An Overview of Deep-Learning-Based Methods for Cardiovascular Risk Assessment with Retinal Images
Abstract
1. Introduction
2. Domain Knowledge
2.1. Classification of Cardiovascular Diseases
- Arrhythmias: Irregular or abnormal heartbeat that can bring on an uneven heartbeat or a heartbeat that is either too slow or too fast.
- Aorta Disease and Marfan Syndrome: This disease is produced when the aorta walls are weak. This can put extra stress on the aorta, which increases the risk of a deadly dissection or rupture.
- Cardiomyopathies: Diseases related to the heart muscle when it is unusually big, thick or stiff. The heart cannot pump blood as well as it should.
- Congenital Heart Disease: Abnormalities in one or more parts of the heart or blood vessels before birth that may appear for different reasons: genetics, virus, alcohol or drug exposure during pregnancy.
- Coronary Artery Disease: Produced when plaque builds up and hardens in the arteries that provide the heart vital oxygen and nutrients. That hardening is also called atherosclerosis.
- Deep Vein Thrombosis and Pulmonary Embolism: When blood clots, normally formed in deep veins, such as the legs, can move in the blood flow to the lungs, provoking blocked points in the bloodstream.
- Heart Failure: It is produced when the heart does not pump as strongly as it should and may provoke swelling and shortness of breath.
- Heart Valve Disease: The valves are located at the exit of each of the four heart chambers. They keep blood flowing through the heart. Examples of heart valve problems include:
- -
- Aortic stenosis: The aortic valve narrows. It slows blood flow from the heart to the rest of the body.
- -
- Mitral valve insufficiency: Caused by a malfunction in the mitral valve that may end up in a lung fluid backup due to blood leaking.
- -
- Mitral valve prolapse: The mitral valve does not close correctly between the left upper and left lower chambers.
- -
- Pericarditis: Often provoked by an infection, the lining around the heart is inflamed.
- -
- Rheumatic Heart Disease: This condition is most common in children. The heart valves are damaged due to rheumatic fever, causing an inflammatory disease.
- -
- Stroke: Reduction or block of the blood to the brain, depriving the correct contribution of oxygen and nutrients. For instance, a blocked artery or a leaking blood vessel can lead to a stroke event.
- Peripheral vascular disease: Involving any abnormality that directly alters the circulatory system, e.g., leg artery diseases may affect blood flow to the brain, ending up in a stroke.
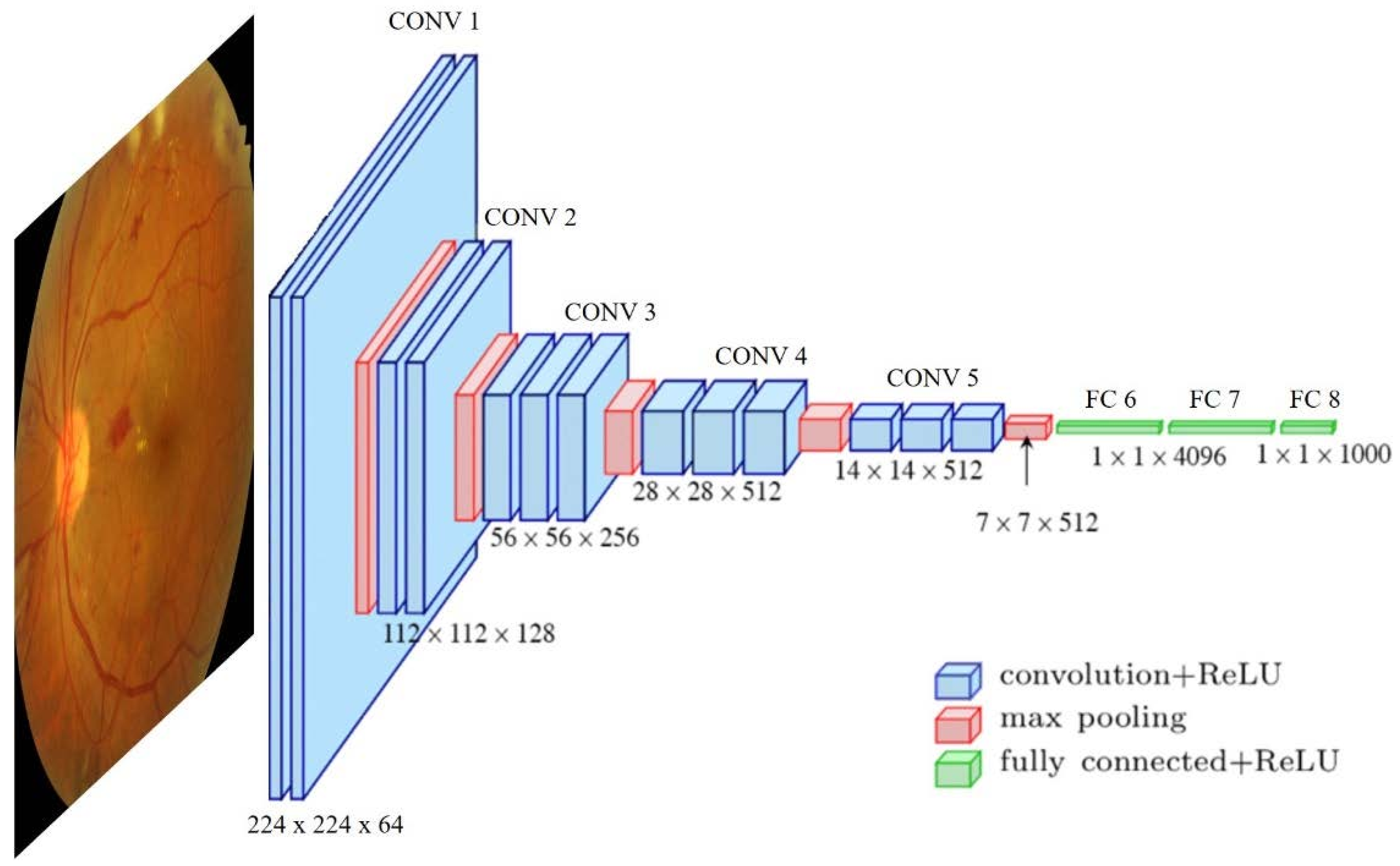
2.2. Deep Learning Approach
- Supervised learning: The quality of deep neural network performance is strongly influenced by the number of labeled/supervised images. The more images are in the training dataset, the higher the accuracy achieved by the model. To solve the problem of a lack of input data, a commonly used option is transfer learning. This approach tackles the small training size problem by pre-training the model using different natural examples, based on the premise that first network layers learn similar features and the later layers are the problem-specialized ones.
