An Alternative Diagnostic Method for C. neoformans: Preliminary Results of Deep-Learning Based Detection Model
Abstract
:1. Introduction
2. Materials and Method
2.1. Data and Data Pre-Processing
2.2. Convolutional Neural Network
2.2.1. Convolution Layer
2.2.2. Pooling Layer
2.2.3. Fully Connected Layer

2.3. VGG16
2.4. Confusion Matrix
- True negative (TN): model has given prediction negative, and the real or actual value was also negative.
- True positive (TP): the model has predicted positive, and the actual value was also positive.
- False negative (FN): the model has predicted negative, but the actual value was positive; it is also called a Type-II error.
- False positive (FP): the model has predicted positive, but the actual value was negative. It is also called a Type-I error.
3. Result and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Negroni, R. Cryptococcosis. Clin. Dermatol. 2012, 30, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Fraser, J.A.; Doering, T.L.; Wang, Z.; Janbon, G.; Idnurm, A.; Bahn, Y.S. Cryptococcus neoformans and Cryptococcus gattii, the Etiologic Agents of Cryptococcosis. Cold. Spring. Harb. Perspect. Med. 2014, 4, a019760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setianingrum, F.; Rautemaa-Richardson, R.; Denning, D.W. Pulmonary cryptococcosis: A review of pathobiology and clinical aspects. Med. Mycol. 2019, 57, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Bahn, Y.S.; Sun, S.; Heitman, J.; Lin, X. Microbe Profile: Cryptococcus neoformans species complex. Microbiology 2020, 166, 797–799. [Google Scholar] [CrossRef]
- Zaragoza, O. Basic principles of the virulence of Cryptococcus. Virulence 2019, 10, 490–501. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/fungal/diseases/cryptococcosis-neoformans/statistics.html (accessed on 1 October 2022).
- Rajasingham, R.; Govender, N.P.; Jordan, A.; Loyse, A.; Shroufi, A.; Denning, D.W.; Meya, D.B.; Chiller, T.M.; Boulware, D.R. The global burden of HIV-associated cryptococcal infection in adults in 2020: A modelling analysis. Lancet Infect. Dis. 2022, 22, 1748–1755. [Google Scholar] [CrossRef]
- Temfack, E.; Rim, J.J.B.; Spijker, R.; Loyse, A.; Chiller, T.; Pappas, P.G.; Perfect, J.; Sorell, T.C.; Harrison, T.S.; Cohen, J.F.; et al. Cryptococcal Antigen in Serum and Cerebrospinal Fluid for Detecting Cryptococcal Meningitis in Adults Living with Human Immunodeficiency Virus: Systematic Review and Meta-Analysis of Diagnostic Test Accuracy Studies. Clin. Infect. Dis. 2021, 72, 1268–1278. [Google Scholar] [CrossRef]
- Lakoh, S.; Rickman, H.; Sesay, M.; Kenneh, S.; Burke, R.; Baldeh, M.; Jiba, D.F.; Tejan, Y.S.; Boyle, S.; Koroma, C.; et al. Prevalence and mortality of cryptococcal disease in adults with advanced HIV in an urban tertiary hospital in Sierra Leone: A prospective study. BMC Infect. Dis. 2020, 20, 141. [Google Scholar] [CrossRef] [Green Version]
- Mustapha, M.; Ozsahin, D.; Ozsahin, I.; Uzun, B. Breast Cancer Screening Based on Supervised Learning and Multi-Criteria Decision-Making. Diagnostics 2022, 12, 1326. [Google Scholar] [CrossRef]
- Ozsahin, I.; Sekeroglu, B.; Musa, M.; Mustapha, M.; Uzun Ozsahin, D. Review on Diagnosis of COVID-19 from Chest CT Images Using Artificial Intelligence. Comput. Math. Methods Med. 2020, 2020, 9756518. [Google Scholar] [CrossRef]
