Endotoxin in Sepsis: Methods for LPS Detection and the Use of Omics Techniques
Abstract
:1. Introduction
1.1. Diagnosis of Sepsis
1.2. Gram-Negative Sepsis and Endotoxin
2. Endotoxin: Structure and Characteristics
3. Endotoxin Removal via Extracorporeal Therapies
4. Omics Techniques for Sepsis
4.1. Proteomics
4.2. Endotoxin Detection
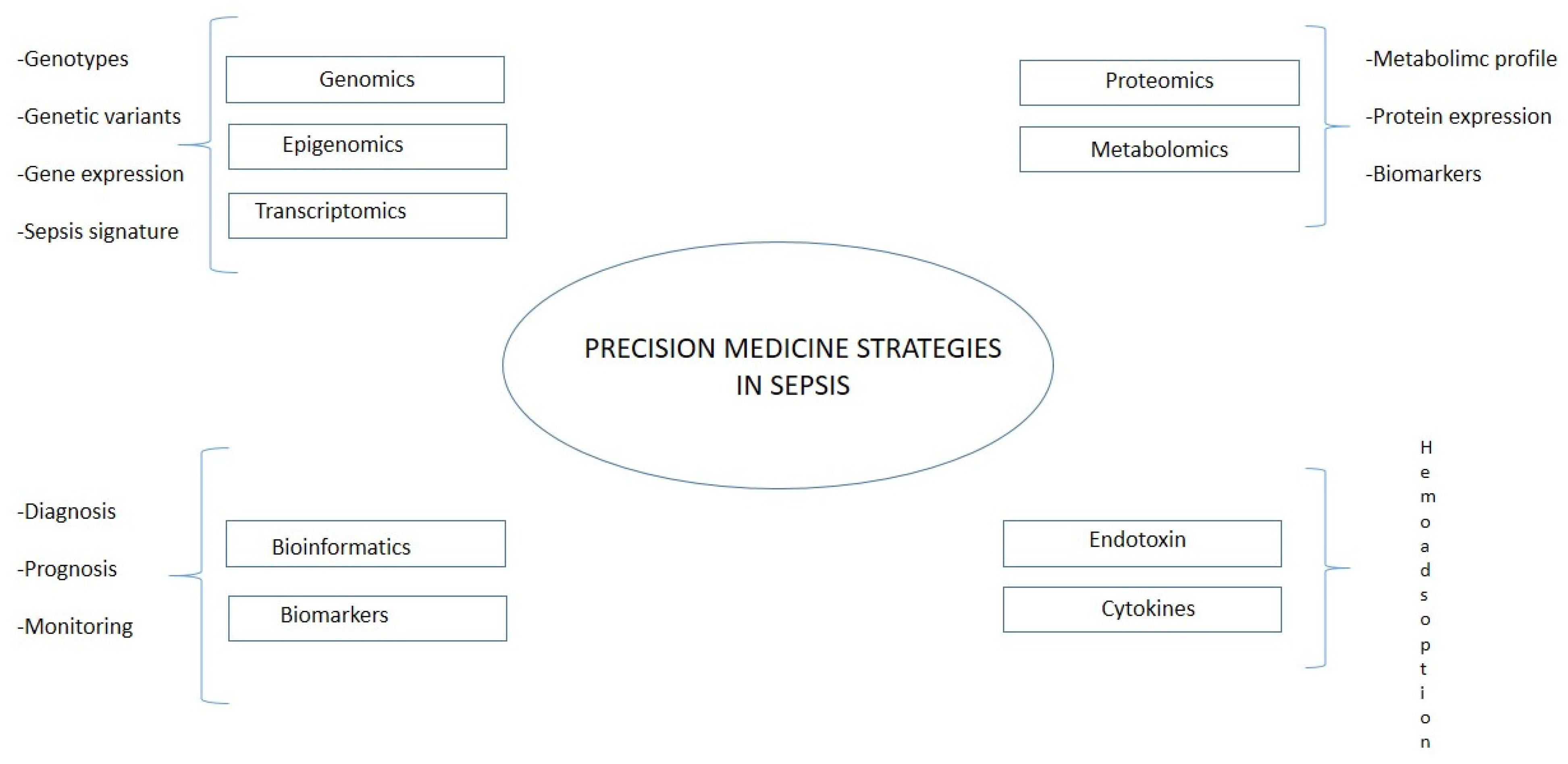
5. Sepsis in the Era of Precision Medicine
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA J. Am. Med. Assoc. 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Riedemann, N.C.; Guo, R.-F.; Ward, P.A. The Enigma of Sepsis. J. Clin. Investig. 2003, 112, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Ianaro, A.; Tersigni, M.; D’Acquisto, F. New Insight in LPS Antagonist. Mini Rev. Med. Chem. 2009, 9, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Sondhi, P.; Maruf, M.H.U.; Stine, K.J. Nanomaterials for Biosensing Lipopolysaccharide. Biosensors 2019, 10, 2. [Google Scholar] [CrossRef] [Green Version]
- Fleischmann, C.; Scherag, A.; Adhikari, N.K.J.; Hartog, C.S.; Tsaganos, T.; Schlattmann, P.; Angus, D.C.; Reinhart, K. Assessment of Global Incidence and Mortality of Hospital-Treated Sepsis. Current Estimates and Limitations. Am. J. Respir. Crit. Care Med. 2016, 193, 259–272. [Google Scholar] [CrossRef]
- Xie, J.; Wang, H.; Kang, Y.; Zhou, L.; Liu, Z.; Qin, B.; Ma, X.; Cao, X.; Chen, D.; Lu, W.; et al. The Epidemiology of Sepsis in Chinese ICUs: A National Cross-Sectional Survey. Crit. Care Med. 2020, 48, e209–e218. [Google Scholar] [CrossRef]
- Miao, H.; Chen, S.; Ding, R. Evaluation of the Molecular Mechanisms of Sepsis Using Proteomics. Front. Immunol. 2021, 12, 733537. [Google Scholar] [CrossRef]
- Virzì, G.M.; Borga, C.; Pasqualin, C.; Pastori, S.; Brocca, A.; de Cal, M.; Nalesso, F.; Zanella, M.; Brendolan, A.; Ronco, C. Direct Effect of Septic Plasma in Human Cell Lines Viability. Blood Purif. 2019, 47, 270–276. [Google Scholar] [CrossRef]
- Virzì, G.M.; Clementi, A.; Brocca, A.; de Cal, M.; Marcante, S.; Ronco, C. Cardiorenal Syndrome Type 5 in Sepsis: Role of Endotoxin in Cell Death Pathways and Inflammation. Kidney Blood Press. Res. 2016, 41, 1008–1015. [Google Scholar] [CrossRef]
- Wang, X.; Quinn, P.J. Lipopolysaccharide: Biosynthetic Pathway and Structure Modification. Prog. Lipid Res. 2010, 49, 97–107. [Google Scholar] [CrossRef]
- Clementi, A.; Virzì, G.M.; Brocca, A.; Ronco, C. The Role of Endotoxin in the Setting of Cardiorenal Syndrome Type 5. Cardiorenal Med. 2017, 7, 276–283. [Google Scholar] [CrossRef]
- Nalesso, F.; Cattarin, L.; Gobbi, L.; Fragasso, A.; Garzotto, F.; Calò, L.A. Evaluating Nephrocheck(®) as a Predictive Tool for Acute Kidney Injury. Int. J. Nephrol. Renovasc. Dis. 2020, 13, 85–96. [Google Scholar] [CrossRef] [Green Version]