- Unsupervised learning: The model learns common associations and structures within the input set. Sometimes there is access to a large dataset of unlabeled data that can be exploited in a semi-supervised or self-supervised way. The main idea is to use these data during the training process to increase the model’s robustness, sometimes even surpassing the supervised cases [18].
- Semi-supervised learning: The data used to perform certain learning tasks are both labeled and unlabeled. It typically incorporates a small size of labeled data in combination with large amounts of unlabeled data. The difficulties here are that unlabeled data can be exploited if they provide information relevant to label estimation that is not present in the labeled data or cannot be easily obtained. This learning method requires the information to be determinable [19].
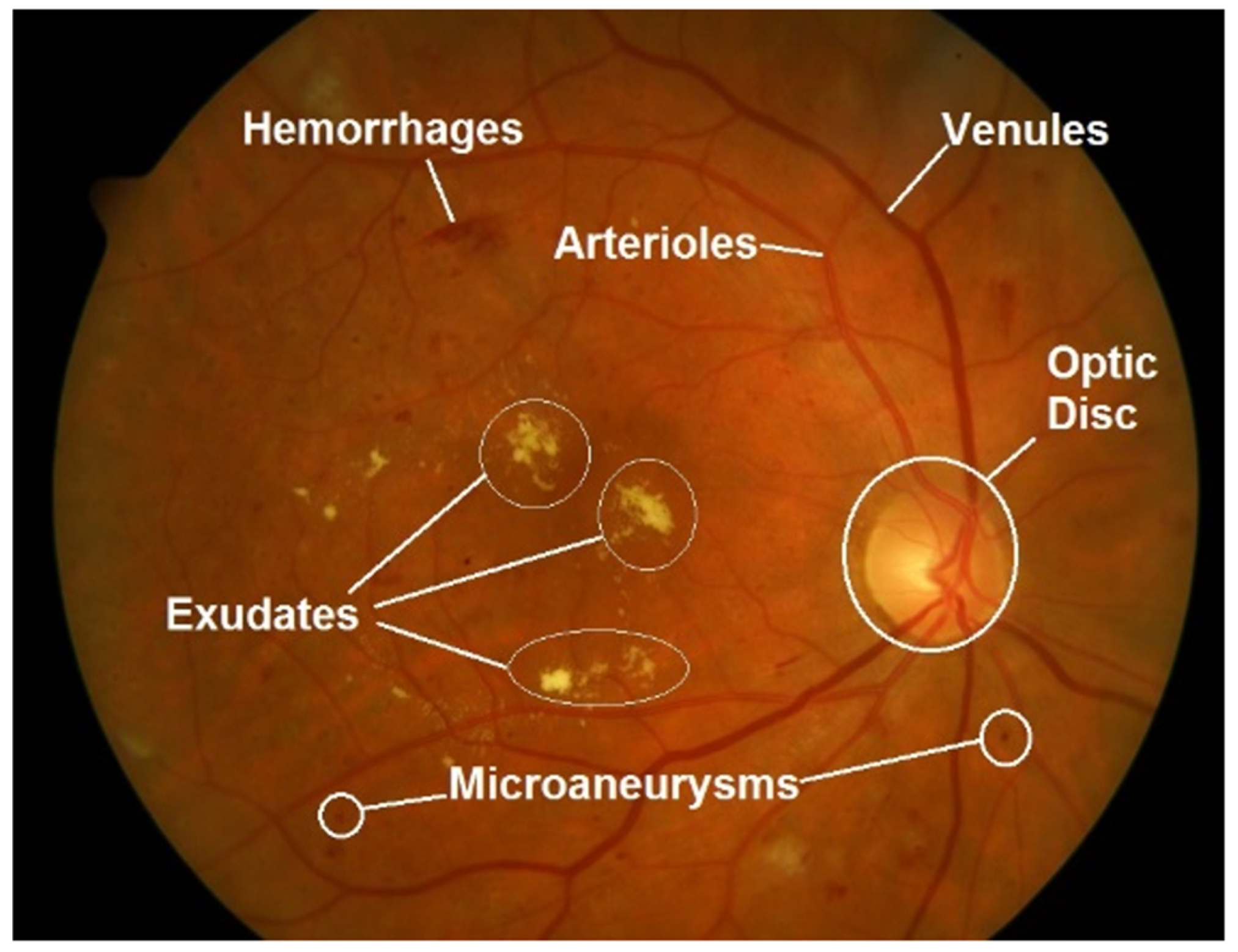
2.3. Retinal Fundus Images
3. Materials and Methods
3.1. Article Search and Selection Strategy
3.2. Datasets
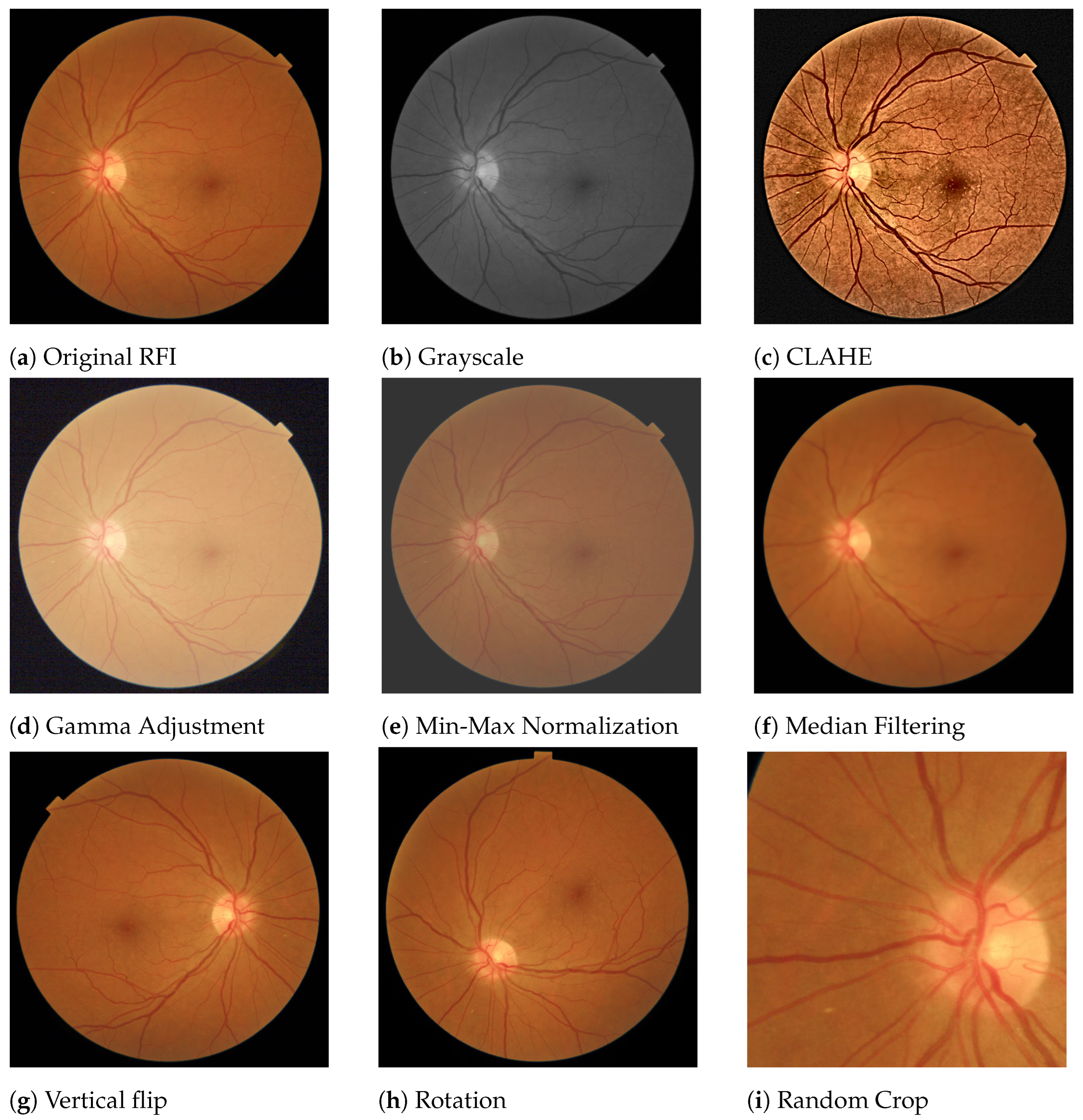
3.3. Pre-Processing Techniques
3.4. Evaluation Metrics
4. Automated Diagnosis of Cardiopathies
- Extracting biomarkers: Methods oriented to cardiovascular anomalies detection but only focused on retinal biomarker extraction.
- Prediction risk factors: Approaches to predict risk factors at the individual (chronological age) or metabolic (coronary artery calcium, hypertension) levels that may result in cardiovascular damage.
- Prediction of cardiovascular events: Direct cardiovascular events (stroke, ictus).
4.1. Automated Methods for Extracting Biomarkers
4.2. Automated Prediction of Cardiovascular Risk Factors
4.3. Automated Prediction of Cardiovascular Events
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| AUPRC | Area Under Precision-Recall Curve |
| AUROC | Area Under Receiver Operating Characteristic |
| BMI | Body Bass Index |
| CAC | Coronary Artery Calcium |
| CLAHE | Contrast Limited Adaptive Histogram Equalization |
| CMR | Cardiac Magnetic Resonance |
| CNN | Convolutional Neural Networks |
| CRVE | Center Retinal Venular Equivalent |
| CRAE | Central Retinal Arteriolar Equivalent |
| CT | Computed Tomography |
| CVD | Cardiovascular Diseases |
| DBN | Deep Belief Networks |
| DL | Deep Learning |
| DNFN | Deep Neuro-Fuzzy Network |
| DR | Diabetic Retinopathy |
| DSA | Digital Subtraction Angiography |
| DXA | Dual-energy X-ray Absorptiometry |
| FD | Fractal Dimension |
| GAN | Generative Adversarial Networks |
| LVEDV | Left Ventricular End-Diastolic Volume |
| LVM | Left Ventricular Mass |
| mcVAE | Multichannel Variational Autoencoder |
| ML | machine learning |
