Endocytoscopic Observation of Esophageal Lesions: Our Own Experience and a Review of the Literature
Abstract
:1. Introduction
2. Adequacy of Magnifying Power for Observation of the Esophagus
3. Classification of Esophageal Squamous Epithelium
4. Principles of EC Observation and Appearance of Various Esophageal Lesions
5. EC Observation of Borderline Lesions (Squamous Intraepithelial Neoplasia (IN) and Squamous Dysplasia)
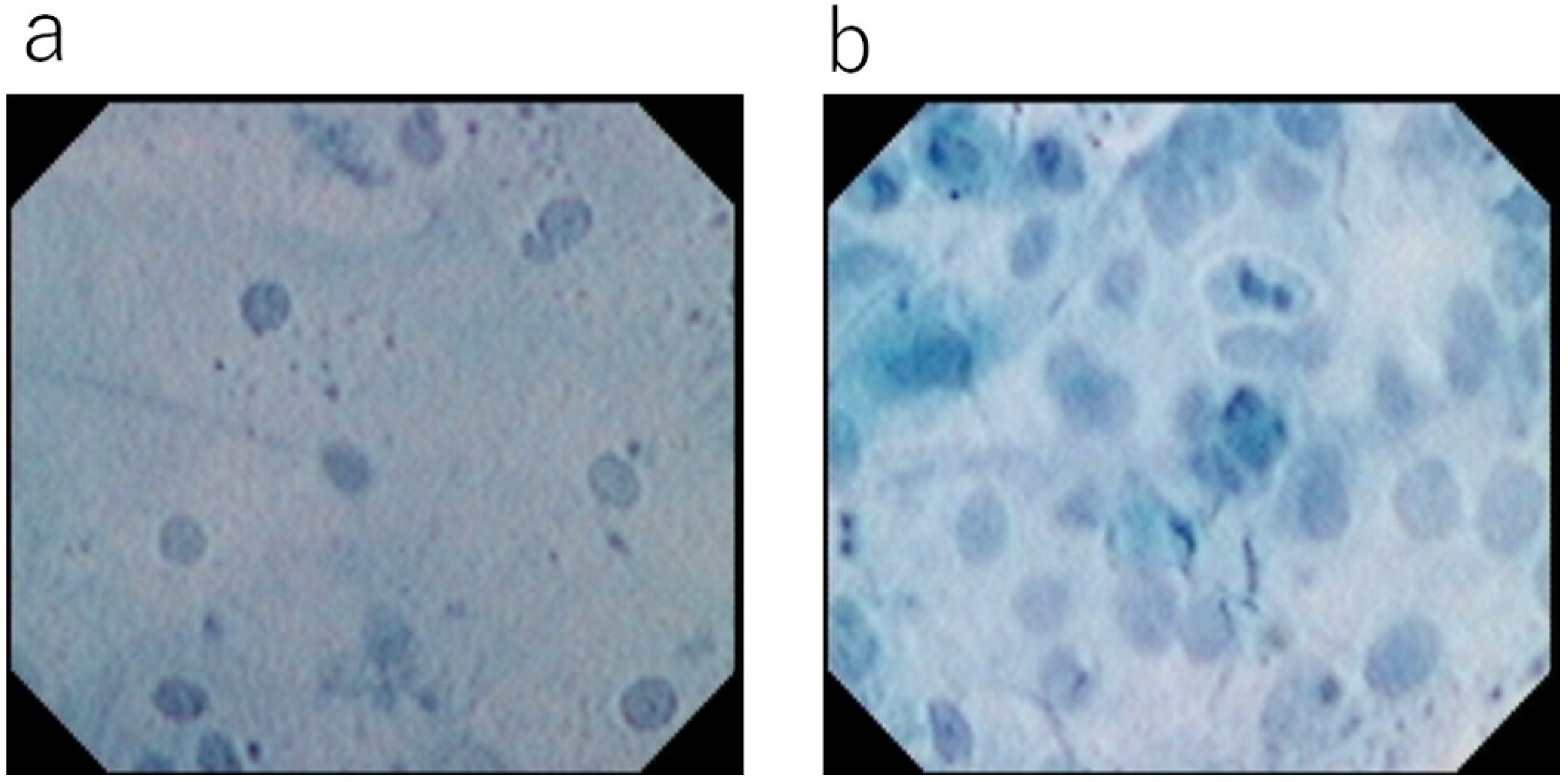
6. Vital Dye Staining of Surface Epithelial Cells
7. Merits of Toluidine Blue Single Staining
8. AI Analysis for Images of Esophageal Cancer Obtained Using EC
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumagai, Y.; Monma, K.; Kawada, K. Magnifying Chromoendoscopy of the esophagus: In-Vivo pathological diagnosis using an endocytoscopy system. Endoscopy 2004, 36, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Muto, M.; Yao, K.; Kaise, M.; Kato, M.; Uedo, N.; Yagi, K.; Tajiri, H. Magnifying endoscopy simple diagnostic algorithm for early gastric cancer (MESDA-G). Dig. Endosc. 2016, 28, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Oyama, T.; Inoue, H.; Arima, M.; Momma, K.; Ishihara, R.; Hirasawa, D.; Takeuchi, M.; Tomori, A.; Goda, K. Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: Magnifying endoscopic classification of the Japan Esophageal Society. Esophagus 2017, 14, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Ohue, M. Morphological diagnosis of mucosal surface at cellular level using contact endoscopy (abstract in Japanese). In Proceedings of the 59th Annual Meeting of Japan Endoscopy Society, Kyoto, Japan, 29–31 May 2000. [Google Scholar]
- Kumagai, Y.; Iida, M.; Yamazaki, S. Ultra-high magnifying observation for superficial esophageal cancer using contact endoscopy with dye-staining (abstract in Japanese). In Proceedings of the 57th Annual Meeting of Japan Esophageal Society, Kyoto, Japan, 27–28 June 2003. [Google Scholar]
- Kumagai, Y.; Takubo, K.; Kawada, K.; Higashi, M.; Ishiguro, T.; Sobajima, J.; Fukuchi, M.; Ishibashi, K.-I.; Mochiki, E.; Aida, J.; et al. A newly developed continuous zoom-focus endocytoscope. Endoscopy 2016, 49, 176–180. [Google Scholar] [CrossRef]
- Inoue, H.; Kazawa, T.; Sato, Y.; Satodate, H.; Sasajima, K.; Kudo, S.-E.; Shiokawa, A. In vivo observation of living cancer cells in the esophagus, stomach, and colon using catheter-type contact endoscope, “Endo-Cytoscopy system”. Gastrointest. Endosc. Clin. N. Am. 2004, 14, 589–594. [Google Scholar] [CrossRef]
