Segmentation of Anatomical Structures of the Left Heart from Echocardiographic Images Using Deep Learning
Abstract
:1. Introduction
2. Related Works
3. Materials and Methods
3.1. Data
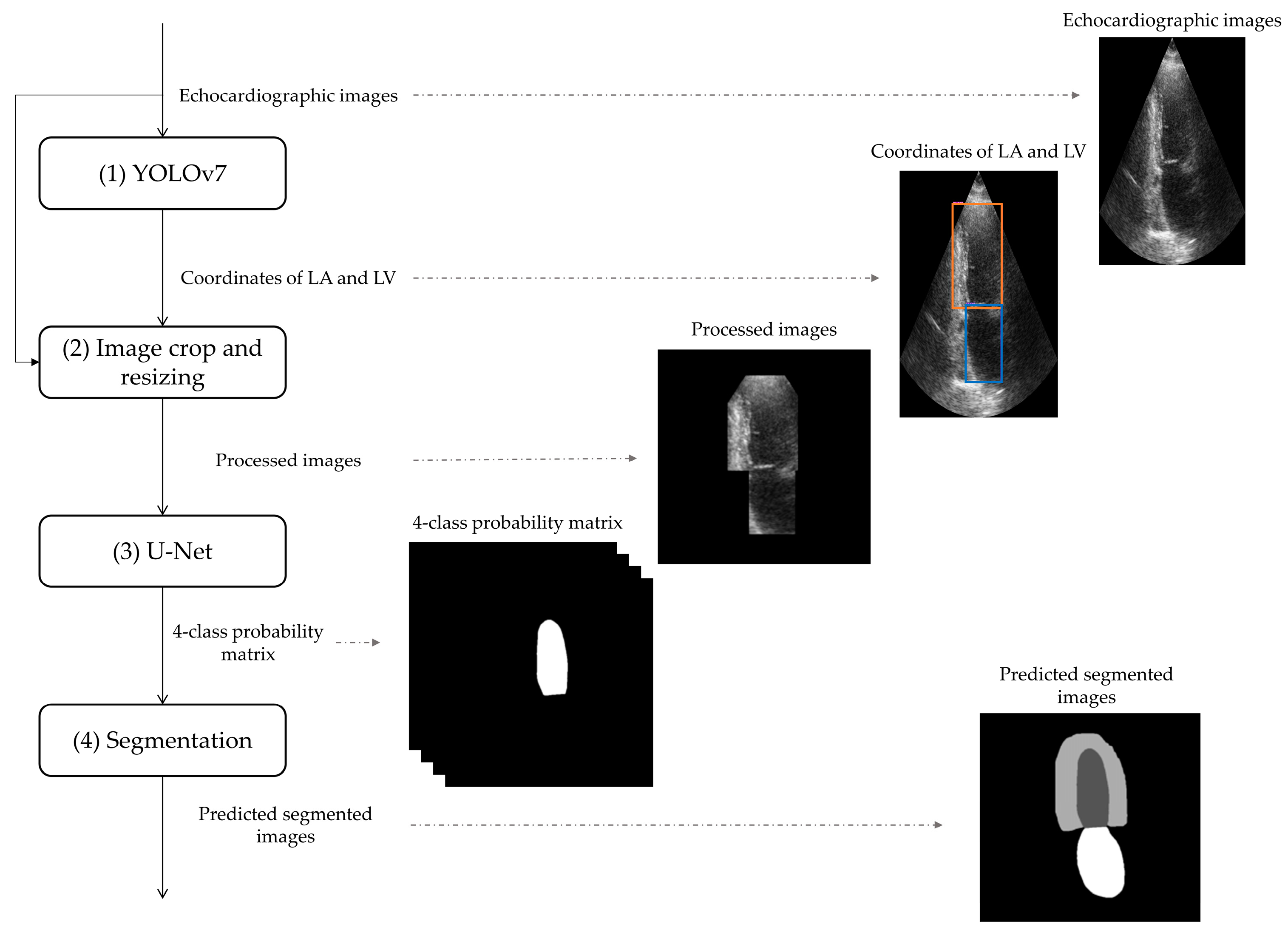
3.2. Deep-Learning-Based Tool for Segmentation of Anatomical Structured of the Left Heart
3.2.1. Detection of Anatomical Structures of the Left Heart by YOLOv7
3.2.2. Image Crop and Resizing
3.2.3. U-Net Application
3.2.4. Segmentation of Anatomical Structures of the Left Heart
3.3. Evaluation Metrics
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pinto, A.; Pinto, F.; Faggian, A.; Rubini, G.; Caranci, F.; Macarini, L.; Genovese, E.A.; Brunese, L. Sources of error in emergency ultrasonography. Crit. Ultrasound J. 2013, 5, S1. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep Learning for Cardiac Image Segmentation: A Review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Peels, C.H.; Visser, C.A.; Kupper, A.J.F.; Visser, F.C.; Roos, J.P. Usefulness of two-dimensional echocardiography for immediate detection of myocardial ischemia in the emergency room. Am. J. Cardiol. 1990, 65, 687–691. [Google Scholar] [CrossRef]
- Baumgartner, H.; Hung, J.; Bermejo, J.; Chambers, J.B.; Evangelista, A.; Griffin, B.P.; Lung, B.; Otto, C.M.; Pellikka, P.A.; Quiñones, M. Erratum: Echocardiographic assessment of valve stenosis: EAE/ASE recommendations for clinical practice (European Journal of Echocardiography 10). Eur. J. Echocardiogr. 2009, 10, 479. [Google Scholar] [CrossRef] [PubMed]
- Foley, T.A.; Mankad, S.V.; Anavekar, N.S.; Bonnichsen, C.R.; Miller, M.F.; Morris, T.D.; Araoz, P.A. Cardiologist Measuring Left Ventricular Ejection Fraction—Techniques and Potential Pitfalls. Eur. Cardiol. Rev. 2012, 8, 108–114. [Google Scholar] [CrossRef]
- Sanches, J.M.; Laine, A.F.; Suri, J.S. Ultrasound Imaging; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Mazaheri, S.; Sulaiman, P.S.B.; Wirza, R.; Khalid, F.; Kadiman, S.; Dimon, M.Z.; Tayebi, R.M. Echocardiography Image Segmentation: A Survey. In Proceedings of the 2013 International Conference on Advanced Computer Science Applications and Technologies, Kuching, Malaysia, 23–24 December 2013; pp. 327–332. [Google Scholar] [CrossRef]
- Jin, K.; Yan, Y.; Wang, S.; Yang, C.; Chen, M.; Liu, X.; Terasaki, H.; Yeo, T.-H.; Singh, N.G.; Wang, Y.; et al. iERM: An Interpretable Deep Learning System to Classify Epiretinal Membrane for Different Optical Coherence Tomography Devices: A Multi-Center Analysis. J. Clin. Med. 2023, 12, 400. [Google Scholar] [CrossRef]