| RCNN | Region-based Convolutional Neural Network |
| RFI | Retina Fundus Image |
| RGB | Red Green Blue |
| RNN | Recurrent Neural Networks |
| ROP | Retinopathy of Prematurity |
| VGG | Visual Geometry Group |
References
- Tang, X. The role of artificial intelligence in medical imaging research. BJR Open 2019, 2, 20190031. [Google Scholar] [CrossRef] [PubMed]
- Kooi, T.; Litjens, G.; Van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Ghafoorian, M.; Karssemeijer, N.; Heskes, T.; van Uder, I.W.M.; de Leeuw, F.E.; Marchiori, E.; van Ginneken, B.; Platel, B. Non-uniform patch sampling with deep convolutional neural networks for white matter hyperintensity segmentation. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1414–1417. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Goutam, B.; Hashmi, M.F.; Geem, Z.W.; Bokde, N.D. A Comprehensive Review of Deep Learning Strategies in Retinal Disease Diagnosis Using Fundus Images. IEEE Access 2022. [Google Scholar] [CrossRef]
- Wagner, S.K.; Fu, D.J.; Faes, L.; Liu, X.; Huemer, J.; Khalid, H.; Ferraz, D.; Korot, E.; Kelly, C.; Balaskas, K.; et al. Insights into systemic disease through retinal imaging-based oculomics. Transl. Vis. Sci. Technol. 2020, 9, 6. [Google Scholar] [CrossRef]
- Kim, B.R.; Yoo, T.K.; Kim, H.K.; Ryu, I.H.; Kim, J.K.; Lee, I.S.; Kim, J.S.; Shin, D.H.; Kim, Y.S.; Kim, B.T. Oculomics for sarcopenia prediction: A machine learning approach toward predictive, preventive, and personalized medicine. EPMA J. 2022, 13, 367–382. [Google Scholar] [CrossRef]
- Harris, G.; Rickard, J.J.S.; Butt, G.; Kelleher, L.; Blanch, R.; Cooper, J.M.; Oppenheimer, P.G. Review: Emerging Oculomics based diagnostic technologies for traumatic brain injury. IEEE Rev. Biomed. Eng. 2022. [Google Scholar] [CrossRef]
- Maldonado García, C.; Bonazzola, R.; Ravikumar, N.; Frangi, A.F. Predicting Myocardial Infarction Using Retinal OCT Imaging. In Medical Image Understanding and Analysis; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2022; Volume 13413, pp. 787–797. [Google Scholar] [CrossRef]
- Sabanayagam, C.; Xu, D.; Ting, D.S.; Nusinovici, S.; Banu, R.; Hamzah, H.; Lim, C.; Tham, Y.C.; Cheung, C.Y.; Tai, E.S.; et al. A deep learning algorithm to detect chronic kidney disease from retinal photographs in community-based populations. Lancet Digit. Health 2020, 2, e295–e302. [Google Scholar] [CrossRef]
- Cheung, C.Y.; Ran, A.R.; Wang, S.; Chan, V.T.T.; Sham, K.; Hilal, S.; Venketasubramanian, N.; Cheng, C.Y.; Sabanayagam, C.; Tham, Y.C.; et al. A deep learning model for detection of Alzheimer’s disease based on retinal photographs: A retrospective, multicentre case-control study. Lancet Digit. Health 2022. [Google Scholar] [CrossRef] [PubMed]
