Evaluation of the EasyScreen™ ESBL/CPO Detection Kit for the Detection of ß-Lactam Resistance Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media Tested
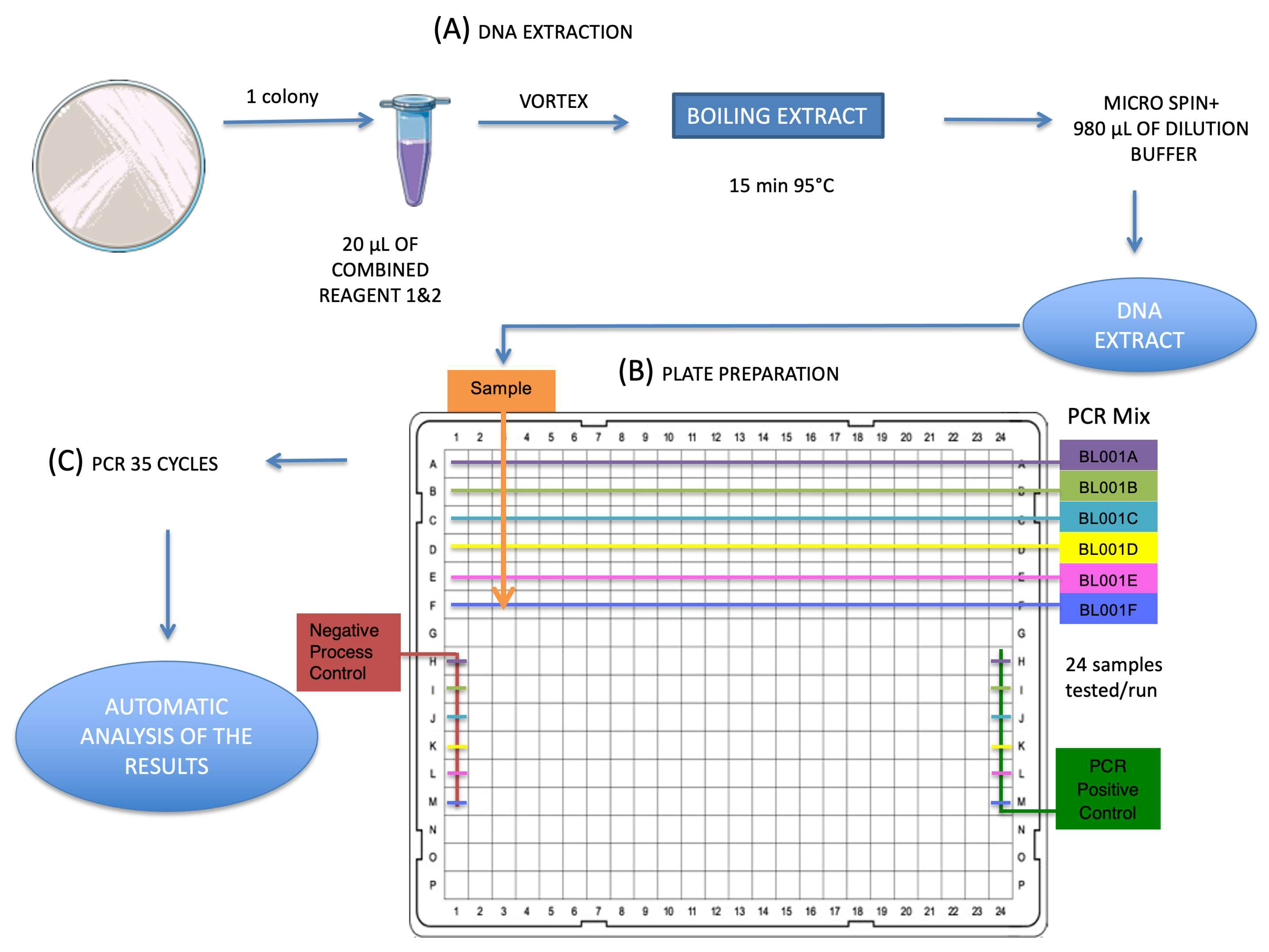
2.2. EasyScreen™ Sample Processing Kit
2.3. EasyScreen™ ESBL/CPO Detection Kit
2.4. Discrepant Results Analysis
2.5. Bacterial Isolates
2.6. Statistical Analysis
3. Results
3.1. Experimental Setup and Evaluation of Culture Media
3.2. Evaluation of the EasyScreenTM ESBL/CPO Detection Kit on Well Characterized Enterobacterales
3.3. Evaluation of the EasyScreenTM ESBL/CPO Detection Kit on Characterized Pseudomonas spp. Isolates
3.4. Evaluation of the EasyScreenTM ESBL/CPO Detection Kit on Characterized A. baumannii Isolates
3.5. Overall Assay Performance for Carbapenemase Detection Extrapolated to the French Epidemiology of CP-GNB
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- WHO Publishes the List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ (accessed on 29 April 2022).