- Xie, B.; Li, S.; Li, M.; Liu, C.H.; Huang, G.; Wang, G. SePiCo: Semantic-Guided Pixel Contrast for Domain Adaptive Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2023; early access. [Google Scholar] [CrossRef]
- Gao, Z.; Pan, X.; Shao, J.; Jiang, X.; Su, Z.; Jin, K.; Ye, J. Automatic interpretation and clinical evaluation for fundus fluorescein angiography images of diabetic retinopathy patients by deep learning. Br. J. Ophthalmol. 2022. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.Y.; Porikli, F.; Plaza, A.J.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 3523–3542. [Google Scholar] [CrossRef]
- Liu, F.; Wang, K.; Liu, D.; Yang, X.; Tian, J. Deep pyramid local attention neural network for cardiac structure segmentation in two-dimensional echocardiography. Med. Image Anal. 2020, 67, 101873. [Google Scholar] [CrossRef]
- Leclerc, S.; Smistad, E.; Pedrosa, J.; Ostvik, A.; Cervenansky, F.; Espinosa, F.; Espeland, T.; Berg, E.A.R.; Jodoin, P.-M.; Grenier, T.; et al. Deep Learning for Segmentation Using an Open Large-Scale Dataset in 2D Echocardiography. IEEE Trans. Med. Imaging 2019, 38, 2198–2210. [Google Scholar] [CrossRef]
- Moradi, S.; Oghli, M.G.; Alizadehasl, A.; Shiri, I.; Oveisi, N.; Oveisi, M.; Maleki, M.; Dhooge, J. MFP-Unet: A novel deep learning based approach for left ventricle segmentation in echocardiography. Phys. Med. 2019, 67, 58–69. [Google Scholar] [CrossRef]
- Kim, T.; Hedayat, M.; Vaitkus, V.V.; Belohlavek, M.; Krishnamurthy, V.; Borazjani, I. Automatic segmentation of the left ventricle in echocardiographic images using convolutional neural networks. Quant. Imaging Med. Surg. 2021, 11, 1763–1781. [Google Scholar] [CrossRef]
- Girum, K.B.; Crehange, G.; Lalande, A. Learning With Context Feedback Loop for Robust Medical Image Segmentation. IEEE Trans. Med. Imaging 2021, 40, 1542–1554. [Google Scholar] [CrossRef]
- Lei, Y.; Fu, Y.; Roper, J.; Higgins, K.; Bradley, J.D.; Curran, W.J.; Liu, T.; Yang, X. Echocardiographic image multi-structure segmentation using Cardiac-SegNet. Med. Phys. 2021, 48, 2426–2437. [Google Scholar] [CrossRef] [PubMed]
- Alam, G.R.; Khan, A.M.; Shejuty, M.F.; Zubayear, S.I.; Shariar, N.; Altaf, M.; Hassan, M.M.; AlQahtani, S.A.; Alsanad, A. Ejection Fraction estimation using deep semantic segmentation neural network. J. Supercomput. 2022, 79, 27–50. [Google Scholar] [CrossRef]
- Saeed, M.; Muhtaseb, R.; Yaqub, M. Is Contrastive Learning Suitable for Left Ventricular Segmentation in Echocardiographic Images? arXiv 2022. [Google Scholar] [CrossRef]