- Mitani, A.; Huang, A.; Venugopalan, S.; Corrado, G.S.; Peng, L.; Webster, D.R.; Hammel, N.; Liu, Y.; Varadarajan, A.V. Detection of anaemia from retinal fundus images via deep learning. Nat. Biomed. Eng. 2020, 4, 18–27. [Google Scholar] [CrossRef] [PubMed]
- WHO. Cardiovascular Diseases (CVDs). 2021. Available online: https://www.who.int/en/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 20 November 2022).
- Wang, H.; Naghavi, M.; Allen, C.; Barber, R.M.; Bhutta, Z.A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; Coates, M.M.; et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef] [PubMed]
- Goff, D.C., Jr.; Lloyd-Jones, D.M.; Bennett, G.; Coady, S.; D’Agostino, R.B.; Gibbons, R.; Greenland, P.; Lackland, D.T.; Levy, D.; O’Donnell, C.J.; et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2013, 129, S49–S73. [Google Scholar] [CrossRef] [PubMed]
- Schmarje, L.; Santarossa, M.; Schröder, S.M.; Koch, R. A survey on semi-, self-and unsupervised learning for image classification. IEEE Access 2021, 9, 82146–82168. [Google Scholar] [CrossRef]
- Van Engelen, J.E.; Hoos, H.H. A survey on semi-supervised learning. Mach. Learn. 2020, 109, 373–440. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Abdullah, M.; Fraz, M.M.; Barman, S.A. Localization and segmentation of optic disc in retinal images using circular Hough transform and grow-cut algorithm. PeerJ 2016, 4, e2003. [Google Scholar] [CrossRef]
- Staal, J.; Abramoff, M.; Niemeijer, M.; Viergever, M.; van Ginneken, B. Ridge based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef]
- Decencière, E.; Zhang, X.; Cazuguel, G.; Lay, B.; Cochener, B.; Trone, C.; Gain, P.; Ordonez, R.; Massin, P.; Erginay, A.; et al. Feedback on a publicly distributed database: The Messidor database. Image Anal. Stereol. 2014, 33, 231–234. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Folk, J.C.; Han, D.P.; Walker, J.D.; Williams, D.F.; Russell, S.R.; Massin, P.; Cochener, B.; Gain, P.; Tang, L.; et al. Automated Analysis of Retinal Images for Detection of Referable Diabetic Retinopathy. JAMA Ophthalmol. 2013, 131, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Hoover, A.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Budai, A.; Bock, R.; Maier, A.; Hornegger, J.; Michelson, G. Robust vessel segmentation in fundus images. Int. J. Biomed. Imaging 2013, 2013, 154860. [Google Scholar] [CrossRef]
- Diabetic Retinopathy Detection | Kaggle. Available online: https://www.kaggle.com/c/diabetic-retinopathy-detection (accessed on 29 November 2022).