- Uzun Ozsahin, D.; Taiwo Mustapha, M.; Saleh Mubarak, A.; Said Ameen, Z.; Uzun, B. Impact of Outliers and Dimensionality Reduction on the Performance of Predictive Models for Medical Disease Diagnosis. In Proceedings of the 2022 International Conference on Artificial Intelligence in Everything (AIE), Kitakyushu, Japan, 19–22 July 2022. [Google Scholar] [CrossRef]
- Ozsahin, D.; Taiwo Mustapha, M.; Mubarak, A.; Said Ameen, Z.; Uzun, B. Impact of feature scaling on machine learning models for the diagnosis of diabetes. In Proceedings of the 2022 International Conference on Artificial Intelligence in Everything (AIE), Kitakyushu, Japan, 19–22 July 2022. [Google Scholar] [CrossRef]
- Uzun Ozsahin, D.; Mustapha, M.T.; Bartholomew Duwa, B.; Ozsahin, I. Evaluating the performance of deep learning frameworks for malaria parasite detection using microscopic images of peripheral blood smears. Diagnostics 2022, 12, 2702. [Google Scholar] [CrossRef]
- Jiang, Y.; Luo, J.; Huang, D.; Liu, Y.; Li, D. Machine Learning Advances in Microbiology: A Review of Methods and Applications. Front. Microbiol. 2022, 13, 925454. [Google Scholar] [CrossRef] [PubMed]
- Qu, K.; Guo, F.; Liu, X.; Lin, Y.; Zou, Q. Application of Machine Learning in Microbiology. Front. Microbiol. 2019, 10, 827. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.P.; Recio, R. Cryptococcus neoformans Meningoencephalitis. Images in Clinical Medicine. N. Engl. J. Med. 2018, 379, 281. [Google Scholar] [CrossRef]
- Doi, A.; Kentaro, I.; Takegawa, H.; Miki, K.; Sono, Y.; Nishioka, H.; Takeshita, J.; Tomii, K.; Haruta, T. Community-acquired pneumonia caused by carbapenem-resistant Streptococcus pneumoniae: Re-examining its prevention and treatment. Int. J. Gen. Med. 2014, 7, 253–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging. 2018, 9, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Alnussairi, M.; İbrahim, A. Malaria parasite detection using deep learning algorithms based on (CNNs) technique. Comput. Electr. Eng. 2022, 103. [Google Scholar] [CrossRef]
- Jeong, Y.; Woo, J.; Kang, A. Malware Detection on Byte Streams of PDF Files Using Convolutional Neural Networks. Secur. Commun. Netw. 2019, 2019, 8485365. [Google Scholar] [CrossRef] [Green Version]
- Saha, S. A Comprehensive Guide to Convolutional Neural Networks—The ELI5 Way. Medium. Available online: https://towardsdatascience.com/a-comprehensive-guide-to-convolutional-neural-networks-the-eli5-way-3bd2b1164a53 (accessed on 10 October 2022).
- Brownlee, J. Available online: https://machinelearningmastery.com/pooling-layers-for-convolutional-neural-networks/#:~:text=Two%20common%20pooling%20methods%20are,presence%20of%20a%20feature%20respectively (accessed on 10 October 2022).
- Alhussainy, A. A New Pooling Layer based on Wavelet Transform for Convolutional Neural Network. JARDCS 2020, 24, 76–85. [Google Scholar] [CrossRef]
- Affine Layer. DeepAI. Available online: https://deepai.org/machine-learning-glossary-and-terms/affine-layer (accessed on 10 October 2022).
- Jiang, G. How the Convolutional Neural Network Work to Identify Numbers? 2021. Available online: https://doi.org/10.14293/s2199-1006.1.sor-.ppaiubj.v1 (accessed on 3 October 2022).
- Fully Connected Layers in Convolutional Neural Networks. IndianTechWarrior. Available online: https://indiantechwarrior.com/fully-connected-layers-in-convolutional-neural-networks/ (accessed on 9 October 2022).