- Bassetti, M.; Peghin, M.; Pecori, D. The management of multidrug-resistant Enterobacteriaceae. Curr. Opin. Infect. Dis. 2016, 29, 583–594. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)-structure and function. J. Enzyme Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J.D.D. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef]
- Tamma, P.D.; Doi, Y.; Bonomo, R.A.; Johnson, J.K.; Simner, P.J. Antibacterial Resistance Leadership Group. A Primer on AmpC β-Lactamases: Necessary Knowledge for an Increasingly Multidrug-resistant World. Clin. Infect. Dis. 2019, 69, 1446–1455. [Google Scholar] [CrossRef]
- Naas, T.; Dortet, L.; Iorga, B.I. Structural and Functional Aspects of Class A Carbapenemases. Curr. Drug Targets 2016, 17, 1006–1028. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Jousset, A.B.; Emeraud, C.; Oueslati, S.; Dortet, L.; Naas, T. Genetic Diversity, Biochemical Properties, and Detection Methods of Minor Carbapenemases in Enterobacterales. Front. Med. 2021, 7, 616490. [Google Scholar] [CrossRef]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microb. Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Lutgring, J.D.; Limbago, B.M. The Problem of Carbapenemase-Producing-Carbapenem-Resistant-Enterobacteriaceae Detection. J. Clin. Microbiol. 2016, 54, 529–534. [Google Scholar] [CrossRef]
- Maurer, F.P.; Castelberg, C.; Quiblier, C.; Bloemberg, G.V.; Hombach, M. Evaluation of Carbapenemase Screening and Confirmation Tests with Enterobacteriaceae and Development of a Practical Diagnostic Algorithm. J. Clin. Microbiol. 2015, 53, 95–104. [Google Scholar] [CrossRef]
- Noster, J.; Thelen, P.; Hamprecht, A. Detection of Multidrug-Resistant Enterobacterales-From ESBLs to Carbapenemases. Antibiotics 2021, 10, 114. [Google Scholar] [CrossRef]
- Dortet, L.; Poirel, P.; Nordmann, P. Detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae from urine samples by use of the ESBL NDP test. J. Clin. Microbiol. 2014, 52, 3701–3706. [Google Scholar] [CrossRef]
- Dortet, L.; Agathine, A.; Naas, T.; Cuzon, G.; Poirel, L.; Nordmann, P. Evaluation of the RAPIDEC CARBA NP, the Rapid CARB Screen and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2015, 70, 3014–3022. [Google Scholar] [CrossRef]
- Rezzoug, I.; Emeraud, C.; Sauvadet, A.; Cotellon, G.; Naas, T.; Dortet, L. Evaluation of a colorimetric test for the rapid detection of carbapenemase activity in Gram negative bacilli: The MAST® PAcE test. Antimicrob. Agents Chemother. 2021, 65, e02351-20. [Google Scholar] [CrossRef]
- Muntean, M.M.; Muntean, A.A.; Gauthier, L.; Creton, E.; Cotellon, G.; Popa, M.I.; Bonnin, R.A.; Naas, T. Evaluation of the rapid carbapenem inactivation method (rCIM): A phenotypic screening test for carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 900–908. [Google Scholar] [CrossRef]
- Bernabeu, S.; Ratnam, K.C.; Boutal, H.; Gonzalez, C.; Vogel, A.; Devilliers, K.; Plaisance, M.; Oueslati, S.; Malhotra-Kumar, S.; Dortet, L.; et al. A Lateral Flow Immunoassay for the Rapid Identification of CTX-M-Producing Enterobacterales from Culture Plates and Positive Blood Cultures. Diagnostics 2020, 10, 764. [Google Scholar] [CrossRef]
- Moguet, C.; Gonzalez, C.; Naas, T.; Simon, S.; Volland, H. Multiplex Lateral Flow Immunoassay for the Detection of Expanded-Spectrum Hydrolysis and CTX-M Enzymes. Diagnostics 2022, 12, 190. [Google Scholar] [CrossRef]
- Boutal, H.; Vogel, A.; Bernabeu, S.; Devilliers, K.; Creton, E.; Cotellon, G.; Plaisance, M.; Oueslati, S.; Dortet, L.; Jousset, A.; et al. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 909–915. [Google Scholar] [CrossRef]
- Dortet, L.; Tandé, D.; de Briel, D.; Bernabeu, S.; Lasserre, C.; Gregorowicz, G.; Jousset, A.B.; Naas, T. MALDI-TOF for the rapid detection of carbapenemase-producing Enterobacteriaceae: Comparison of the commercialized MBT STAR-Carba IVD Kit with two in-house MALDI-TOF techniques and the RAPIDEC CARBA NP. J. Antimicrob. Chemother. 2018, 73, 2352–2359. [Google Scholar] [CrossRef]