- Decencière, E.; Cazuguel, G.; Zhang, X.; Thibault, G.; Klein, J.C.; Meyer, F.; Marcotegui, B.; Quellec, G.; Lamard, M.; Danno, R.; et al. TeleOphta: Machine learning and image processing methods for teleophthalmology. IRBM 2013, 34, 196–203. [Google Scholar] [CrossRef]
- Zheng, Y.; Cheng, C.Y.; Lamoureux, E.L.; Chiang, P.P.C.; Rahman Anuar, A.; Wang, J.J.; Mitchell, P.; Saw, S.M.; Wong, T.Y. How much eye care services do Asian populations need? Projection from the Singapore Epidemiology of Eye Disease (SEED) study. Investig. Ophthalmol. Vis. Sci. 2013, 54, 2171–2177. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Cheng, J.; Xu, Y.; Zhang, C.; Wong, D.W.K.; Liu, J.; Cao, X. Disc-aware ensemble network for glaucoma screening from fundus image. IEEE Trans. Med. Imaging 2018, 37, 2493–2501. [Google Scholar] [CrossRef] [PubMed]
- Majithia, S.; Tham, Y.C.; Chee, M.L.; Teo, C.L.; Chee, M.L.; Dai, W.; Kumari, N.; Lamoureux, E.L.; Sabanayagam, C.; Wong, T.Y.; et al. Singapore Chinese Eye Study: Key findings from baseline examination and the rationale, methodology of the 6-year follow-up series. Br. J. Ophthalmol. 2020, 104, 610–615. [Google Scholar] [CrossRef]
- Foong, A.W.; Saw, S.M.; Loo, J.L.; Shen, S.; Loon, S.C.; Rosman, M.; Aung, T.; Tan, D.T.; Tai, E.S.; Wong, T.Y. Rationale and methodology for a population-based study of eye diseases in Malay people: The Singapore Malay eye study (SiMES). Ophthalmic Epidemiol. 2007, 14, 25–35. [Google Scholar] [CrossRef]
- Jonas, J.B.; Xu, L.; Wang, Y.X. The Beijing Eye Study. Acta Ophthalmol. 2009, 87, 247–261. [Google Scholar] [CrossRef]
- Zuiderveld, K. Contrast limited adaptive histogram equalization. In Academic Press Graphics Gems Series: Graphics Gems IV; Academic Press: London, UK, 1994; pp. 474–485. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Cheung, C.Y.L.; Ikram, M.K.; Sabanayagam, C.; Wong, T.Y. Retinal microvasculature as a model to study the manifestations of hypertension. Hypertension 2012, 60, 1094–1103. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Lin, Z.; Wang, W.; Tan, Z.; Shang, X.; Zhang, X.; Meng, W.; Ge, Z.; He, M. A Deep Learning System for Fully Automated Retinal Vessel Measurement in High Throughput Image Analysis. Front. Cardiovasc. Med. 2022, 9, 823436. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Yu, F.; Zhao, J.; Gong, Y.; Wang, Z.; Li, Y.; Yang, F.; Dong, B.; Li, Q.; Zhang, L. Annotation-Free Cardiac Vessel Segmentation via Knowledge Transfer from Retinal Images. arXiv 2019, arXiv:1907.11483. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Yin, X. Prediction Algorithm of Young Students’ Physical Health Risk Factors Based on Deep Learning. J. Healthc. Eng. 2021, 2021, 9049266. [Google Scholar] [CrossRef]
- Zekavat, S.M.; Raghu, V.K.; Trinder, M.; Ye, Y.; Koyama, S.; Honigberg, M.C.; Yu, Z.; Pampana, A.; Urbut, S.; Haidermota, S.; et al. Deep Learning of the Retina Enables Phenome- and Genome-Wide Analyses of the Microvasculature. Circulation 2022, 145, 134–150. [Google Scholar] [CrossRef]
- Hoque, M.E.; Kipli, K. Deep Learning in Retinal Image Segmentation and Feature Extraction: A Review. Int. J. Online Biomed. Eng. 2021, 17, 103–118. [Google Scholar] [CrossRef]
- Budoff, M.J.; Raggi, P.; Beller, G.A.; Berman, D.S.; Druz, R.S.; Malik, S.; Rigolin, V.H.; Weigold, W.G.; Soman, P.; the Imaging Council of the American College of Cardiology. Noninvasive cardiovascular risk assessment of the asymptomatic diabetic patient: The Imaging Council of the American College of Cardiology. JACC Cardiovasc. Imaging 2016, 9, 176–192. [Google Scholar] [CrossRef]
- Simó, R.; Bañeras, J.; Hernández, C.; Rodríguez-Palomares, J.; Valente, F.; Gutierrez, L.; González-Alujas, T.; Ferreira, I.; Aguadé-Bruix, S.; Montaner, J.; et al. Diabetic retinopathy as an independent predictor of subclinical cardiovascular disease: Baseline results of the PRECISED study. BMJ Open Diabetes Res. Care 2019, 7, e000845. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA J. Am. Med. Assoc. 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Gargeya, R.; Leng, T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology 2017, 124, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Routledge: London, UK, 2017. [Google Scholar]
- Shetkar, A.; Mai, C.K.; Yamini, C. Diabetic symptoms prediction through retinopathy. In Machine Learning Technologies and Applications; Springer: Berlin/Heidelberg, Germany, 2021; pp. 13–20. [Google Scholar]
- Ting, D.S.W.; Cheung, C.Y.L.; Lim, G.; Tan, G.S.W.; Quang, N.D.; Gan, A.; Hamzah, H.; Garcia-Franco, R.; Yeo, I.Y.S.; Lee, S.Y.; et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA J. Am. Med. Assoc. 2017, 318, 2211–2223. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.C.S.; Kiew, G.; Jeon, S.; Ting, D. Singapore Eye Lesions Analyzer (SELENA): The Deep Learning System for Retinal Diseases. In Artificial Intelligence in Ophthalmology; Grzybowski, A., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 177–185. [Google Scholar] [CrossRef]