- Guan, Q.; Wang, Y.; Ping, B.; Li, D.; Du, J.; Qin, Y.; Lu, H.; Wan, X.; Xiang, J. Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: A pilot study. J. Cancer 2019, 10, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Rohini, G. Everything You Need to Know about VGG16. Medium. Available online: https://medium.com/@mygreatlearning/everything-you-need-to-know-about-vgg16-7315defb5918#:~:text=VGG16%20is%20object%20detection%20and,to%20use%20with%20transfer%20learning (accessed on 10 October 2022).
- Bansal, M.; Kumar, M.; Sachdeva, M.; Mittal, A. Transfer learning for image classification using VGG19: Caltech-101 image data set. J. Ambient. Intell. Humaniz. Comput. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Fatemi, M.; Monirujjaman, K.M.; Kaur, M.; Zaguia, A. Comparative Analysis of Skin Cancer (Benign vs. Malignant) Detection Using Convolutional Neural Networks. J. Healthc. Eng. 2021, 2021, 5895156. [Google Scholar] [CrossRef] [PubMed]
- Narkhede, S. Understanding Confusion Matrix. Medium. Available online: https://towardsdatascience.com/understanding-confusion-matrix-a9ad42dcfd62 (accessed on 9 October 2022).
- Towards Data Science; Feng, V. An Overview of ResNet and Its Variants. Medium. Available online: https://towardsdatascience.com/an-overview-of-resnet-and-its-variants-5281e2f56035 (accessed on 10 October 2022).
- Paperspace Blog; Kurama, V. A Guide to ResNet, Inception v3, and SqueezeNet. Available online: https://blog.paperspace.com/popular-deep-learning-architectures-resnet-inceptionv3-squeezenet/ (accessed on 10 October 2022).
- Huston, S.M.; Mody, C.H. Cryptococcosis: An Emerging Respiratory Mycosis. Clin. Chest. Med. 2009, 30, 253–264. [Google Scholar] [CrossRef]
- Rathore, S.S.; Sathiyamoorthy, J.; Lalitha, C.; Ramakrishnan, J. A holistic review on Cryptococcus neoformans. Microb. Pathog. 2022, 166, 105521. [Google Scholar] [CrossRef]
- Bermas, A. Geddes-McAlister. Combatting the evaluation of antifungal resistance in Cryptococcus neoformans. Mol. Microbiol. 2020, 114, 721–734. [Google Scholar] [CrossRef]
- Iyer, K.R.; Revie, N.M.; Fu, C.; Robbins, N.; Cowen, L.E. Treatment strategies for cryptococcal infection: Challenges, advances, and future outlook. Nat. Rev. Microbiol. 2021, 19, 454–466. [Google Scholar] [CrossRef]
- ITPC. Available online: https://itpcglobal.org/blog/resource/ending-cryptococcal-meningitis-deaths-by-2030/#:~:text=Treatment%20with%20fluconazole%20alone%2C%20most,end%20all%20HIV%2Drelated%20deaths. (accessed on 13 October 2022).