- Bogaerts, P.; Oueslati, S.; Meunier, D.; Nonhoff, C.; Yunus, S.; Massart, M.; Denis, O.; Woodford, N.; Hopkins, K.L.; Naas, T.; et al. Multicentre evaluation of the BYG Carba v2.0 test, a simplified electrochemical assay for the rapid laboratory detection of carbapenemase-producing Enterobacteriaceae. Sci. Rep. 2017, 7, 9937. [Google Scholar] [CrossRef]
- Renvoisé, A.; Decré, D.; Amarsy-Guerle, R.; Huang, T.D.; Jost, C.; Podglajen, I.; Raskine, L.; Genel, N.; Bogaerts, P.; Jarlier, V.; et al. Evaluation of the β LACTA™ test, a rapid test detecting resistance to third-generation cephalosporins in clinical strains of Enterobacteriaceae. J. Clin. Microbiol. 2013, 51, 4012–4017. [Google Scholar] [CrossRef]
- Bernabeu, S.; Dortet, L.; Naas, T. Evaluation of the beta-CARBA test, a colorimetric test for the rapid detection of carbapenemase activity in Gram-negative bacilli. J. Antimicrob. Chemother. 2017, 72, 1646–1658. [Google Scholar] [CrossRef]
- Hoyos-Mallecot, Y.; Ouzani, S.; Dortet, L.; Fortineau, N.; Naas, T. Performance of the Xpert Carba-R v2 in the daily workflow of a hygiene unit in a country with a low prevalence of carbapenemase-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2017, 49, 774–777. [Google Scholar] [CrossRef]
- Anjum, M.F.; Zankari, E.; Hasman, H. Molecular Methods for Detection of Antimicrobial Resistance. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Girlich, D.; Bogaerts, P.; Bouchahrouf, W.; Bernabeu, S.; Langlois, I.; Begasse, C.; Arangia, N.; Dortet, L.; Huang, T.D.; Glupczynski, Y.; et al. Evaluation of the Novodiag CarbaR+, a Novel Integrated Sample to Result Platform for the Multiplex Qualitative Detection of Carbapenem and Colistin Resistance Markers. Microb. Drug Resist. 2021, 27, 170–178. [Google Scholar] [CrossRef]
- Girlich, D.; Oueslati, S.; Bernabeu, S.; Langlois, I.; Begasse, C.; Arangia, N.; Creton, E.; Cotellon, G.; Sauvadet, A.; Dortet, L.; et al. Evaluation of the BD MAX Check-Points CPO Assay for the Detection of Carbapenemase Producers Directly from Rectal Swabs. J. Mol. Diagn. 2020, 22, 294–300. [Google Scholar] [CrossRef]
- Girlich, D.; Laguide, M.; Dortet, L.; Naas, T. Evaluation of the Revogene Carba C Assay for Detection and Differentiation of Carbapenemase-Producing Gram-Negative Bacteria. J. Clin. Microbiol. 2020, 5, e01927-19. [Google Scholar] [CrossRef] [Green Version]
- Oueslati, S.; Girlich, D.; Dortet, L.; Naas, T. Evaluation of the Amplidiag CarbaR+VRE Kit for Accurate Detection of Carbapenemase-Producing Bacteria. J. Clin. Microbiol. 2018, 56, e01092-17. [Google Scholar] [CrossRef]
- Girlich, D.; Bernabeu, S.; Fortineau, N.; Dortet, L.; Naas, T. Evaluation of the CRE and ESBL ELITe MGB® kits for the accurate detection of carbapenemase- or CTX-M–producing bacteria. Diagn. Microbiol. Infect. Dis. 2018, 92, 1–7. [Google Scholar] [CrossRef]
- Palmeira, J.D.; Haenni, M.; Metayer, V.; Madec, J.Y.; Ferreira, H.M.N. Epidemic spread of IncI1/pST113 plasmid carrying the Extended-Spectrum Beta-Lactamase (ESBL) blaCTX-M-8 gene in Escherichia coli of Brazilian cattle. Vet. Microbiol. 2020, 243, 108629. [Google Scholar] [CrossRef]
- Sennati, S.; Santella, G.; Di Conza, J.; Pallecchi, L.; Pino, M.; Ghiglione, B.; Rossolini, G.M.; Radice, M.; Gutkind, G. Changing epidemiology of extended-spectrum β-lactamases in Argentina: Emergence of CTX-M-15. Antimicrob. Agents Chemother. 2012, 56, 6003–6005. [Google Scholar] [CrossRef]
- Centre National de Référence de la Résistance aux Antibiotiques. Rapport d’activité 2019–2020. Available online: https://www.cnr-resistance-antibiotiques.fr/ressources/pages/Rapport_CNR_RA_2019_2020v2.pdf (accessed on 7 September 2022).
