Automatic Bone Segmentation from MRI for Real-Time Knee Tracking in Fluoroscopic Imaging
Abstract
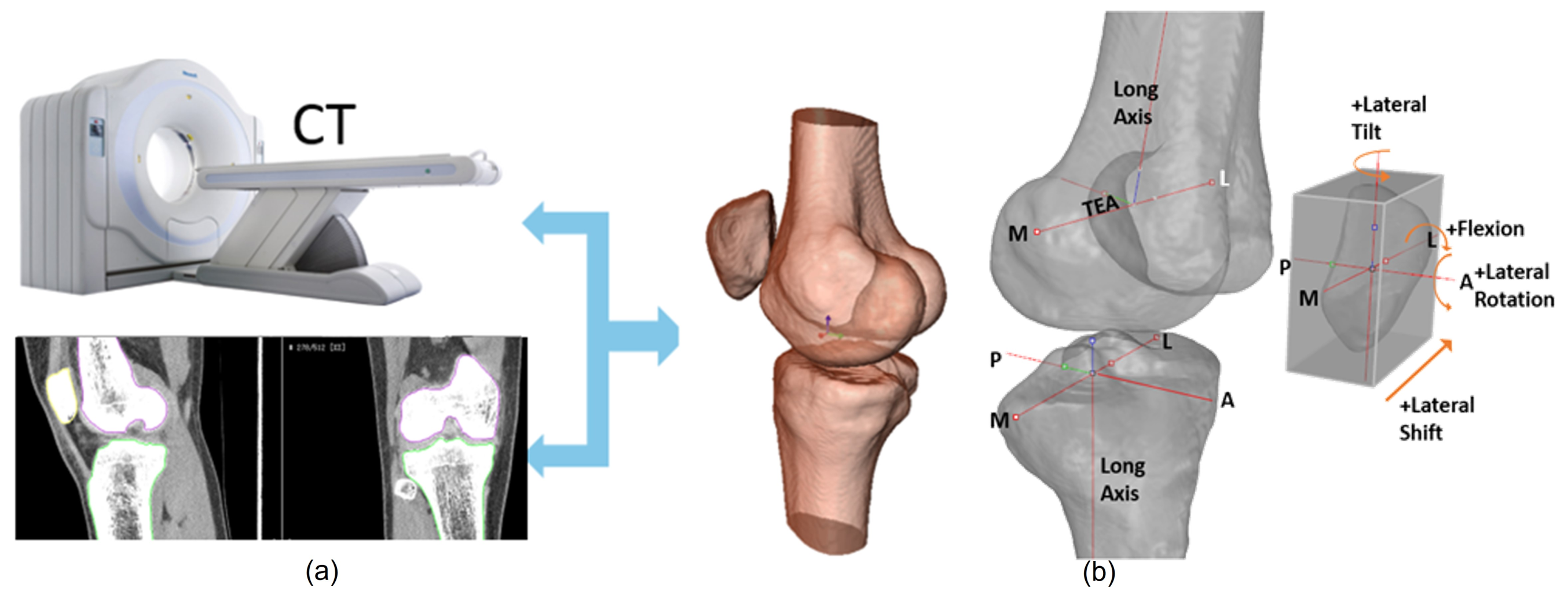
:1. Introduction
1.1. Contributions of the Paper
1.2. Organization of the Paper
2. Related Work
- The average surface distance between the boundary and for one MRI n is defined as:
- The root mean square surface distance for one MRI n is defined as:where is the number of 3D points located on the segmented bone surface boundary . The calculation of the distance between 3D points and corresponds to the minimum distance between a 3D point and a point located on reference surface boundary . Statistics are represented as ASD = and RSD = , where is the average value and is the variance for a group of M test MRIs.
3. Method
3.1. SKI10 Training Datasets
3.2. Segmenting Bone MRI Using 2.5D U-Net
- Ease of implementation on a GPU with limited global memory;
- Significant reduction in training and prediction time;
- Leveraging of the higher spatial resolution in the axial direction.
- Batch size: The mini-batch size is the number of sub-samples given to the network, after which a parameter update happens.
- Learning rate: The learning rate defines how quickly a network updates its parameters.
- Momentum: Momentum helps to know the direction of the next step with the knowledge of the previous steps.
- Loss/cost function: The loss function measures how wrong the model is.
- Activation function: Activation functions are used to introduce nonlinearity to models, which allows deep learning models to learn nonlinear prediction boundaries.
- Optimiser: An optimization algorithm is used.
- Architecture: This indicates the number of hidden layers and units.
- Dropout: Dropout is a regularization technique for avoiding overfitting (increasing the validation accuracy), thus increasing the generalization power.