- Zieliński, B.; Sroka-Oleksiak, A.; Rymarczyk, D.; Piekarczyk, A.; Brzychczy-Włoch, M. Deep learning approach to describe and classify fungi microscopic images. PLoS ONE 2020, 15, e0234806. [Google Scholar] [CrossRef]
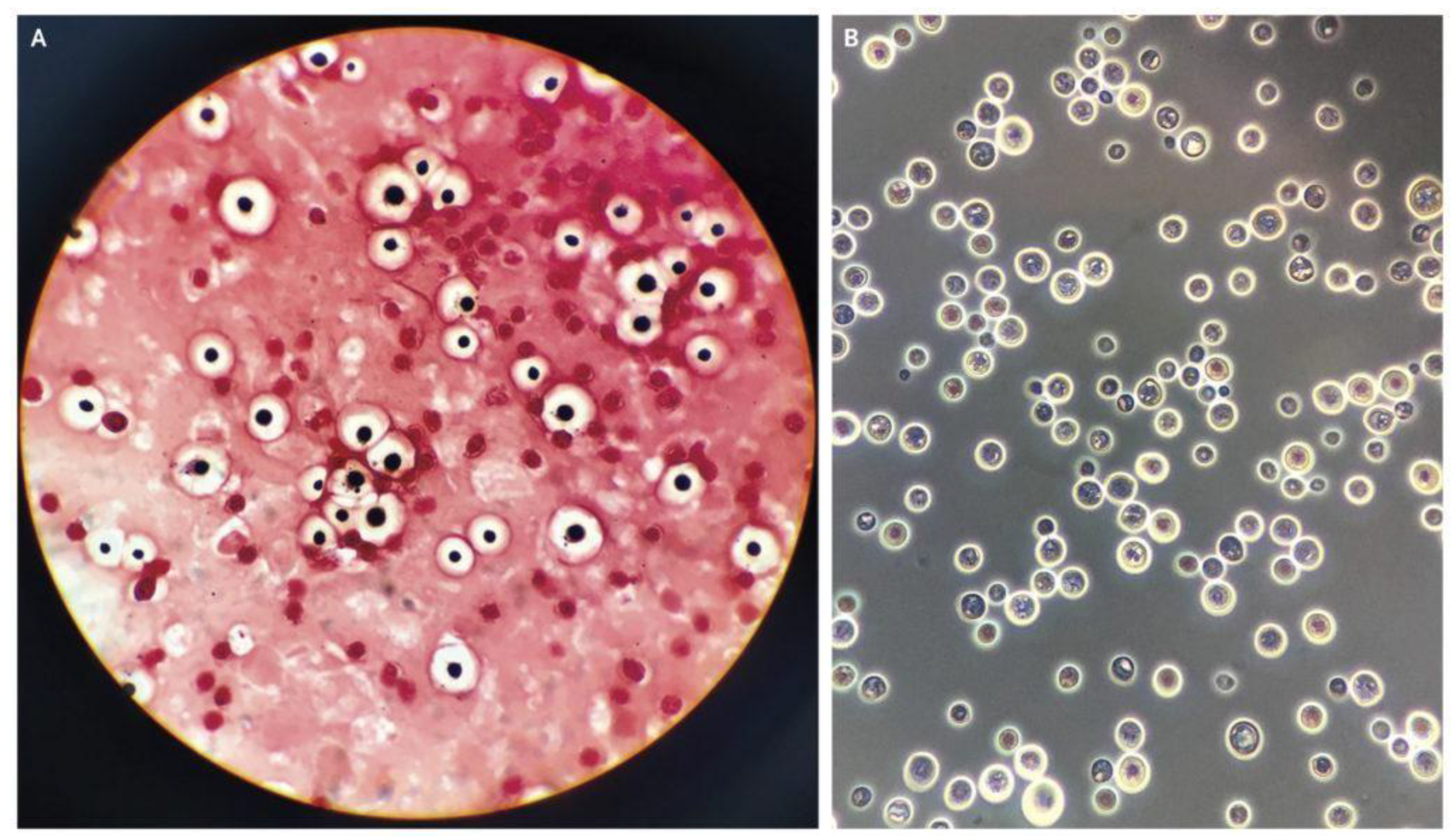
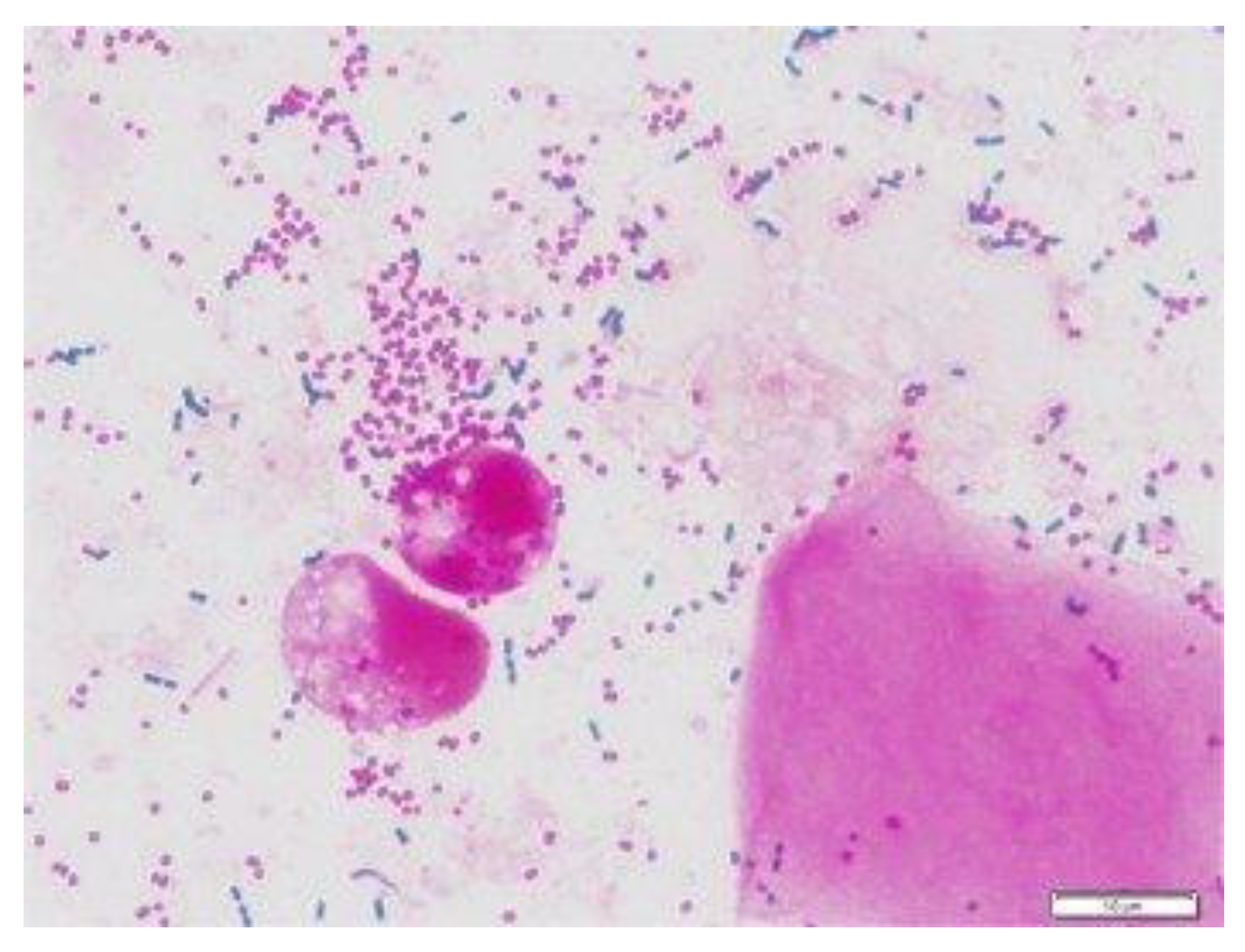







| Original Number of Dataset | Augmented Number of Dataset | ||
|---|---|---|---|
| 1 | Positive | 63 | 1000 |
| 2 | Negative | 63 | 1000 |
| Layers | Outcome Structure | Number of Parameters |
|---|---|---|
| Conv1 | (224 × 224 × 64) | 1792 |
| Conv2 | (224 × 224 × 64) | 36,928 |
| MaxPool | (112 × 112 × 64) | - |
| Conv1 | (112 × 112 × 128) | 73,856 |
| Conv2 | (112 × 112 × 128) | 147,584 |
| MaxPool | (56 × 56 × 128) | - |
| Conv1 | (56 × 56 × 256) | 295,168 |
| Conv2 | (56 × 56 × 256) | 590,080 |
| Conv3 | (56 × 56 × 256) | 590,080 |
| MaxPool | (28 × 28 × 256) | - |
| Conv1 | (28 × 28 × 512) | 1,180,160 |
| Conv2 | (28 × 28 × 512) | 2,359,808 |
| Conv3 | (28 × 28 × 512) | 2,359,808 |
| MaxPool | (14 × 14 × 512) | - |
| Conv1 | (14 × 14 × 512) | 2,359,808 |
| Conv2 | (14 × 14 × 512) | 2,359,808 |