- Jousset, A.B.; Emeraud, C.; Bonnin, R.A.; Naas, T.; Dortet, L. Characteristics and evolution of carbapenemase-producing enterobacterales in france, 2012–2020. BEH 2021, 18–19, 351–358. [Google Scholar]
- Lombes, A.; Bonnin, R.A.; Laurent, F.; Guet-Revillet, H.; Bille, E.; Cattoir, V.; Fangous, M.S.; Le Brun, C.; Fihman, V.; Janvier, F.; et al. GMC Study Group. High Prevalence of OXA-23 Carbapenemase-Producing Proteus mirabilis among Amoxicillin-Clavulanate-Resistant Isolates in France. Antimicrob. Agents Chemother. 2022, 66, e0198321. [Google Scholar] [CrossRef]
- Hopkins, K.L.; Findlay, J.; Meunier, D.; Cummins, M.; Curtis, S.; Kustos, I.; Mustafa, N.; Perry, C.; Pike, R.; Woodford, N. Serratia marcescens producing SME carbapenemases: An emerging resistance problem in the UK? J. Antimicrob. Chemother. 2017, 72, 1535–1537. [Google Scholar]
- Bush, K.; Pannell, M.; Lock, J.L.; Queenan, A.M.; Jorgensen, J.H.; Lee, R.M.; Lewis, J.S.; Jarrett, D. Detection systems for carbapenemase gene identification should include the SME serine carbapenemase. Int. J. Antimicrob. Agents 2013, 41, 1–4. [Google Scholar] [CrossRef]
- Ellington, M.J.; Davies, F.; Jauneikaite, E.; Hopkins, K.L.; Turton, J.F.; Adams, G.; Pavlu, J.; Innes, A.J.; Eades, C.; Brannigan, E.T.; et al. Multispecies Cluster of GES-5 Carbapenemase-Producing Enterobacterales Linked by a Geographically Disseminated Plasmid. Clin. Infect. Dis. 2020, 71, 2553–2560. [Google Scholar] [CrossRef]
- Kanayama, A.; Kawahara, R.; Yamagishi, T.; Goto, K.; Kobaru, Y.; Takano, M.; Morisada, K.; Ukimura, A.; Kawanishi, F.; Tabuchi, A.; et al. Successful control of an outbreak of GES-5 extended-spectrum β-lactamase-producing Pseudomonas aeruginosa in a long-term care facility in Japan. J. Hosp. Infect. 2016, 93, 35–41. [Google Scholar] [CrossRef]
- Ibrahim, M.E. Prevalence of Acinetobacter baumannii in Saudi Arabia: Risk factors, antimicrobial resistance patterns and mechanisms of carbapenem resistance. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 1. [Google Scholar] [CrossRef]
- Hishinuma, T.; Tada, T.; Kuwahara-Arai, K.; Yamamoto, N.; Shimojima, M.; Kirikae, T. Spread of GES-5 carbapenemase-producing Pseudomonas aeruginosa clinical isolates in Japan due to clonal expansion of ST235. PLoS ONE 2018, 13, e0207134. [Google Scholar] [CrossRef]
- Naas, T.; Poirel, L.; Nordmann, P. Minor extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 2008, 14 (Suppl. S1), 42–52. [Google Scholar] [CrossRef] [Green Version]
| Detection Channel | Mix A | Mix B | Mix C | Mix D | Mix E | Mix F |
|---|---|---|---|---|---|---|
| # 1 | TEM | GES | OXA-48 | MCR-1 | VIM | CMY |
| # 2 | EC a | EC a | EC a | EC a | EC a | EC a |
| # 3 | CTX-M | KPC | OXA-23-like | DHA | IMI | IMP |
| # 4 | Not used | NDM | SHV | OXA-51-like | SME | Not used |
| Species | Resistance Mechanism | EasyScreenTM ESBL/CPO Detection Kit Results | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KPC | OXA-48 | NDM | VIM | IMP | SME | IMI | GES | CTX-M | TEM | SHV | DHA | CMY | OXA-23 | OXA-51 | Mcr-1 | ||