- Trivedi, A.; Desbiens, J.; Gross, R.; Gupta, S.; Dodhia, R.; Ferres, J.L. Retinal Microvasculature as Biomarker for Diabetes and Cardiovascular Diseases. arXiv 2021, arXiv:2107.13157. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Redd, T.K.; Campbell, J.P.; Brown, J.M.; Kim, S.J.; Ostmo, S.; Chan, R.V.P.; Dy, J.; Erdogmus, D.; Ioannidis, S.; Kalpathy-Cramer, J.; et al. Evaluation of a deep learning image assessment system for detecting severe retinopathy of prematurity. Br. J. Ophthalmol. 2019, 103, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Poplin, R.; Varadarajan, A.V.; Blumer, K.; Liu, Y.; McConnell, M.V.; Corrado, G.S.; Peng, L.; Webster, D.R. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2018, 2, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Gerrits, N.; Elen, B.; Craenendonck, T.V.; Triantafyllidou, D.; Petropoulos, I.N.; Malik, R.A.; Boever, P.D. Age and sex affect deep learning prediction of cardiometabolic risk factors from retinal images. Sci. Rep. 2020, 10, 9432. [Google Scholar] [CrossRef] [PubMed]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Cheung, C.Y.; Xu, D.; Cheng, C.Y.; Sabanayagam, C.; Tham, Y.C.; Yu, M.; Rim, T.H.; Chai, C.Y.; Gopinath, B.; Mitchell, P.; et al. A deep-learning system for the assessment of cardiovascular disease risk via the measurement of retinal-vessel calibre. Nat. Biomed. Eng. 2021, 5, 498–508. [Google Scholar] [CrossRef]
- Rim, T.H.; Lee, G.; Kim, Y.; Tham, Y.C.; Lee, C.J.; Baik, S.J.; Kim, Y.A.; Yu, M.; Deshmukh, M.; Lee, B.K.; et al. Prediction of systemic biomarkers from retinal photographs: Development and validation of deep-learning algorithms. Lancet Digit. Health 2020, 2, e526–e536. [Google Scholar] [CrossRef]
- Nusinovici, S.; Rim, T.H.; Yu, M.; Lee, G.; Tham, Y.C.; Cheung, N.; Chong, C.C.Y.; Soh, Z.D.; Thakur, S.; Lee, C.J.; et al. Retinal photograph-based deep learning predicts biological age, and stratifies morbidity and mortality risk. Age Ageing 2022, 51. [Google Scholar] [CrossRef]
- Zhu, Z.; Chen, Y.; Wang, W.; Wang, Y.; Hu, W.; Shang, X.; Liao, H.; Shi, D.; Huang, Y.; Ha, J.; et al. Association of Retinal Age Gap with Arterial Stiffness and Incident Cardiovascular Disease. medRxiv 2022. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Diederichsen, A.; Mickley, H. Coronary Artery Calcium Score and Cardiovascular Event Prediction. JAMA 2010, 304, 741–742. [Google Scholar] [CrossRef] [PubMed]
- Greenland, P.; Blaha, M.J.; Budoff, M.J.; Erbel, R.; Watson, K.E. Coronary calcium score and cardiovascular risk. J. Am. Coll. Cardiol. 2018, 72, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Son, J.; Shin, J.Y.; Chun, E.J.; Jung, K.H.; Park, K.H.; Park, S.J. Predicting high coronary artery calcium score from retinal fundus images with deep learning algorithms. Transl. Vis. Sci. Technol. 2020, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rim, T.H.; Lee, C.J.; Tham, Y.C.; Cheung, N.; Yu, M.; Lee, G.; Kim, Y.; Ting, D.S.; Chong, C.C.Y.; Choi, Y.S.; et al. Deep-learning-based cardiovascular risk stratification using coronary artery calcium scores predicted from retinal photographs. Lancet Digit. Health 2021, 3, e306–e316. [Google Scholar] [CrossRef]
- Barriada, R.G.; Simó-Servat, O.; Planas, A.; Hernández, C.; Simó, R.; Masip, D. Deep Learning of Retinal Imaging: A Useful Tool for Coronary Artery Calcium Score Prediction in Diabetic Patients. Appl. Sci. 2022, 12, 1401. [Google Scholar] [CrossRef]
- Hubbard, L.D.; Brothers, R.J.; King, W.N.; Clegg, L.X.; Klein, R.; Cooper, L.S.; Sharrett, A.R.; Davis, M.D.; Cai, J.; Atherosclerosis Risk in Communities Study Group; et al. Methods for evaluation of retinal microvascular abnormalities associated with hypertension/sclerosis in the Atherosclerosis Risk in Communities Study. Ophthalmology 1999, 106, 2269–2280. [Google Scholar] [CrossRef]
- Zhang, L.; Yuan, M.; An, Z.; Zhao, X.; Wu, H.; Li, H.; Wang, Y.; Sun, B.; Li, H.; Ding, S.; et al. Prediction of hypertension, hyperglycemia and dyslipidemia from retinal fundus photographs via deep learning: A cross-sectional study of chronic diseases in central China. PLoS ONE 2020, 15, e0233166. [Google Scholar] [CrossRef]
- Srilakshmi, V.; Anuradha, K.; Shoba Bindu, C. Intelligent decision support system for cardiovascular risk prediction using hybrid loss deep joint segmentation and optimized deep learning. Adv. Eng. Softw. 2022, 173, 103198. [Google Scholar] [CrossRef]
- Chang, J.; Ko, A.; Park, S.M.; Choi, S.; Kim, K.; Kim, S.M.; Yun, J.M.; Kang, U.; Shin, I.H.; Shin, J.Y.; et al. Association of Cardiovascular Mortality and Deep Learning-Funduscopic Atherosclerosis Score derived from Retinal Fundus Images. Am. J. Ophthalmol. 2020, 217, 121–130. [Google Scholar] [CrossRef]
- Huang, F.; Lian, J.; Ng, K.S.; Shih, K.C.; Vardhanabhuti, V. Predicting CT-Based Coronary Artery Disease Using Vascular Biomarkers Derived from Fundus Photographs with a Graph Convolutional Neural Network. Diagnostics 2022, 12, 1390. [Google Scholar] [CrossRef]
- Lim, G.; Lim, Z.W.; Xu, D.; Ting, D.S.; Wong, T.Y.; Lee, M.L.; Hsu, W. Feature isolation for hypothesis testing in retinal imaging: An ischemic stroke prediction case study. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January 2019; Volume 33, pp. 9510–9515. [Google Scholar]