- Kumagai, Y.; Kawada, K.; Higashi, M.; Ishiguro, T.; Sobajima, J.; Fukuchi, M.; Ishibashi, K.; Baba, H.; Mochiki, E.; Aida, J.; et al. Endocytoscopic observation of various esophageal lesions at ×600: Can nuclear abnormality be recognized? Dis. Esophagus 2015, 28, 269–275. [Google Scholar] [CrossRef]
- Kumagai, Y.; Kawada, K.; Yamazaki, S.; Iida, M.; Odajima, H.; Ochiai, T.; Kawano, T.; Takubo, K. Current status and limitations of the newly developed endocytoscope GIF-Y0002 with reference to its diagnostic performance for common esophageal lesions. J. Dig. Dis. 2012, 13, 393–400. [Google Scholar] [CrossRef]
- Kawada, K.; Momma, K.; Kawachi, H.; Fujiwara, J.; Funada, N.; Kumagai, Y.; Yoshida, M. Endoscopic diagnosis of iodine unstained areas observed by endo-cytoscopy system. Endo-Cytoscopy 2006, 41, 225–232. [Google Scholar] [CrossRef]
- Kumagai, Y.; Kawada, K.; Yamazaki, S.; Iida, M.; Momma, K.; Odajima, H.; Kawachi, H.; Nemoto, T.; Kawano, T.; Takubo, K. Endocytoscopic observation for esophageal squamous cell carcinoma: Can biopsy histology be omitted? Dis. Esophagus 2009, 22, 505–512. [Google Scholar] [CrossRef]
- Inoue, H.; Sasajima, K.; Kaga, M.; Sugaya, S.; Sato, Y.; Wada, Y.; Inui, M.; Satodate, H.; Kudo, S.-E.; Kimura, S.; et al. Endoscopic in vivo evaluation of tissue atypia in the esophagus using a newly designed integrated endocytoscopy: A pilot trial. Endoscopy 2006, 38, 891–895. [Google Scholar] [CrossRef]
- Shimamura, Y.; Inoue, H.; de Santiago, E.R.; Abad, M.R.A.; Fujiyoshi, Y.; Toshimori, A.; Tanabe, M.; Sumi, K.; Iwaya, Y.; Ikeda, H.; et al. Diagnostic yield of fourth-generation endocytoscopy for esophageal squamous lesions using a modified endocytoscopic classification. Dig. Endosc. 2020, 33, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Takubo, K.; Sato, T.; Ishikawa, H.; Yamamoto, E.; Ishiguro, T.; Hatano, S.; Toyomasu, Y.; Kawada, K.; Matsuyama, T.; et al. AI analysis and modified type classification for endocytoscopic observation of esophageal lesions. Dis. Esophagus, 2022; online ahead of print. [Google Scholar] [CrossRef]
- Kumagai, Y.; Kawada, K.; Yamazaki, S.; Iida, M.; Ochiai, T.; Momma, K.; Odajima, H.; Kawachi, H.; Nemoto, T.; Kawano, T.; et al. Endocytoscopic observation of esophageal squamous cell carcinoma. Dig. Endosc. 2010, 22, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Takubo, K.; Kawada, K.; Higashi, M.; Ishiguro, T.; Sobajima, J.; Fukuchi, M.; Ishibashi, K.; Mochiki, E.; Aida, J.; et al. Endocytoscopic observation of various types of esophagitis. Esophagus 2016, 13, 200–207. [Google Scholar] [CrossRef]
- Solcia, E.; Villani, L.; Luinetti, O.; Trespi, E.; Strada, E.; Tinelli, C.; Fiocca, R. Altered intercellular glycoconjugates and dilated intercellular spaces of esophageal epithelium in reflux disease. Virchows Arch. 2000, 436, 207–216. [Google Scholar] [CrossRef]