- Initialiser: This is the network weight initialization.
- Validation split: This is the ratio of validation data.
- Test split: This is the ratio of test data.
- Epoch number: The number of epochs is the number of times the whole set of training data is shown to the network while training.
3.3. Geometric Validation of the Segmentation
- Generate a 3D mesh template of one of the knee bone structures using a marching cube algorithm from the SKI10 ground truth;
- Generate a 3D mesh template of one of the knee bone structures using a marching cube algorithm from the segmentation produced by 2.5D U-Net;
- Register the MRI and SKI10 3D mesh templates for each patient using a robust ICP algorithm [29];
- Compute the Euclidean distance between the MRI and SKI10 3D mesh templates;
- Display colour-coded differences in the 3D rendering of the mesh geometry.
3.4. Pre-processing the Training Datasets
4. Experimental Results
Improving Geometric Accuracy by Optimising Hyper-Parameters
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomas, M.J.; Wood, L.; Selfe, J.; Peat, G. Anterior knee pain in younger adults as a precursor to subsequent patellofe- moral osteoarthritis: A systematic review. BMC Musculoskelet. Disord. 2010, 11, 201. [Google Scholar] [CrossRef]
- Crossley, K.M. Is patellofemoral osteoarthritis a common sequela of patellofemoral pain? Br. J. Sports Med. 2014, 48, 409–410. [Google Scholar] [CrossRef] [PubMed]
- Connolly, K.D.; Ronsky, J.L.; Westover, L.M.; Küpper, J.C.; Frayne, R. Differences in patellofemoral contact mechanics associated with patellofemoral pain syndrome. J. Biomech. 2009, 42, 2802–2807. [Google Scholar] [CrossRef] [PubMed]
- Fulkerson, J.P. The etiology of patellofemoral pain in young, active patients: A prospective study. Clin. Orthop. Relat. Res. 1983, 179, 129–133. [Google Scholar] [CrossRef]
- Stagni, R.; Fantozzi, S.; Cappello, A.; Leardini, A. Quantification of soft tissue artefact in motion analysis by combining 3D fluoroscopy and stereophotogrammetry: A study on two subjects. Clin. Biomech. 2005, 20, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Esfandiarpour, F.; Lebrun, C.M.; Dhillon, S.; Boulanger, P. In-Vivo patellar tracking in individuals with patellofemoral pain and healthy individuals. J. Orthop. Res. 2018, 36, 2193–2201. [Google Scholar] [CrossRef]
- Lin, C.C.; Lu, T.W.; Li, J.D.; Kuo, M.Y.; Kuo, C.C.; Hsu, H.C. An Automated Three-Dimensional Bone Pose Tracking Method Using Clinical Interleaved Biplane Fluoroscopy Systems: Application to the Knee. Appl. Sci. 2020, 10, 8426. [Google Scholar] [CrossRef]
- Zhang, L.; Lai, W.Z.; Shah, M.A. Construction of 3D model of knee joint motion based on MRI image registration. J. Intell. Syst. 2021, 31, 15–26. [Google Scholar] [CrossRef]
- Nardini, F.; Belvedere, C.; Sancisi, N.; Conconi, M.; Leardini, A.; Durante, S.; Parenti-Castelli, V. An Anatomical-Based Subject-Specific Model of In-Vivo Knee Joint 3D Kinematics from Medical Imaging. Appl. Sci. 2020, 10, 2100. [Google Scholar] [CrossRef]
- Sun, Y.; Teo, E.C.; Zhang, Q.H. Discussions of Knee joint segmentation. In Proceedings of the International Conference on Biomedical and Pharmaceutical Engineering, Singapore, Singapore, 11–14 December 2006. [Google Scholar]
- Peterfy, C.G.; Genant, H.K. Emerging applications of magnetic resonance imaging in the evaluation of articular cartilage. Radiol. Clin. N. Am. 1996, 34, 195–213. [Google Scholar] [CrossRef]
- Pham, D.L.; Xu, C.; Prince, J.L. Current methods in medical image segmentation. Annu. Rev. Biomed. Eng. 2000, 2, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Kapur, T.; Beardsley, P.B.; Gibson, S.F.; Grimson, W.; Wells, W.M. Model-based segmentation of clinical knee MRI. In Proceedings of the IEEE International Workshop on Model-Based 3D Image Analysis, Bombay, India, 3 January 1998; pp. 97–106. [Google Scholar]
- Minerny, T.; Terzopoulos, D. Deformable models in medical image analysis: A survey. Med. Image Anal. 1996, 1, 91–108. [Google Scholar] [CrossRef]