| Non-targeted β-lactamase producers (17)a | |||||||||||||||||
| E. coli (2) a, P. mirabilis (2), E. cloacae (3) H. alvei (2), S. marcescens (1) | ↗↗↗ b Case (7) a, pACC-1 (3) | - c | - | - | - | - | - | - | - | - | 1/1 | - | - c | - | - | - | - |
| E. cloacae (3), P. mirabilis (1), C. freundii (2) | GIM-1, LMB-1, TMB-1, OXA-372, OXA-58 | - | - | - | - | - | - | - | - | - | 1/1 | - | - | 0/2 | - | - | - |
| Targeted AmpC producers | |||||||||||||||||
| E. coli (3), K. pneumoniae (2), M. morganii (1) | DHA (5), CMY-136 | - | - | - | - | - | - | - | - | - | 1/1 | 3/3 | 5/5 | 1/1 | - | - | - |
| Targeted ESBLproducers | |||||||||||||||||
| E. coli (32), K. pneumoniae (13), C. freundii (2), C. koseri (1), M. morganii (1), E. cloacae (4), P. mirabilis (1) | CTX-M-1(5), -2(3), -3(2), -8(2), -9(1), -10(1), -13(1), -14(3), -15(16), -17(1), -18(1), -19(1), -24(1), -27(2), -32(2), -37(1), -55(2), -57(1), -65(1), -71(2), -82(1), -93(1), 101(1), 182(1) | - | - | - | - | - | - | 52/54 | 29/29 | 11/13 | 1/1 | 1/3 | - | - | - | ||
| K. pneumoniae | GES-1, | - | - | - | - | - | - | 1/1 | - | - | 1/1 | - | - | - | - | - | |
| K. pneumoniae, E. cloacae, S. marcescens | OXA-163 (2), OXA-405 | - | 3/3 | - | - | - | - | - | - | - | 2/2 | 2/2 | - | - | - | - | - |
| K. pneumoniae (6), C. freundii | SHV-2a (2), -11, -12, -36, -38, -99 | - | - | - | - | - | - | - | - | 1/1 | 3/3 | 7/7 | - | 1/1 | - | - | - |
| E. coli (1), E. cloacae (1), K. aerogenes (2), C. freundii (2), K. pneumoniae (1), P. mirabilis | TEM-3 (4), -24(2), -52 (2) | - | - | - | - | - | - | - | - | - | 8/8 | 1/1 | - | 0/2 | - | - | - |
| Targeted carbapenemase producers | |||||||||||||||||
| P. mirabilis | OXA-23 | - | - | - | - | - | - | - | - | - | - | - | 1/1 | - | - | ||
| S. marcescens (3) | Sme-1, Sme-2, Sme-4 | - | - | - | - | 3/3 | - | - | - | - | - | - | - | - | - | ||
| E. cloacae complex (6) | IMI-1, IMI-2 (2), IMI-3, IMI-17, NMCA | - | - | - | - | - | 6/6 | - | - | - | - | - | - | - | - | ||
| K. pneumoniae (1), E. cloacae (2),) | GES-5 (2), GES-6 | - | - | - | - | - | - | 3/3 | 1/1 | - | 2/2 | - | - | - | - | ||
| E. coli (11), K. pneumoniae (4), E. cloacae (2), C. freundii, S. marcescens | KPC-2 (7), KPC-3 (4), KPC-5, KPC-6, KPC-7, KPC-14, KPC-28, KPC-31, KPC-33, KPC-39 | 19/19 | - | - | - | - | - | - | 4/4 | 9/9 | 4/4 | 0/1 | - | - | - | ||
| E. coli (9), K. pneumoniae (1), S. enterica | NDM-1 (5), NDM-4, NDM-6, NDM-6, NDM-9, NDM-19 (2) | - | - | 11/11 | - | - | - | - | 8/8 | 9/9 | 1/1 | 2/2 | 4/4 | - | - | ||
| E. coli (2), K. pneumoniae (3), E. cloacae, C. freundii (2) | VIM-1 (4), VIM-2 (2), VIM-4, VIM-19 | - | - | - | 8/8 | - | - | - | - | 1/1 | 3/3 | 4/4 | - | 1/3 | - | - | |
| E. coli (3), K. pneumoniae (3), E. cloacae, S. marcescens | IMP-1 (2), IMP-8 (4), IMP-10, IMP-IMP-14 | - | - | - | - | 8/8 | - | - | - | - | 6/6 | 6/6 | - | - | - | - | |