- Shimamura, Y.; Goda, K.; Hirooka, S.; Inoue, H. Observation of bilobed nucleus sign by endocytoscopy in eosinophilic esophagitis. Gastrointest. Endosc. 2021, 93, 259–260. [Google Scholar] [CrossRef]
- Shcherbynina, M.; Itabashi, M.; Soloviova, N. Histological criteria for “intraepithelial squamous cell carcinoma” of the esophagus: Continued dialogue between Ukrainian and Japanese pathologists. Exp. Oncol. 2020, 42, 314–317. [Google Scholar] [CrossRef]
- The Japan Esophageal Society. Japanese classification of esophageal cancer, eleventh edition: Part II and III. Esophagus 2017, 14, 50–51. [Google Scholar]
- Takubo, K.; Fujii, S. Oesophageal squamous dysplasia. In WHO Classification of Tumours, Digestive System Tumours, 5th ed.; WHO Classification of Tumours Editorial Board, Ed.; IARC: Lyon, France, 2019; pp. 36–37. [Google Scholar]
- Shimoda, Y.; Shimizu, Y.; Takahashi, H.; Okahara, S.; Miyake, T.; Ichihara, S.; Tanaka, I.; Inoue, M.; Kinowaki, S.; Ono, M.; et al. Optical biopsy for esophageal squamous cell neoplasia by using endocytoscopy. BMC Gastroenterol. 2022, 22, 1–9. [Google Scholar] [CrossRef]
- Kodashima, S.; Fujishiro, M.; Takubo, K.; Kammori, M.; Nomura, S.; Kakushima, N.; Muraki, Y.; Tateishi, A.; Kaminishi, M.; Omata, M. Ex-vivo study of high-magnification chromoendoscopy in the gastrointestinal tract to determine the optimal staining conditions for endocytoscopy. Endoscopy 2006, 38, 1115–1121. [Google Scholar] [CrossRef]
- Minami, H.; Inoue, H.; Yokoyama, A.; Ikeda, H.; Satodate, H.; Hamatani, S.; Haji, A.; Kudo, S. Recent advancement of observing living cells in the esophagus using CM double staining: Endocytoscopic atypia classification. Dis. Esophagus 2012, 25, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Olliver, J.; Wild, C.; Sahay, P.; Dexter, S.; Hardie, L. Chromoendoscopy with methylene blue and associated DNA damage in Barrett’s oesophagus. Lancet 2003, 362, 373–374. [Google Scholar] [CrossRef]
- National Toxicology Program. Toxicology and carcinogenesis studies of methylene blue trihydrate (Cas No. 7220-79-3) in F344/N rats and B6C3F1 mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 2008, 540, 1–224. [Google Scholar]
- Bibault, J.-E.; Giraud, P.; Burgun, A. Big Data and machine learning in radiation oncology: State of the art and future prospects. Cancer Lett. 2016, 382, 110–117. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Yoshida, H.; Shimazu, T.; Kiyuna, T.; Marugame, A.; Yamashita, Y.; Cosatto, E.; Taniguchi, H.; Sekine, S.; Ochiai, A. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer 2018, 21, 249–257. [Google Scholar] [CrossRef]