- Tavener, S.A.; Long, E.M.; Robbins, S.M.; McRae, K.M.; Van Remmen, H.; Kubes, P. Immune Cell Toll-like Receptor 4 Is Required for Cardiac Myocyte Impairment during Endotoxemia. Circ. Res. 2004, 95, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Angus, D.C.; van der Poll, T. Severe Sepsis and Septic Shock. N. Engl. J. Med. 2013, 369, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Steinhagen, F.; Schmidt, S.V.; Schewe, J.-C.; Peukert, K.; Klinman, D.M.; Bode, C. Immunotherapy in Sepsis-Brake or Accelerate? Pharmacol. Ther. 2020, 208, 107476. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.R.S.; Picco, N.; Morgan, B.P.; Ghazal, P. Sepsis Target Validation for Repurposing and Combining Complement and Immune Checkpoint Inhibition Therapeutics. Expert Opin. Drug Discov. 2021, 16, 537–551. [Google Scholar] [CrossRef]
- American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: Definitions for Sepsis and Organ Failure and Guidelines for the Use of Innovative Therapies in Sepsis. Crit. Care Med. 1992, 20, 864–874. [CrossRef]
- Vincent, J.L.; Moreno, R.; Takala, J.; Willatts, S.; De Mendonça, A.; Bruining, H.; Reinhart, C.K.; Suter, P.M.; Thijs, L.G. The SOFA (Sepsis-Related Organ Failure Assessment) Score to Describe Organ Dysfunction/Failure. On Behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996, 22, 707–710. [Google Scholar] [CrossRef]
- Levy, M.M.; Fink, M.P.; Marshall, J.C.; Abraham, E.; Angus, D.; Cook, D.; Cohen, J.; Opal, S.M.; Vincent, J.-L.; Ramsay, G. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Intensive Care Med. 2003, 29, 530–538. [Google Scholar] [CrossRef]
- Marshall, J.C.; Cook, D.J.; Christou, N.V.; Bernard, G.R.; Sprung, C.L.; Sibbald, W.J. Multiple Organ Dysfunction Score: A Reliable Descriptor of a Complex Clinical Outcome. Crit. Care Med. 1995, 23, 1638–1652. [Google Scholar] [CrossRef]
- Grattard, F.; Allegra, S.; Morel, J.; Court-Fortune, I.; Auboyer, C.; Pozzetto, B.; Berthelot, P. Septic Shock Due to Legionella Pneumophila Serogroup 2: Usefulness of Molecular Biology for Diagnosis, Treatment and Epidemiological Investigation. Intensive Care Med. 2010, 36, 1439–1440. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; De Simone, G.; Boccia, G.; De Caro, F.; Pagliano, P. Sepsis and Septic Shock: New Definitions, New Diagnostic and Therapeutic Approaches. J. Glob. Antimicrob. Resist. 2017, 10, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Janosevic, D.; Myslinski, J.; McCarthy, T.W.; Zollman, A.; Syed, F.; Xuei, X.; Gao, H.; Liu, Y.-L.; Collins, K.S.; Cheng, Y.-H.; et al. The Orchestrated Cellular and Molecular Responses of the Kidney to Endotoxin Define a Precise Sepsis Timeline. eLife 2021, 10, 62270. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R. Biochemistry of Endotoxins. Annu. Rev. Biochem. 1990, 59, 129–170. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, K.; Takeuchi, O.; Kawai, T.; Sanjo, H.; Ogawa, T.; Takeda, Y.; Takeda, K.; Akira, S. Cutting Edge: Toll-like Receptor 4 (TLR4)-Deficient Mice Are Hyporesponsive to Lipopolysaccharide: Evidence for TLR4 as the Lps Gene Product. J. Immunol. 1999, 162, 3749–3752. [Google Scholar] [CrossRef]
