 1.5T_T2_6channel_1mm_smallQR | - | - | - | - | - |
| 1.5T_T2_6channel_1mm_largeQR | 1/3 | - | - | 2/3 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veres, G.; Kiss, J.; Vas, N.F.; Kallos-Balogh, P.; Máthé, N.B.; Lassen, M.L.; Berényi, E.; Balkay, L. Phantom Study on the Robustness of MR Radiomics Features: Comparing the Applicability of 3D Printed and Biological Phantoms. Diagnostics 2022, 12, 2196. https://doi.org/10.3390/diagnostics12092196
Veres G, Kiss J, Vas NF, Kallos-Balogh P, Máthé NB, Lassen ML, Berényi E, Balkay L. Phantom Study on the Robustness of MR Radiomics Features: Comparing the Applicability of 3D Printed and Biological Phantoms. Diagnostics. 2022; 12(9):2196. https://doi.org/10.3390/diagnostics12092196
Chicago/Turabian StyleVeres, Gergő, János Kiss, Norman Félix Vas, Piroska Kallos-Balogh, Nóra Beatrix Máthé, Martin Lyngby Lassen, Ervin Berényi, and László Balkay. 2022. "Phantom Study on the Robustness of MR Radiomics Features: Comparing the Applicability of 3D Printed and Biological Phantoms" Diagnostics 12, no. 9: 2196. https://doi.org/10.3390/diagnostics12092196