- Misawa, M.; Kudo, S.-E.; Mori, Y.; Takeda, K.; Maeda, Y.; Kataoka, S.; Nakamura, H.; Kudo, T.; Wakamura, K.; Hayashi, T.; et al. Accuracy of computer-aided diagnosis based on narrow-band imaging endocytoscopy for diagnosing colorectal lesions: Comparison with experts. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 757–766. [Google Scholar] [CrossRef]
- Mori, Y.; Neumann, H.; Misawa, M.; Kudo, S.; Bretthauer, M. Artificial intelligence in colonoscopy—Now on the market. What’s next? J. Gastroenterol. Hepatol. 2021, 36, 7–11. [Google Scholar] [CrossRef]
- Shichijo, S.; Nomura, S.; Aoyama, K.; Nishikawa, Y.; Miura, M.; Shinagawa, T.; Takiyama, H.; Tanimoto, T.; Ishihara, S.; Matsuo, K.; et al. Application of Convolutional Neural Networks in the Diagnosis of Helicobacter pylori Infection Based on Endoscopic Images. eBioMedicine 2017, 25, 106–111. [Google Scholar] [CrossRef]
- Hirasawa, T.; Aoyama, K.; Tanimoto, T.; Ishihara, S.; Shichijo, S.; Ozawa, T.; Ohnishi, T.; Fujishiro, M.; Matsuo, K.; Fujisaki, J.; et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018, 21, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Horie, Y.; Yoshio, T.; Aoyama, K.; Yoshimizu, S.; Horiuchi, Y.; Ishiyama, A.; Hirasawa, T.; Tsuchida, T.; Ozawa, T.; Ishihara, S.; et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest. Endosc. 2019, 89, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xiao, X.; Wu, C.; Zeng, X.; Zhang, Y.; Du, J.; Bai, S.; Xie, J.; Zhang, Z.; Li, Y.; et al. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest. Endosc. 2020, 91, 41–51. [Google Scholar] [CrossRef]
- Ohmori, M.; Ishihara, R.; Aoyama, K.; Nakagawa, K.; Iwagami, H.; Matsuura, N.; Shichijo, S.; Yamamoto, K.; Nagaike, K.; Nakahara, M.; et al. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest. Endosc. 2020, 91, 301–309.e1. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H.; Ishihara, R.; Kato, Y.; Matsunaga, T.; Nishida, T.; Yamada, T.; Ogiyama, H.; Horie, M.; Kinoshita, K.; Tada, T. Comparison of performances of artificial intelligence versus expert endoscopists for real-time assisted diagnosis of esophageal squamous cell carcinoma (with video). Gastrointest. Endosc. 2020, 92, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, K.; Ishihara, R.; Aoyama, K.; Ohmori, M.; Nakahira, H.; Matsuura, N.; Shichijo, S.; Nishida, T.; Yamada, T.; Yamaguchi, S.; et al. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest. Endosc. 2019, 90, 407–414. [Google Scholar] [CrossRef]