| E. coli (12), K. pneumoniae (11), E. cloacae, C. koseri, C. freundii (2), Shewanella sp. | OXA-48 (8), OXA-162, OXA-181 (2), OXA-204 (5), OXA-232 (2), OXA-244 (4), OXA-370, OXA-484, OXA-517, OXA-519, OXA-535, OXA-793 | 28/28 | 16/16 | 15/15 | 14/14 | 8/8 | |||||||||||
| Targeted multiple carbapenemase producers | |||||||||||||||||
| E. coli (3), K. pneumoniae (4), E. cloacae, C. freundii (2) | OXA-48-like + NDM-Like (6); OXA-48-like + VIM-like (3); OXA-48-like + KPC-28 | 1/1 | 10/10 | 6/6 | 3/3 | 8/8 | 7/7 | 5/5 | 0/2 | 1/1 | |||||||
| K. pneumoniae | OXA-48-like + NDM-5 + VIM-1 | 1/1 | 1/1 | 1/1 | 1/1 | ||||||||||||
| K. pneumoniae (2) | NDM + KPC (2) | 2/2 | 2/2 | 1/1 | 2/2 | 1/2 | |||||||||||
| E. coli | NDM-1 + VIM-2 | 1/1 | 1/1 | 1/1 | |||||||||||||
| K. pneumoniae | KPC-2 +VIM-1 | 1/1 | 1/1 | 1/1 | 1/1 | ||||||||||||
| Plasmid-encoded colistin resistance | |||||||||||||||||
| E. coli (6), K. pneumoniae, Salmonella (4) | mgrB mutation (6), mcr-3.2, mcr-4, mcr-5(2), mcr-2. | 1/1 | 3/3 | 1/1 | - | ||||||||||||
| E. coli (6), K. pneumoniae (3), Salmonella sp. | Mcr-1 | - | 2/2 | 1/1 | - | - | - | - | - | 5/5 | 6/6 | 3/3 | - | - | - | - | 10/10 |
| Total | 23/23 | 44/44 d | 22/22 | 14/14 | 8/8 | 3/3 | 6/6 | 4/4 e | 98/100 f | 107/107 g | 65/68 h | 8/8 | 16/27 i | 1/1 | 0/0 | 10/10 | |
| Sensitivity | 100% (85.2% to 100%) | 100% (91.4% to 100%) | 100% (84.6% to 100%) | 100% (76.8% to 100%) | 100% (63.1% to 100%) | 100% (29.2% to 100%) | 100% (54.1% to 100%) | 100% (29.2% to 100%) | 98.0% (92.9% to 99.8%) | 7.5% (3.3% to 14.2% | 10.3% (4.24% to 20.1%) | 100% (63.1% to 100%) | 59.3% (38.8% to 77.6%) | 100% (2.5% to 100%) | 100% (69.2% to 100%) | ||
| Specificity | 100% (98.1% to 100% | 98.3% (95.2% to 99.6% | 100% (98.1% to 100% | 100% (98.2% to 100% | 100% (98.3% to 100% | 100% (98.3% to 100% | 100% (98.3% to 100% | 99.5% (97.5% to 100% | 100% (96.9% to 100% | 100% (96.7% to 100% | 100% (97.6% to 100% | 100% (98.3% to 100% | 100% (98.1% to 100% | 100% (98.3% to 100% | 100% (98.3% to 100% | 100% (98.3% to 100% | |
| Species | β-Lactamase Content | EasyScreenTM ESBL/CPO Detection Kit Results | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KPC | OXA-48 | NDM | VIM | IMP | SME | IMI | GES | CTX-M | TEM | SHV | DHA | CMY | OXA-23 | OXA-51 | Mcr-1 | ||
| Non-targeted β-lactamase producers (22) a | |||||||||||||||||
| P. aeruginosa (2) a | Mex A/B-OprM, Mex C/D-OprJ | - c | - | - | - | - | - | - | - | - | - | - | - c | - | - | ||
| P. aeruginosa (4) | ↗↗↗ b Case + OprD deficient | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| P. aeruginosa (9) | BEL-1, VEB-1, PER-1, OXA-10, OXA-13, OXA-14, OXA-18/20, OXA-32, CARB-2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| P. aeruginosa (7) | GIM-1, AIM-1, SPM-1, DIM-1 (2), PME-1, OXA-198 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Targeted ESBLproducers | |||||||||||||||||
| P. aeruginosa | GES-9, | - | - | - | - | - | - | 1/1 | - | - | - | - | - | - | - | - | |
| P. aeruginosa | CTX-M-2 | + | - | - | - | - | - | - | - | 1/1 | - | - | - | - | - | - | - |