- González-Bello, C. The Inhibition of Lipid A Biosynthesis—The Antidote Against Superbugs? Adv. Ther. 2019, 2, 1800117. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide Endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [Green Version]
- Joiner, K.A. Complement Evasion by Bacteria and Parasites. Annu. Rev. Microbiol. 1988, 42, 201–230. [Google Scholar] [CrossRef]
- Monard, C.; Rimmelé, T.; Ronco, C. Extracorporeal Blood Purification Therapies for Sepsis. Blood Purif. 2019, 47, 2–15. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Laterre, P.-F.; Cohen, J.; Burchardi, H.; Bruining, H.; Lerma, F.A.; Wittebole, X.; De Backer, D.; Brett, S.; Marzo, D.; et al. A Pilot-Controlled Study of a Polymyxin B-Immobilized Hemoperfusion Cartridge in Patients with Severe Sepsis Secondary to Intra-Abdominal Infection. Shock 2005, 23, 400–405. [Google Scholar] [CrossRef]
- Cruz, D.N.; Antonelli, M.; Fumagalli, R.; Foltran, F.; Brienza, N.; Donati, A.; Malcangi, V.; Petrini, F.; Volta, G.; Bobbio Pallavicini, F.M.; et al. Early Use of Polymyxin B Hemoperfusion in Abdominal Septic Shock: The EUPHAS Randomized Controlled Trial. JAMA 2009, 301, 2445–2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payen, D.M.; Guilhot, J.; Launey, Y.; Lukaszewicz, A.C.; Kaaki, M.; Veber, B.; Pottecher, J.; Joannes-Boyau, O.; Martin-Lefevre, L.; Jabaudon, M.; et al. Early Use of Polymyxin B Hemoperfusion in Patients with Septic Shock Due to Peritonitis: A Multicenter Randomized Control Trial. Intensive Care Med. 2015, 41, 975–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dellinger, R.P.; Bagshaw, S.M.; Antonelli, M.; Foster, D.M.; Klein, D.J.; Marshall, J.C.; Palevsky, P.M.; Weisberg, L.S.; Schorr, C.A.; Trzeciak, S.; et al. Effect of Targeted Polymyxin B Hemoperfusion on 28-Day Mortality in Patients With Septic Shock and Elevated Endotoxin Level: The EUPHRATES Randomized Clinical Trial. JAMA 2018, 320, 1455–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipcsey, M.; Tenhunen, J.; Sjölin, J.; Frithiof, R.; Bendel, S.; Flaatten, H.; Kawati, R.; Kuitunen, A.; Tønnessen, T.I.; Rubertsson, S. Abdominal Septic Shock-Endotoxin Adsorption Treatment (ASSET)-Endotoxin Removal in Abdominal and Urogenital Septic Shock with the Alteco® LPS Adsorber: Study Protocol for a Double-Blinded, Randomized Placebo-Controlled Trial. Trials 2016, 17, 587. [Google Scholar] [CrossRef] [Green Version]