- Tokai, Y.; Yoshio, T.; Aoyama, K.; Horie, Y.; Yoshimizu, S.; Horiuchi, Y.; Ishiyama, A.; Tsuchida, T.; Hirasawa, T.; Sakakibara, Y.; et al. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus 2020, 17, 250–256. [Google Scholar] [CrossRef]
- Uema, R.; Hayashi, Y.; Tashiro, T.; Saiki, H.; Kato, M.; Amano, T.; Tani, M.; Yoshihara, T.; Inoue, T.; Kimura, K.; et al. Use of a convolutional neural network for classifying microvessels of superficial esophageal squamous cell carcinomas. J. Gastroenterol. Hepatol. 2021, 36, 2239–2246. [Google Scholar] [CrossRef]
- Kumagai, Y.; Takubo, K.; Kawada, K.; Aoyama, K.; Endo, Y.; Ozawa, T.; Hirasawa, T.; Yoshio, T.; Ishihara, S.; Fujishiro, M.; et al. Diagnosis using deep-learning artificial intelligence based on the endocytoscopic observation of the esophagus. Esophagus 2019, 16, 180–187. [Google Scholar] [CrossRef]



| XEC120U 1st Generation | XEC300F 1st Generation | GIF-Y0001 2nd Generation | GIF-Y0002 3rd Generation | GIF-H290EC 4th Generation | |
|---|---|---|---|---|---|
| Type of scope | probe | probe | scope (2 lens integrated) | scope | scope |
| Magnification power | ×1125 | ×450 | ×450 | Optical: ×380 | Optical: ×500 |
| With electrical: ×700 | With electrical: ×900 | ||||
| Observation field | 120 μm × 120 μm | 300 μm × 300 μm | 400 μm × 400 μm | Optical: 700 μm× 600 μm | Optical: 570 μm × 500 μm |
| With electrical: 440 μm × 380 μm | With electrical: 360 μm × 310 μm | ||||
| Magnification method | fix | fix | fix | Continuous zoom | Continuous zoom |
| Outer diameter | 3.4 mm | 3.4 mm | 11.6 mm | 10.7 mm | 9.7 mm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumagai, Y.; Takubo, K.; Kawada, K.; Ohue, M.; Higashi, M.; Ishiguro, T.; Hatano, S.; Toyomasu, Y.; Matsuyama, T.; Mochiki, E.; et al. Endocytoscopic Observation of Esophageal Lesions: Our Own Experience and a Review of the Literature. Diagnostics 2022, 12, 2222. https://doi.org/10.3390/diagnostics12092222
Kumagai Y, Takubo K, Kawada K, Ohue M, Higashi M, Ishiguro T, Hatano S, Toyomasu Y, Matsuyama T, Mochiki E, et al. Endocytoscopic Observation of Esophageal Lesions: Our Own Experience and a Review of the Literature. Diagnostics. 2022; 12(9):2222. https://doi.org/10.3390/diagnostics12092222
Chicago/Turabian StyleKumagai, Youichi, Kaiyo Takubo, Kenro Kawada, Masayuki Ohue, Morihiro Higashi, Toru Ishiguro, Satoshi Hatano, Yoshitaka Toyomasu, Takatoshi Matsuyama, Erito Mochiki, and et al. 2022. "Endocytoscopic Observation of Esophageal Lesions: Our Own Experience and a Review of the Literature" Diagnostics 12, no. 9: 2222. https://doi.org/10.3390/diagnostics12092222