| Conv3 | (14 × 14 × 512) | 2,359,808 |
| MaxPool | (7 × 7 × 512) | - |
| Flatten-Layer | (25,088) | - |
| Fully connected layer Dense-1 | (4096) | 102,764,544 |
| Fully connected layer Dense-2 | (4096) | 16,781,312 |
| Dropout | (4096) | - |
| Softmax | (1) | 4097 |
| Large Number | Parameter | Test Accuracy | Test Loss |
|---|---|---|---|
| 16 | 134,264,641 | 86.88% | 0.36203 |
| Precision | Sensitivity | F1 Score | TP | TN | FP | FN | |
|---|---|---|---|---|---|---|---|
| Positive | 90.00% | 83.00% | 86.00% | 114 | 131 | 24 | 13 |
| Negative | 85.00% | 91.00% | 88.00% |
| Prec. % | Sens. % | F1 Score % | TP | TN | FP | FN | Acc. % | Loss | ||
|---|---|---|---|---|---|---|---|---|---|---|
| VGG16 | Pos. | 90.00 | 83.00 | 86.00 | 114 | 131 | 24 | 13 | 86.88 | 0.36203 |
| Neg. | 85.00 | 91.00 | 88.00 | |||||||
| ResNet50 | Pos. | 86.00 | 79.00 | 82.00 | 105 | 92 | 33 | 52 | 83.33 | 0.39312 |
| Neg. | 81.00 | 88.00 | 84.00 | |||||||
| InceptionV3 | Pos. | 67.00 | 76.00 | 71.00 | 109 | 126 | 29 | 18 | 69.86 | 0.55700 |