- Broman, M.E.; Hansson, F.; Vincent, J.-L.; Bodelsson, M. Endotoxin and Cytokine Reducing Properties of the OXiris Membrane in Patients with Septic Shock: A Randomized Crossover Double-Blind Study. PLoS ONE 2019, 14, e0220444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, E.T.-Y.; Ong, V.; Remani, D.; Wong, W.-K.; Haroon, S.; Lau, T.; Nyeo, H.-Q.; Mukhopadhyay, A.; Tan, B.-H.; Chua, H.-R. Filter Life and Safety of Heparin-Grafted Membrane for Continuous Renal Replacement Therapy-A Randomized Controlled Trial. Semin. Dial. 2021, 34, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Andrei, S.; Nguyen, M.; Berthoud, V.; Morgant, M.-C.; Bouhemad, B.; Guinot, P.-G. Evaluation of the Oxiris Membrane in Cardiogenic Shock Requiring Extracorporeal Membrane Oxygenation Support: Study Protocol for a Single Center, Single-Blind, Randomized Controlled Trial. Front. Cardiovasc. Med. 2021, 8, 738496. [Google Scholar] [CrossRef] [PubMed]
- Abou-Arab, O.; Huette, P.; Haye, G.; Guilbart, M.; Touati, G.; Diouf, M.; Beyls, C.; Dupont, H.; Mahjoub, Y. Effect of the OXiris Membrane on Microcirculation after Cardiac Surgery under Cardiopulmonary Bypass: Study Protocol for a Randomised Controlled Trial (OXICARD Study). BMJ Open 2021, 11, e044424. [Google Scholar] [CrossRef]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: The Re-Emerging Antibiotic for Multidrug-Resistant Gram-Negative Bacterial Infections. Lancet Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Payen, D. Haemoperfusion with Polymyxin B Membrane: Recent Results for an Old Debate! Anaesth. Crit. Care Pain Med. 2019, 38, 3–4. [Google Scholar] [CrossRef]
- Klein, D.J.; Foster, D.; Schorr, C.A.; Kazempour, K.; Walker, P.M.; Dellinger, R.P. The EUPHRATES Trial (Evaluating the Use of Polymyxin B Hemoperfusion in a Randomized Controlled Trial of Adults Treated for Endotoxemia and Septic Shock): Study Protocol for a Randomized Controlled Trial. Trials 2014, 15, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.; Tu, Y.-K.; Lee, C.-T.; Chao, A.; Huang, C.-H.; Wang, M.-J.; Yeh, Y.-C. Effects of Polymyxin B Hemoperfusion on Mortality in Patients With Severe Sepsis and Septic Shock: A Systemic Review, Meta-Analysis Update, and Disease Severity Subgroup Meta-Analysis. Crit. Care Med. 2017, 45, e858–e864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamik, B.; Zielinski, S.; Smiechowicz, J.; Kübler, A. Endotoxin Elimination in Patients with Septic Shock: An Observation Study. Arch. Immunol. Ther. Exp. 2015, 63, 475–483. [Google Scholar] [CrossRef] [Green Version]
- Kellum, J.A.; Dishart, M.K. Effect of Hemofiltration Filter Adsorption on Circulating IL-6 Levels in Septic Rats. Crit. Care 2002, 6, 429–433. [Google Scholar] [CrossRef]
- Su, W.; Ding, X. Methods of Endotoxin Detection. J. Lab. Autom. 2015, 20, 354–364. [Google Scholar] [CrossRef]
- Cooper, J.F.; Levin, J.; Wagner, H.N.J. Quantitative Comparison of in Vitro and in Vivo Methods for the Detection of Endotoxin. J. Lab. Clin. Med. 1971, 78, 138–148. [Google Scholar] [PubMed]