- Ma, Y.; Xiong, J.; Zhu, Y.; Ge, Z.; Hua, R.; Fu, M.; Li, C.; Wang, B.; Dong, L.; Zhao, X.; et al. Development and validation of a deep learning algorithm using fundus photographs to predict 10-year risk of ischemic cardiovascular diseases among Chinese population. medRxiv 2021. [Google Scholar] [CrossRef]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Revathi, T.K.; Sathiyabhama, B.; Sankar, S. Diagnosing Cardio Vascular Disease (CVD) using Generative Adversarial Network (GAN) in Retinal Fundus Images. Ann. Rom. Soc. Cell Biol. 2021, 25, 2563–2572. [Google Scholar]
- Al-Absi, H.R.H.; Islam, M.T.; Refaee, M.A.; Chowdhury, M.E.H.; Alam, T. Cardiovascular Disease Diagnosis from DXA Scan and Retinal Images Using Deep Learning. Sensors 2022, 22, 4310. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Pinto, A.; Ravikumar, N.; Attar, R.; Suinesiaputra, A.; Zhao, Y.; Levelt, E.; Dall’Armellina, E.; Lorenzi, M.; Chen, Q.; Keenan, T.D.; et al. Predicting myocardial infarction through retinal scans and minimal personal information. Nat. Mach. Intell. 2022, 4, 55–61. [Google Scholar] [CrossRef]
- Antelmi, L.; Ayache, N.; Robert, P.; Lorenzi, M. Sparse multi-channel variational autoencoder for the joint analysis of heterogeneous data. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 302–311. [Google Scholar]

| Name | #Images | Size | Labels | Application |
|---|---|---|---|---|
| Public Access | ||||
| DRIVE [22] | 40 | 33—Normal, 7—Mild early DR | Vessel segmentation, DR diagnosis | |
| Messidor-1 [23] | 1200 | , , | Macular Edema, DR diagnosis | |
| Messidor-2 [24] | 1748 | , , | 0-None, 1-Mild, 2-Moderate, 3-Severe, 4-Proliferative | DR prediction |
| STARE [25] | 400 | Vessel segmentation | ||
| HRF [26] | 45 | 15—healthy, 15— DR, 15—glaucoma | Vessel segmentation, DR diagnosis, Glaucoma assessment | |
| Kaggle/EyePACS [27] | 9963 | DR scale of 0–4 | DR diagnosis | |
| Restricted Access | ||||
| E-Optha [28] | 463 | , , | 7—exudates and 35—no lesion 148—microaneurysms/ small hemorrhages and 233—no lesion | DR diagnosis |
| SEED [29] | 235 | 43—Glaucomatous | Glaucoma assessment | |
| SiNDI [30] | 5783 | 5670—Healthy, 113—Glaucomatous | Glaucoma assessment | |
| SCES [31] | 1751 | CVD assessment | ||
| SiMES [32] | 1488 | CVD assessment | ||
| BES [33] | 8585 | CVD assessment |
| Metric | Formula | Description |
|---|---|---|
| Accuracy | Indicates the global ratio of correct predictions either positive or negative | |
| Precision/PPV | Also known as Positive Predictive Value. Is the average of the retrieved samples that were relevant | |
| Sensitivity/Recall/TPR | Also know as True Positive Ratio, indicates the ratio of the relevant samples that are successfully identified. | |
| Specificity/FPR | Also know as False Positive Ratio, is the ratio of identified negative samples that are actually negative. | |
| F1-measure | Represents the harmonic mean of the precision and recall | |
| AUROC/AUC | Area Under Receiver Operating Characteristic. Relates TPR against FPR. It depicts the prediction capability of a classifier system as its discrimination threshold is varied | |
| AUPRC | Area Under Precision-Recall Curve. Relates Precision and Recall providing a single number that summarizes the information in the PR curve | |
| Sørensen–Dice coefficient | The Sørensen–Dice coefficient, or simply Dice score index, is a statistic metric used to compare the similarity of two samples | |
| R² | The coefficient of determination, or R squared, is a statistical variable that represents the ratio of the variation in the dependent variable that is predictable from the independent variable(s) | |
| CRAE | Center Retinal Venular Equivalent. Expressed in | |
| CRVE | Central Retinal Arteriolar Equivalent. Expressed in |
| Retinal Parameter | Change | Outcome | |
|---|---|---|---|
| Tortuosity | Retinal arteriolar tortuosity | Increased | Current blood pressure and early kidney disease |
| Decreased | Current blood pressure and ischemic heart disease | ||
| Retinal venular tortuosity | Increased | Current blood pressure | |
| Fractal dimension (FD) | Retinal vascular FD | Increased | Acute lacunar stroke |
| Decreased | Current blood pressure, lacunar and incident stroke | ||
| Suboptimal | Chronic kidney disease and coronary heart disease | ||
| Bifurcation | Retinal arteriolar branching angle | Decreased | Current blood pressure |
| Retinal arteriolar branching asymmetry ratio | Increased | Current blood pressure | |
| Retinal arteriolar length: diameter ratio | Increased | Current blood pressure, hypertension and stroke | |
| Retinal arteriolar branching coefficient (optimal ratio) | Increased | ischemic heart disease | |
| Retinal arteriolar optimal parameter (deviation of junction exponent) | Decreased | Peripheral vascular disease. |
| Ref. | Application | Architectures | Metrics | Result | Dataset | Pre-Processing |
|---|---|---|---|---|---|---|
| [37] | Vessel segmentation | U-net | AUC | >0.90 (AUC) | UK Biobank (420), 21 datasets (4015), but filtering images with labels: Macular edema, Hypertensive, pathologic myopia and DR | CLAHE |
| [39] | Vessel segmentation | SC-GAN and U-Net | Acc, Precision, recall, Dice score | 0.953 (Acc) | DRIVE | Grayscale, Median-filtering, CLAHE |
| [41] | Vessel segmentation | Custom U-Net-based | AUC, Acc, sensitivity, specificity | 0.98 (AUC) | DRIVE, STARE | |
| [42] | Retinal vessel detection | U-Net | AUC, Acc (DICE score) | 0.99 (AUC) in FD, 0.88 (AUC) in vascular density | UK BioBank (97895) | |