| P. aeruginosa (2) | SHV-2a, SHV-5 | + | - | - | - | - | - | - | - | - | - | 2/2 | - | - | - | - | - |
| P. aeruginosa | TEM-4 | + | - | - | - | - | - | - | - | - | 1/1 | - | - | - | - | - | - |
| Targeted carbapenemase producers | |||||||||||||||||
| P. aeruginosa (3) | GES-2, GES-5, GES-14 | - | - | - | - | - | - | 3/3 | - | - | - | - | - | - | - | ||
| P. aeruginosa (4) | KPC-2 (4) | 4/4 | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| P. aeruginosa (2) | NDM-1 (2) | - | - | 2/2 | - | - | - | - | - | - | - | - | - | - | |||
| P. aeruginosa (7), P. putida 2), P. stutzeri, P. fluorescens | VIM-1 (2), VIM-2 (7), VIM-4 (1), VIM-5 (1) | - | - | - | 11/11 | - | - | - | - | - | 2/2 | - | - | - | - | - | |
| P. aeruginosa (12), P. putida (2), P. stutzeri | IMP-1 (3), IMP-2(2), IMP-7, IMP-15, IMP-19, IMP-26, IMP-31, IMP-39, IMP-46, IMP-56, IMP-63, IMP-71 | - | - | - | - | 15/15 | - | - | - | - | 2/2 | - | - | - | - | ||
| P. aeruginosa (3), | IMP-13 (2), IMP-29 | - | - | - | - | 0/3 | - | - | - | - | - | - | - | - | - | - | |
| Total | 4/4 | - | 2/2 | 11/11 | 15/18 d | 4/4 e | 1/1 | 5/5 g | 2/2 f | - | - | - | - | - | |||
| Sensitivity | 100% (39.8% to 100%) | 100% (15.8% to 100%) | 100% (71.5% to 100%) | 83.3% (58.6% to 96.4%) | 100% (29.2% to 100%) | 100% (2.5% to 100%) | 100% (2.5% to 100%) | 100% (15.8% to 100%) | |||||||||
| Specificity | 100% (94.1% to 100%) | 100% (94.5% to 100%) | 100% (94.3% to 100%) | 100% (93.4% to 100%) | 100% (92.4% to 100%) | 100% (94.5% to 100%) | 100% (94.5% to 100%) | 98.4% (91.3% to 99.9%) | 100% (94.4% to 100%) | 93.8% (84.8% to 98.3%) | 100% (94.3% to 100%) | 100% (94.5% to 100%) | 100% (94.5% to 100%) | 100% (94.5% to 100%) | 100% (94.5% to 100%) | 100% (94.5% to 100%) | |
| Species | β-Lactamase Content | EasyScreenTM ESBL/CPO Detection Kit Results | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KPC | OXA-48 | NDM | VIM | IMP | SME | IMI | GES | CTX-M | TEM | SHV | DHA | CMY | OXA-23 | OXA-51 | Mcr-1 | ||
| Non-targeted β-lactamase producers (15) a | |||||||||||||||||
| A. baumannii (8) a | WT (2), OXA-21, PER-1, VEB-1 (2), SCO-1, RTG-4, | - b | - | - | - | - | - | - | - | - | 2/2 | - | - c | - | - | 2/8 | - |
| A. baumannii (7) | SIM-1, OXA-143, OXA-253, OXA-24/40, OXA-72, OXA-58, OXA-97 | - | - | - | - | - | - | - | - | - | 1/1 | - | - | - | - | 4/7 | - |
| Targeted ESBLproducers (5) | |||||||||||||||||
| A. baumannii | GES-12, | - | - | - | - | - | - | - | 1/1 | - | - | - | - | - | 1/1 | - | |
| A. baumannii (2) | CTX-M-2, CTX-M-15 | - | - | - | - | - | - | - | - | 2/2 | 2/2 | - | - | - | - | 2/2 | - |
| A. baumannii | SHV-5 | - | - | - | - | - | - | - | - | - | 1/1 | 1/1 | - | - | - | 1/1 | - |
| A. baumannii | TEM-1 | - | - | - | - | - | - | - | - | - | 1/1 | - | - | - | - | 1/1 | - |
| Targeted carbapenemase producers (36) | |||||||||||||||||
| A. baumannii (6) | GES-11 (3), GES-14 (3) | - | - | - | - | - | - | 7/7 | - | 1/1 | - | - | 1/1 | 7/7 | - | ||