- Muta, T.; Oda, T.; Iwanaga, S. Horseshoe Crab Coagulation Factor B. A Unique Serine Protease Zymogen Activated by Cleavage of an Ile-Ile Bond. J. Biol. Chem. 1993, 268, 21384–21388. [Google Scholar] [CrossRef]
- Nachum, R.; Berzofsky, R.N. Chromogenic Limulus Amoebocyte Lysate Assay for Rapid Detection of Gram-Negative Bacteriuria. J. Clin. Microbiol. 1985, 21, 759–763. [Google Scholar] [CrossRef] [Green Version]
- Qiao, J.; Cui, L. Multi-Omics Techniques Make It Possible to Analyze Sepsis-Associated Acute Kidney Injury Comprehensively. Front. Immunol. 2022, 13, 905601. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.V.; Hu, Y.-J. Integrative Analysis of Multi-Omics Data for Discovery and Functional Studies of Complex Human Diseases. Adv. Genet. 2016, 93, 147–190. [Google Scholar] [CrossRef]
- Bahassi, E.M.; Stambrook, P.J. Next-Generation Sequencing Technologies: Breaking the Sound Barrier of Human Genetics. Mutagenesis 2014, 29, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Henry, V.J.; Bandrowski, A.E.; Pepin, A.-S.; Gonzalez, B.J.; Desfeux, A. OMICtools: An Informative Directory for Multi-Omic Data Analysis. Database 2014, 2014, bau069. [Google Scholar] [CrossRef] [Green Version]
- van Karnebeek, C.D.M.; Wortmann, S.B.; Tarailo-Graovac, M.; Langeveld, M.; Ferreira, C.R.; van de Kamp, J.M.; Hollak, C.E.; Wasserman, W.W.; Waterham, H.R.; Wevers, R.A.; et al. The Role of the Clinician in the Multi-Omics Era: Are You Ready? J. Inherit. Metab. Dis. 2018, 41, 571–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langston, J.C.; Rossi, M.T.; Yang, Q.; Ohley, W.; Perez, E.; Kilpatrick, L.E.; Prabhakarpandian, B.; Kiani, M.F. Omics of Endothelial Cell Dysfunction in Sepsis. Vasc. Biol. 2022, 4, R15–R34. [Google Scholar] [CrossRef] [PubMed]
- Seki, N.; Muta, T.; Oda, T.; Iwaki, D.; Kuma, K.; Miyata, T.; Iwanaga, S. Horseshoe Crab (1,3)-Beta-D-Glucan-Sensitive Coagulation Factor G. A Serine Protease Zymogen Heterodimer with Similarities to Beta-Glucan-Binding Proteins. J. Biol. Chem. 1994, 269, 1370–1374. [Google Scholar] [CrossRef]
- Guntupalli, R.; Hu, J.; Lakshmanan, R.S.; Huang, T.S.; Barbaree, J.M.; Chin, B.A. A Magnetoelastic Resonance Biosensor Immobilized with Polyclonal Antibody for the Detection of Salmonella Typhimurium. Biosens. Bioelectron. 2007, 22, 1474–1479. [Google Scholar] [CrossRef]
- Yang, M.; Kostov, Y.; Bruck, H.A.; Rasooly, A. Carbon Nanotubes with Enhanced Chemiluminescence Immunoassay for CCD-Based Detection of Staphylococcal Enterotoxin B in Food. Anal. Chem. 2008, 80, 8532–8537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, N.K.; Salomao, R. Sepsis Through the Eyes of Proteomics: The Progress in the Last Decade. Shock 2017, 47, 17–25. [Google Scholar] [CrossRef]