- Zhuang, Z.; Jin, P.; Raj, A.N.J.; Yuan, Y.; Zhuang, S. Automatic Segmentation of Left Ventricle in Echocardiography Based on YOLOv3 Model to Achieve Constraint and Positioning. Comput. Math. Methods Med. 2021, 2021, 3772129. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Bochkovskiy, A.; Liao, H. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. arXiv 2022. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014. [Google Scholar] [CrossRef]
- He, X.; Zhao, K.; Chu, X. AutoML: A survey of the state-of-the-art. Knowl. -Based Syst. 2021, 212, 106622. [Google Scholar] [CrossRef]
- Qadri, S.F.; Lin, H.; Shen, L.; Ahmad, M.; Qadri, S.; Khan, S.; Khan, M.; Zareen, S.S.; Akbar, M.A.; Bin Heyat, B.; et al. CT-Based Automatic Spine Segmentation Using Patch-Based Deep Learning. Int. J. Intell. Syst. 2023, 2023, 2345835. [Google Scholar] [CrossRef]


| Training Dataset | Validation Dataset | Testing Dataset | |||
|---|---|---|---|---|---|
| Number of patients | 270 | 45 | 135 | ||
| Number of images | 1080 | 180 | 540 | ||
| Overall | LA | DSC (%) | 95.12 ± 3.91 | 93.76 ± 7.36 | 87.57 ± 13.48 |
| JAC(%) | 90.90 ± 5.41 | 88.86 ± 8.85 | 79.75 ± 15.76 | ||
| HD (pixels) | 3.60 ± 0.83 | 3.68 ± 0.82 | 4.07 ± 1.08 | ||
| LVepi | DSC (%) | 91.79 ± 2.47 | 89.08 ± 3.29 | 85.59 ± 7.14 | |
| JAC(%) | 84.93 ± 4.12 | 80.47 ± 5.25 | 75.38 ± 9.17 | ||
| HD (pixels) | 4.32 ± 0.73 | 4.62 ± 0.81 | 4.96 ± 1.09 | ||
| LVendo | DSC (%) | 95.18 ± 2.24 | 92.76 ± 4.64 | 92.63 ± 6.60 | |
| JAC(%) | 90.89 ± 3.90 | 86.81 ± 7.24 | 86.76 ± 8.40 | ||
| HD (pixels) | 3.41 ± 0.81 | 3.66 ± 0.87 | 3.81 ± 1.09 | ||
| Ref. | Dataset (Patients/Images) | Dataset Split | View | Classes | Method | Performance (on CAMUS Dataset) | ||
|---|---|---|---|---|---|---|---|---|
| LA | LVepi | LVendo | ||||||
| [13] | CAMUS (406/1624) | 10-fold cross-validation | Two and four chamber | LVendo and LVepi | U-Net | n.a. | ED: DSC = 95.4 ±2.3 HD = 6.0 ± 3.4 ES: DSC = 94.5 ±3.9 HD = 6.1 ±4.6 | ED: DSC = 93.9 ± 4.3 HD = 5.3 ± 3.6 ES: DSC = 91.6 ±6.1 HD = 5.5 ±3.8 |
| [14] | (1) CAMUS (500/n.a.) (2) custom dataset (137/n.a.) | 5-fold cross-validation | Four chambers | LV | MFP-U-Net | n.a. | DSC = 95.3 ± 1.9 HD = 3.5 ± 0.9 | |
| [15] | (1) custom dataset (8/1649) (2) CAMUS (450/1800) | n.a. | Two and four chambers | LVepi and LVendo | SegAN | n.a. | DSC = 85.9 ± 6.4 HD = 6.2 ± 1.2 | DSC = 91.7 ± 7.1 HD = 5.1 ± 1.7 |