| Neg. | 74.00 | 64.00 | 68.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seyer Cagatan, A.; Taiwo Mustapha, M.; Bagkur, C.; Sanlidag, T.; Ozsahin, D.U. An Alternative Diagnostic Method for C. neoformans: Preliminary Results of Deep-Learning Based Detection Model. Diagnostics 2023, 13, 81. https://doi.org/10.3390/diagnostics13010081
Seyer Cagatan A, Taiwo Mustapha M, Bagkur C, Sanlidag T, Ozsahin DU. An Alternative Diagnostic Method for C. neoformans: Preliminary Results of Deep-Learning Based Detection Model. Diagnostics. 2023; 13(1):81. https://doi.org/10.3390/diagnostics13010081
Chicago/Turabian StyleSeyer Cagatan, Ayse, Mubarak Taiwo Mustapha, Cemile Bagkur, Tamer Sanlidag, and Dilber Uzun Ozsahin. 2023. "An Alternative Diagnostic Method for C. neoformans: Preliminary Results of Deep-Learning Based Detection Model" Diagnostics 13, no. 1: 81. https://doi.org/10.3390/diagnostics13010081
APA StyleSeyer Cagatan, A., Taiwo Mustapha, M., Bagkur, C., Sanlidag, T., & Ozsahin, D. U. (2023). An Alternative Diagnostic Method for C. neoformans: Preliminary Results of Deep-Learning Based Detection Model. Diagnostics, 13(1), 81. https://doi.org/10.3390/diagnostics13010081