- Hermann, T.; Patel, D.J. Adaptive Recognition by Nucleic Acid Aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [Green Version]
- Petricoin, E.F.; Zoon, K.C.; Kohn, E.C.; Barrett, J.C.; Liotta, L.A. Clinical Proteomics: Translating Benchside Promise into Bedside Reality. Nat. Rev. Drug Discov. 2002, 1, 683–695. [Google Scholar] [CrossRef]
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and Their Applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Schneiderhan-Marra, N.; Joos, T.O. Protein Microarrays for Personalized Medicine. Clin. Chem. 2010, 56, 376–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, K.R.; Hummon, A.B. Mass Spectrometry for the Discovery of Biomarkers of Sepsis. Mol. Biosyst. 2017, 13, 648–664. [Google Scholar] [CrossRef] [Green Version]
- Blangy-Letheule, A.; Persello, A.; Rozec, B.; De Waard, M.; Lauzier, B. New Approaches to Identify Sepsis Biomarkers: The Importance of Model and Sample Source for Mass Spectrometry. Oxid. Med. Cell. Longev. 2020, 2020, 6681073. [Google Scholar] [CrossRef] [PubMed]
- Tektonidou, M.G. Antiphospholipid Syndrome Nephropathy: From Pathogenesis to Treatment. Front. Immunol. 2018, 9, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA Sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Cheng, G.; He, P.; Fang, Y. A Review: Electrochemical Aptasensors with Various Detection Strategies. Electroanalysis 2009, 21, 1251–1259. [Google Scholar] [CrossRef]
- Ikeda, T.; Kamohara, H.; Suda, S.; Nagura, T.; Tomino, M.; Sugi, M.; Wajima, Z. Comparative Evaluation of Endotoxin Activity Level and Various Biomarkers for Infection and Outcome of ICU-Admitted Patients. Biomedicines 2019, 7, 47. [Google Scholar] [CrossRef] [Green Version]
- Denning, N.-L.; Aziz, M.; Gurien, S.D.; Wang, P. DAMPs and NETs in Sepsis. Front. Immunol. 2019, 10, 2536. [Google Scholar] [CrossRef] [Green Version]
- Arina, P.; Singer, M. Pathophysiology of Sepsis. Curr. Opin. Anaesthesiol. 2021, 34, 77–84. [Google Scholar] [CrossRef]
- Gotts, J.E.; Matthay, M.A. Sepsis: Pathophysiology and Clinical Management. BMJ 2016, 353, i1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, A.P. Tech.Sight. Biochemistry. Biosensors--Sense and Sensitivity. Science 2000, 290, 1315–1317. [Google Scholar] [CrossRef]
- Juzgado, A.; Soldà, A.; Ostric, A.; Criado, A.; Valenti, G.; Rapino, S.; Conti, G.; Fracasso, G.; Paolucci, F.; Prato, M. Highly Sensitive Electrochemiluminescence Detection of a Prostate Cancer Biomarker. J. Mater. Chem. B 2017, 5, 6681–6687. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T.; Cullum, B. Biosensors and Biochips: Advances in Biological and Medical Diagnostics. Fresenius. J. Anal. Chem. 2000, 366, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Valenti, G.; Rampazzo, E.; Biavardi, E.; Villani, E.; Fracasso, G.; Marcaccio, M.; Bertani, F.; Ramarli, D.; Dalcanale, E.; Paolucci, F.; et al. An Electrochemiluminescence-Supramolecular Approach to Sarcosine Detection for Early Diagnosis of Prostate Cancer. Faraday Discuss. 2015, 185, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Welch, E.C.; Powell, J.M.; Clevinger, T.B.; Fairman, A.E.; Shukla, A. Advances in Biosensors and Diagnostic Technologies Using Nanostructures and Nanomaterials. Adv. Funct. Mater. 2021, 31, 2104126. [Google Scholar] [CrossRef]