| [43] | Vessel detection | Fast RCNN | Sensitivity, Positive Predictive Value (PPV) | 0.92 (sensitivity) | HRF, DRIVE, STARE, MESSIDOR, (450) | CLAHE |
| Ref. | Application | Architectures | Metrics | Result | Dataset | Pre-Processing |
|---|---|---|---|---|---|---|
| [46] | DR, Macular edema prediction | InceptionV3 | AUC, sensitivity and specificity | 0.99 (AUC) | EyePACS (128175): train, EyePACS (9963) and Messidor-2 (1748): test | |
| [48] | DR screening | Custom +Decision Tree | AUC, sensitivity and specificity | 0.95 (AUC) | Total EyePACS MESSIDOR, E-Optha, (75137 in total) | Crop, brightness and contrast adjustment, DA: rotations |
| [50] | DR assessment | Custom CNN | Acc | 0.89 (Acc) | Custom (150) | Min–Max normalization |
| [51] | DR, Glaucoma, AMD | Adapted VGG | AUC, sensitivity and specificity | 0.95 (AUC), 0.94 (AUC), 0.93 (AUC) | Several datasets with more than 300,000 RFI just for train | |
| [52] | DR, Glaucoma, AMD | VGG19 | AUC, Sensitivity and Specificity | 0.93 (AUC) | 10 diff studies: (76,730): train, (112648): test | |
| [53] | DR assessment | U-Net, ResNet101 | AUC | 0.93 (AUC) | IRIS | Grayscale, CLAHE, Gamma-Adjustment |
| [55] | Retinopathy of prematurity detection | U-Net | AUC, sensitivity and specificity | 0.96 (AUC) | i-ROP Study (4861) | |
| [57] | CVD diagnosis | Inception-v3 + Ensembling of 10 iterations | AUC | 0.70 (AUC) | Biobank (48,101), EyePACs (23,6234): train, Bionbank (12,026), EyePacs (999): test | |
| [58] | Biomarker prediction | MobileNet-V2 | MAE, AUC, | 0.97 (AUC), 0.78 (AUC) | Qatar Biobank (12,000) | Gaussian filter, Crop, DA: flip, random rotation, shift |
| [61] | CVD risk, blood presure, body-mass index, total cholesterol, and glycated-hemoglobin level. | SIVA-DLS | , | SEED (Singapore Epidemiology of Eye Disease), BES (Beijing Eye Study), UK Biobank, Kangbuk Samsung Health (KSH), The Austin Health Study (Austin study). | ||
| [62] | 47 biomarkers prediction | VGG16 | AUC, | 0.90 (AUC) | Two health screening centers in South Korea, the BES, SEED, UK Biobank (236,257) | CLAHE, DA: random crop, flip up-down, rotation, brightness, and saturation |
| [63] | BA prediction | VGG- | c-index, sensitivity, specificity | 0.76 (sensitivity) | KHS (129,236): train, UK Biobank: test | |
| [64] | Retinal Age prediction | Xception | p-value and MSA | 0.80 (p < 0.001), 3.55 (MSA) | UK Biobank (19,200) | |
| [68] | CAC assessment | InceptionV3 | AUC | 0.83(AUC) | Seoul National University Bundang Hospital (44,184) | Crop DA: flip, rotation |
| [69] | CAC assessment | Custom+ EfficientNet | AUC | 0.74 (AUC) | Biobank, Shouth Korean Datasets (216,152) | CLAHE, DA: random crop, random rotation |
| [70] | CAC assessment | VGG16 | Acc, Rec, Pre, F1, CM | 0.78 (Acc), 0.75 (Rec), 0.91 (Pre) | Endocrinology Department, Vall d’Hebron University Hospital (152) | Crop |
| [72] | Proxys: hypertension, hyperglycemia and dyslipidemia | InceptionV3 | AUC, Acc | 0.76 (AUC), 0.88 (AUC), 0.70 (AUC) | China dataset (1222) | DA in minority classes |
| [73] | Hypertension prediction | DNFN | Acc, Rec, Pre | 0.91(Acc) | fundusimage1000 | Grayscale |
| [74] | Atherosclerosis assessment | Xception | AUROC, AUPRC, accuracy, sensitivity, specificity, positive and negative predictive values | 0.71 (AUC) | Health Promotion Center of Seoul National University Hospital (15,408) | DA: Random zoom, horizontal flip |
| [75] | Coronary Artery Disease prediction | GraphSAGE | Acc, Rec, Pre, AUC | 0.69 (AUC) | Custom |
| Ref. | Application | Architectures | Metrics | Result | Dataset | Pre-Processing |
|---|---|---|---|---|---|---|
| [76] | Stroke risk prediction | U-Net, VGG19 | AUC, sensitivity and specificity | 0.96 (AUC) | Singapore, Sydney and Melbourne and SCES, SiMES, SiNDI, DMPMelb, SP2 | DA: random flipping and rotation |
| [77] | ICVD Diagnosis | Inception-ResNet-V2 | AUC and | 0.97 (AUC) | China and Beijing Research on Aging, BRAVE (411518) | |
| [79] | CVD diagnosis | InceptionV3 | Acc, F1 | 0.91 (Acc) | Biobank and EyePACS, STARE | |
| [80] | CVD diagnosis | CNN-ResNet+Custom | Acc, Rec, Pre | 0.75 (Acc) | Qatar BioBank (1805) | Crop, Mean-filtering |
| [81] | Myocardial infarction prediction | Multichannel variational autoencoder, CNN-ResNet50 | AUC, Sensitivity and Specificity | 0.80 (AUC) | UK Biobank (71515) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barriada, R.G.; Masip, D. An Overview of Deep-Learning-Based Methods for Cardiovascular Risk Assessment with Retinal Images. Diagnostics 2023, 13, 68. https://doi.org/10.3390/diagnostics13010068
Barriada RG, Masip D. An Overview of Deep-Learning-Based Methods for Cardiovascular Risk Assessment with Retinal Images. Diagnostics. 2023; 13(1):68. https://doi.org/10.3390/diagnostics13010068
Chicago/Turabian StyleBarriada, Rubén G., and David Masip. 2023. "An Overview of Deep-Learning-Based Methods for Cardiovascular Risk Assessment with Retinal Images" Diagnostics 13, no. 1: 68. https://doi.org/10.3390/diagnostics13010068
APA StyleBarriada, R. G., & Masip, D. (2023). An Overview of Deep-Learning-Based Methods for Cardiovascular Risk Assessment with Retinal Images. Diagnostics, 13(1), 68. https://doi.org/10.3390/diagnostics13010068