- Fripp, J.; Warfield, S.K.; Crozier, S.; Ourselin, S. Automatic segmentation of the knee bones using 3d active shape models. In Proceedings of the IEEE 18th International Conference on Pattern Recognition, Hong Kong, China, 20–24 August 2006; Volume 52, pp. 167–170. [Google Scholar]
- Folkesson, J.; Carballido-Gamio, J.; Eckstein, F.; Link, L.M.; Majumdar, S. Local bone enhancement fuzzy clustering for segmentation of MR trabecular bone images. Med. Phys. 2010, 37, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Folkesson, J.; Olsen, O.F.; Pettersen, P.; Dam, E.; Christiansen, C. Combining Binary Classifiers for Automatic Cartilage Segmentation in Knee MRI. In Computer Vision for Biomedical Image Applications; Liu, Y., Jiang, T., Zhang, C., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2005; Volume 3765. [Google Scholar]
- Ebrahimkhani, S.; Jaward, M.H.; Cicuttini, F.M.; Dharmaratne, A.; Wang, Y.; de Herrera, A.G.S. Review on segmentation of knee articular cartilage: From conventional methods towards deep learning. Artif. Intell. Med. 2020, 106, 101851. [Google Scholar] [CrossRef] [PubMed]
- Tamez-Peña, J.G.; Farber, J.; González, P.C.; Schreyer, E.; Schneider, E.; Totterman, S. Unsupervised Segmentation and Quantification of Anatomical Knee Features: Data From the Osteoarthritis Initiative. IEEE Trans. Biomed. Eng. 2012, 59, 1177–1186. [Google Scholar] [CrossRef]
- Shan, L.; Zach, C.; Charles, C.; Niethammer, M. Automatic atlas-based three-label cartilage segmentation from MR knee images. Med. Image Anal. 2014, 18, 1233–1246. [Google Scholar] [CrossRef]
- Liu, F.; Zhou, Z.; Jang, H.; Samsonov, A.; Zhao, G.; Kijowski, R. Deep convolutional neural network and 3d deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn. Reson. Med. 2018, 79, 2379–2391. [Google Scholar] [CrossRef]
- Ambellan, F.; Tack, A.; Ehlke, M.; Zachow, S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the osteoarthritis initiative. Med. Image Anal. 2019, 52, 109–118. [Google Scholar] [CrossRef]
- Almajalid, R.; Zhang, M.; Shan, J. Fully Automatic Knee Bone Detection and Segmentation on Three-Dimensional MRI. Diagnostics 2022, 12, 123. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, N.; Tan, T.; Kang, Y.; Sun, C.; Xie, G.; Verdonschot, N.; Sprengers, A. Knee Bone and Cartilage Segmentation Based on a 3D Deep Neural Network Using Adversarial Loss for Prior Shape Constraint. Front. Med. 2022, 9, 792900. [Google Scholar] [CrossRef]
- Ciresan, D.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Deep Neural Networks Segment Neuronal Membranes in Electron Microscopy Images. Adv. Neural Inf. Process. Syst. 2012, 25, 2843–2851. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI; Navab, N., Hornegger, J., Wells, W., Frangi, A., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Volume 9351. [Google Scholar]
- Cicek, O.; Abdulkadir, A.; Lienkamp, S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, MICCAI; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2016; Volume 9901. [Google Scholar]
- Angermann, C.; Haltmeier, M.; Steiger, R.; Pereverzyev, S.; Gizewski, E. Projection-Based 2.5D U-net Architecture for Fast Volumetric Segmentation. In Proceedings of the 2019 13th International Conference on Sampling Theory and Applications (SampTA), Bordeaux, France, 8–12 July 2019. [Google Scholar]
- Besl, P.; McKay, N.D. A Method for Registration of 3-D Shapes. IEEE Trans. Pattern Anal. Mach. Intell. 1992, 14, 239–256. [Google Scholar] [CrossRef] [Green Version]


















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robert, B.; Boulanger, P. Automatic Bone Segmentation from MRI for Real-Time Knee Tracking in Fluoroscopic Imaging. Diagnostics 2022, 12, 2228. https://doi.org/10.3390/diagnostics12092228
Robert B, Boulanger P. Automatic Bone Segmentation from MRI for Real-Time Knee Tracking in Fluoroscopic Imaging. Diagnostics. 2022; 12(9):2228. https://doi.org/10.3390/diagnostics12092228
Chicago/Turabian StyleRobert, Brenden, and Pierre Boulanger. 2022. "Automatic Bone Segmentation from MRI for Real-Time Knee Tracking in Fluoroscopic Imaging" Diagnostics 12, no. 9: 2228. https://doi.org/10.3390/diagnostics12092228