- Guo, X.; Wen, F.; Zheng, N.; Saive, M.; Fauconnier, M.-L.; Wang, J. Aptamer-Based Biosensor for Detection of Mycotoxins. Front. Chem. 2020, 8, 195. [Google Scholar] [CrossRef] [Green Version]
- Pais de Barros, J.-P.; Gautier, T.; Sali, W.; Adrie, C.; Choubley, H.; Charron, E.; Lalande, C.; Le Guern, N.; Deckert, V.; Monchi, M.; et al. Quantitative Lipopolysaccharide Analysis Using HPLC/MS/MS and Its Combination with the Limulus Amebocyte Lysate Assay. J. Lipid Res. 2015, 56, 1363–1369. [Google Scholar] [CrossRef] [Green Version]
- Munford, R.S. Endotoxemia-Menace, Marker, or Mistake? J. Leukoc. Biol. 2016, 100, 687–698. [Google Scholar] [CrossRef] [Green Version]
- McDonagh, B. Detection of ROS Induced Proteomic Signatures by Mass Spectrometry. Front. Physiol. 2017, 8, 470. [Google Scholar] [CrossRef]
- Virzì, G.M.; Breglia, A.; Castellani, C.; Ankawi, G.; Bolin, C.; de Cal, M.; Cianci, V.; Angelini, A.; Vescovo, G.; Ronco, C. Lipopolysaccharide in Systemic Circulation Induces Activation of Inflammatory Response and Oxidative Stress in Cardiorenal Syndrome Type 1. J. Nephrol. 2019, 32, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.C.; Patel, K.; Vunikili, R.D.; Johnson, K.W.; Abdu, F.; Belman, S.K.; Glicksberg, B.S.; Tandale, P.; Fontanez, R.; Mathew, O.K.; et al. Sepsis in the Era of Data-Driven Medicine: Personalizing Risks, Diagnoses, Treatments and Prognoses. Brief. Bioinform. 2020, 21, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Von Groote, T.; Meersch-Dini, M. Biomarkers for the Prediction and Judgement of Sepsis and Sepsis Complications: A Step towards Precision Medicine? J. Clin. Med. 2022, 11, 5782. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.E.; Shidham, A.; Wong, H.R.; Khatri, P. A Comprehensive Time-Course-Based Multicohort Analysis of Sepsis and Sterile Inflammation Reveals a Robust Diagnostic Gene Set. Sci. Transl. Med. 2015, 7, 287ra71. [Google Scholar] [CrossRef] [PubMed]


| Filter | Study | Clinical Setting | Patients (n) | Endpoints/Outcomes | Status | Year, Place, Reference |
|---|---|---|---|---|---|---|
| Toraymyxin® | Multicenter (6) RCT | Severe sepsis after abdominal surgery | 17 PMX-B vs. 19 SoC |
| Completed | 2005, Europe [30] |
| Toraymyxin® | Multicenter (10) RCT [EUPHAS] | Severe sepsis after abdominal surgery | 119 PMX-B vs. 113 SoC |
| Completed | 2004–2007, Italy [31] |
| Toraymyxin® | Multicenter (18) RCT [ABDOMIX] | Severe sepsis after abdominal surgery | 34 PMX-B vs. 30 SoC |