| [16] | CAMUS (450/1800) | Static data division | Two and four chambers | LA, LVepi and LVendo | LFB-Net | Four-chamber view: DSC = 92.0 ± 4.0 HD = 5.2 ± 3.5 Two-chamber view: DSC = 92.0 ± 5.0 HD = 4.8 ± 2.8 | Four-chamber view: DSC = 86.0 ± 6.0 HD = 6.7 ± 3.0 Two-chamber view: DSC = 88.0 ± 4.0 HD = 7.1 ± 3.9 | Four-chamber view: DSC = 94.0 ± 3.0 HD = 5.0 ± 2.8 Two-chamber view: DSC = 94.0 ± 3.0 HD = 5.6 ± 3.2 |
| [12] | (1) EchoNet-Dynamic (2500/5000) (2) CAMUS (500/2000) | Static data divison | Two and four chambers | LVepi and LVendo | PLANet | n.a. | ED: DSC = 96.2 ± 1.2 HD = 4.6 ± 1.5 ES: DSC = 95.6 ± 1.4 HD = 4.6 ± 1.4 | ED: DSC = 95.1 ± 1.8 HD = 4.2 ± 1.4 ES: DSC = 93.1 ± 3.2 HD = 4.3 ± 1.5 |
| [17] | CAMUS (450/1800) | 5-fold cross-validation, | Two and four chambers | LA, LVepi and LVendo | Cardiac- SegNet | ED: DSC = 89.5 ± 8.5 HD = 2.2 ± 4.1 ES: DSC = 92.2 ± 5.5 HD = 2.7 ± 3.5 | ED: DSC = 96.0 ± 1.6 HD = 2.9 ± 2.1 ES: DSC = 95.3 ± 2.2 HD = 2.8 ± 2.2 | ED: DSC = 94.8 ± 2.4 HD = 2.3 ± 1.8 ES: DSC = 92.7 ± 4.3 HD = 2.3 ± 2.3 |
| [18] | custom dataset (380/380) | Static data division | Four chambers | LV | Deep Res-U-Net | n.a. | ES: DSC = 82.1 ±0.8 JAC = 66.9 ± 6.4 HD = 23.8 ± 0.1 ED: DSC = 86.5 ± 1.1 JAC = 63.7 ± 9.6 HD = 19.7 ± 0.2 | |
| [19] | (1) EchoNet-Dynamic (10,024/20,048) (2) CAMUS (400/800) | Static data division | Four chambers | LV | DeepLabV3 | n.a. | DSC = 93.1 ± 0.04 | |
| [20] | custom dataset | n.a. | n.a. | LVendo | YOLOv3 (Darknet53) | n.a. | n.a. | DSC = 93.6 ± 2.0 HD = 6.7 ± 1.8 |
| This study | CAMUS (450/1800) | Static data division | Two and four chambers | LA, LVepi and LVendo | YOLOv7 and U-Net | DSC = 87.6± 13.5 JAC = 79.8 ± 15.8 HD = 4.1 ± 1.1 | DSC = 85.6 ± 7.1 JAC = 75.4 ± 9.2 HD = 5.0 ± 1.1 | DSC = 92.6 ± 6.6 JAC = 86.8 ± 8.4 HD = 3.8 ± 1.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mortada, M.J.; Tomassini, S.; Anbar, H.; Morettini, M.; Burattini, L.; Sbrollini, A. Segmentation of Anatomical Structures of the Left Heart from Echocardiographic Images Using Deep Learning. Diagnostics 2023, 13, 1683. https://doi.org/10.3390/diagnostics13101683
Mortada MJ, Tomassini S, Anbar H, Morettini M, Burattini L, Sbrollini A. Segmentation of Anatomical Structures of the Left Heart from Echocardiographic Images Using Deep Learning. Diagnostics. 2023; 13(10):1683. https://doi.org/10.3390/diagnostics13101683
Chicago/Turabian StyleMortada, MHD Jafar, Selene Tomassini, Haidar Anbar, Micaela Morettini, Laura Burattini, and Agnese Sbrollini. 2023. "Segmentation of Anatomical Structures of the Left Heart from Echocardiographic Images Using Deep Learning" Diagnostics 13, no. 10: 1683. https://doi.org/10.3390/diagnostics13101683