| A. baumannii (8) | ISAba1/OXA-51-like (3), ISAba1/OXA-51-like + SHV-5, ISAba1/OXA-51-like + GES-12 (4) | - | - | - | - | - | - | 4/4 | - | 3/3 | 1/1 | - | - | 2/8 | - | ||
| A. baumannii (7) | OXA-23 (5), OXA-23 + GES-1, OXA-23 +GES-14 | - | - | - | - | - | - | 1/1 | - | 4/4 | - | 6/6 | 6/6 | - | |||
| A. baumannii (11) | NDM-1 (4), NDM-1 + OXA-23, NDM-1 (4) + ISAba1/OXA-51-like, NDM-2, NDM-9 | - | - | 11/11 | - | - | - | - | - | 2/2 | - | - | 6/6 | 11/11 | - | ||
| A. baumannii (3) | IMP-1 (2), IMP-4 + OXA-58 | - | - | - | - | 3/3 | - | - | - | - | 3/3 | - | - | - | - | 0/3 | |
| A. genomospecies 16 | VIM-4 | - | - | - | 1/1 | - | - | - | - | - | - | - | - | - | - | 0/1 | - |
| Total | - | - | 11/11 | 1/1 | 3/3 | - | - | 13/13 c | 2/2 | 23/23 d | 2/2 e | - | - | 13/13 | 37/55 f | ||
| Sensitivity | 100% (71.5% to 100%) | 100% (2.5% to 100%) | 100% (29.2% to 100%) | 100% (63.1% to 100%) | 100% (15.8% to 100%) | 100% (75.3% to 100%) | 67.3% (53.3% to 79.3%) | ||||||||||
| Specificity | 100% (93.6% to 100%) | 100% (93.6% to 100%) | 100% (92.1% to 100%) | 100% (93.5% to 100%) | 100% (93.3% to 100%) | 100% (93.6% to 100%) | 100% (93.6% to 100%) | 89.6% (77.3% to 96.5%) | 100% (93.4% to 100%) | 64.6% (51.7% to 76.1%) | 96.9% 8(9.3% to 99.6%) | 100% (93.6% to 100%) | 100% (93.6% to 100%) | 100% (91.8% to 100%) | 100% (93.6% to 100%) | ||
| EasyScreenTM ESBL/CPO Detection Kit | |||
|---|---|---|---|
| Global French CPE Epidemiology a | Global French CP-Pa Epidemiology b | Global French CP-Ab Epidemiology c | |
| (2012–2018) | 2018 | 2018 | |
| Isolates received at the F-NRCs | 19,600 | 678 | 379 |
| Carbapenemase positive isolates | 9468 (6798 OXA-48, 284 KPC, 1676 NDM, 492 VIM, 123 IMP, 54 IMI; 1 FRI-1; 2 GES; 10 OXA-23; 2 TMB-1; and 1 SME-4) | 169 (127 VIM, 1 NDM, 0 KPC, 18 IMP, 20 GES, 3 DIM) | 366 (284 OXA-23, 38 OXA-24/40 et 5 OXA-58; 65 NDM) |
| Sensitivity | 99.97% (95%CI: 99.91–99.99) | 98.22% (95%CI: 94.9–99.6) | 88.25% (95%CI: 84.5–91.4) |
| Specificity | 99.99% (95%CI: 99.95–100) | 100% (95%CI: 99.3–100) | 100% (95%CI: 75.3–100) |
| Expected False positive | 1 OXA-405 | None | None |
| Expected False negative | 1 FRI, 2 TMB-1 => 3 isolates | 3 DIM, =>3 isolates | 38 OXA-24/40 and 5 OXA-58-producers => 43 isolates |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez, C.; Oueslati, S.; Biez, L.; Dortet, L.; Naas, T. Evaluation of the EasyScreen™ ESBL/CPO Detection Kit for the Detection of ß-Lactam Resistance Genes. Diagnostics 2022, 12, 2223. https://doi.org/10.3390/diagnostics12092223
Gonzalez C, Oueslati S, Biez L, Dortet L, Naas T. Evaluation of the EasyScreen™ ESBL/CPO Detection Kit for the Detection of ß-Lactam Resistance Genes. Diagnostics. 2022; 12(9):2223. https://doi.org/10.3390/diagnostics12092223
Chicago/Turabian StyleGonzalez, Camille, Saoussen Oueslati, Laura Biez, Laurent Dortet, and Thierry Naas. 2022. "Evaluation of the EasyScreen™ ESBL/CPO Detection Kit for the Detection of ß-Lactam Resistance Genes" Diagnostics 12, no. 9: 2223. https://doi.org/10.3390/diagnostics12092223