| Completed | 2010–2013, France [32] |
| Toraymyxin® | Multicenter (55) RCT [EUPHRATES] | Severe sepsis after abdominal surgery | 233 PMX-B vs. 226 SoC |
| Completed | 2010–2016, North America [33] |
| Alteco® LPS adsorber | Multicenter (5) RCT [ASSET] | Severe sepsis of abdominal (20) or urogenital (12) origin | 16 LPS Adsorber vs. 16 SoC | Early termination due to patient recruitment issue | Early termination | 2015–2016, Northern Europe [34] |
| oXiris® | Monocentric cross over RCT | Septic shock and endotoxin levels > 0.03 EU/mL | 10 oXiris vs. 10 SoC |
| Completed | 2016–2018, Belgium [35] |
| oXiris® | Monocentric RCT | Critically ill patients with bleeding risk who underwent anticoagulation-free CRRT | 11 oXiris vs. 9 SoC |
| Completed | 2012–2016 Singapore [36] |
| oXiris® | Monocentric RCT [ECMORIX] | Cardiogenic shock requiring VA-ECMO | 40 oXiris vs. 40 SoC |
| Ongoing | 2021–2024 (NCT04886180) [37] |
| oXiris® | Monocentric RCT [OXICARD] | Elective cardiac surgery under CPB | 35 oXiris vs. 35 SoC |
| Ongoing | France, 2019 (NCT04201119) [38] |
| Methods | Principle | Advantages | Limitations | |
|---|---|---|---|---|
| Rabbit pyrogen test | Increase in rabbit’s temperature after exposition to pyrogenic molecules | First method approved by US Food and Drug Administration |
| [13] |
| Limulus amebocyte lysate test (LAL) | Clot formation after exposure of amoebocytes to LPS |
|
| [14,15,65] |
| Antibody-based biosensors | Highly specific antigen/antibody affinity (lock and key fit mechanism) |
|
| [13,57] |
| Aptames-based biosensors | Base pairing of ss-DNA or RNA forming an aptamer/target complex |
|
| [66,67] |
| Endotoxin activity assay (EAA) | Monoclonal antibody against LPS (activity measured through oxidative burst of primed neutrophils) |
| [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Virzì, G.M.; Mattiotti, M.; de Cal, M.; Ronco, C.; Zanella, M.; De Rosa, S. Endotoxin in Sepsis: Methods for LPS Detection and the Use of Omics Techniques. Diagnostics 2023, 13, 79. https://doi.org/10.3390/diagnostics13010079
Virzì GM, Mattiotti M, de Cal M, Ronco C, Zanella M, De Rosa S. Endotoxin in Sepsis: Methods for LPS Detection and the Use of Omics Techniques. Diagnostics. 2023; 13(1):79. https://doi.org/10.3390/diagnostics13010079
Chicago/Turabian StyleVirzì, Grazia Maria, Maria Mattiotti, Massimo de Cal, Claudio Ronco, Monica Zanella, and Silvia De Rosa. 2023. "Endotoxin in Sepsis: Methods for LPS Detection and the Use of Omics Techniques" Diagnostics 13, no. 1: 79. https://doi.org/10.3390/diagnostics13010079
APA StyleVirzì, G. M., Mattiotti, M., de Cal, M., Ronco, C., Zanella, M., & De Rosa, S. (2023). Endotoxin in Sepsis: Methods for LPS Detection and the Use of Omics Techniques. Diagnostics, 13(1), 79. https://doi.org/10.3390/diagnostics13010079







