Exploring the Utility of Cardiovascular Magnetic Resonance Radiomic Feature Extraction for Evaluation of Cardiac Sarcoidosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Subject Selection
2.3. Imaging Protocol
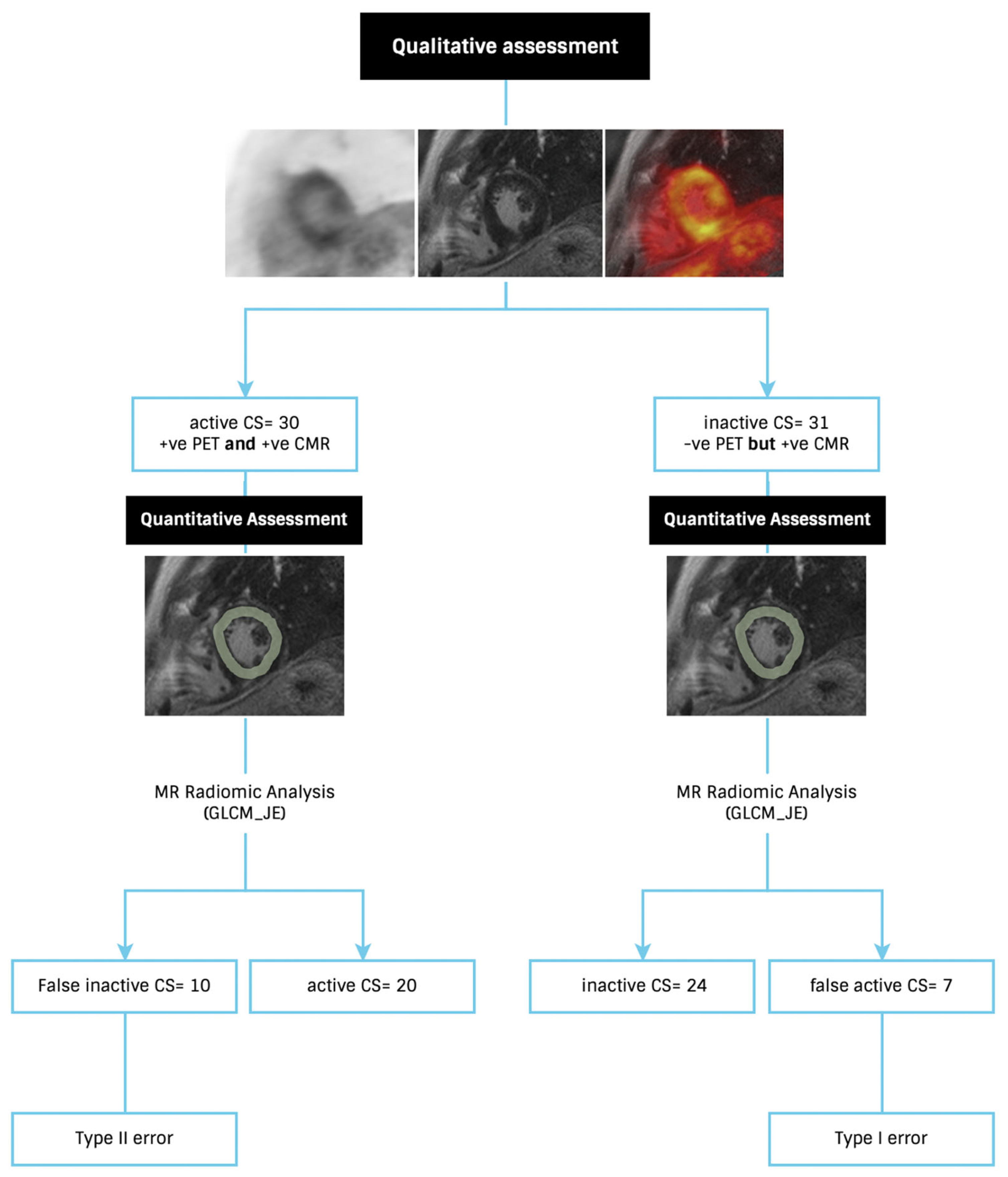
2.4. Patient Classification
2.5. Segmentation
2.6. Feature Extraction
2.7. Statistical Analysis
3. Results
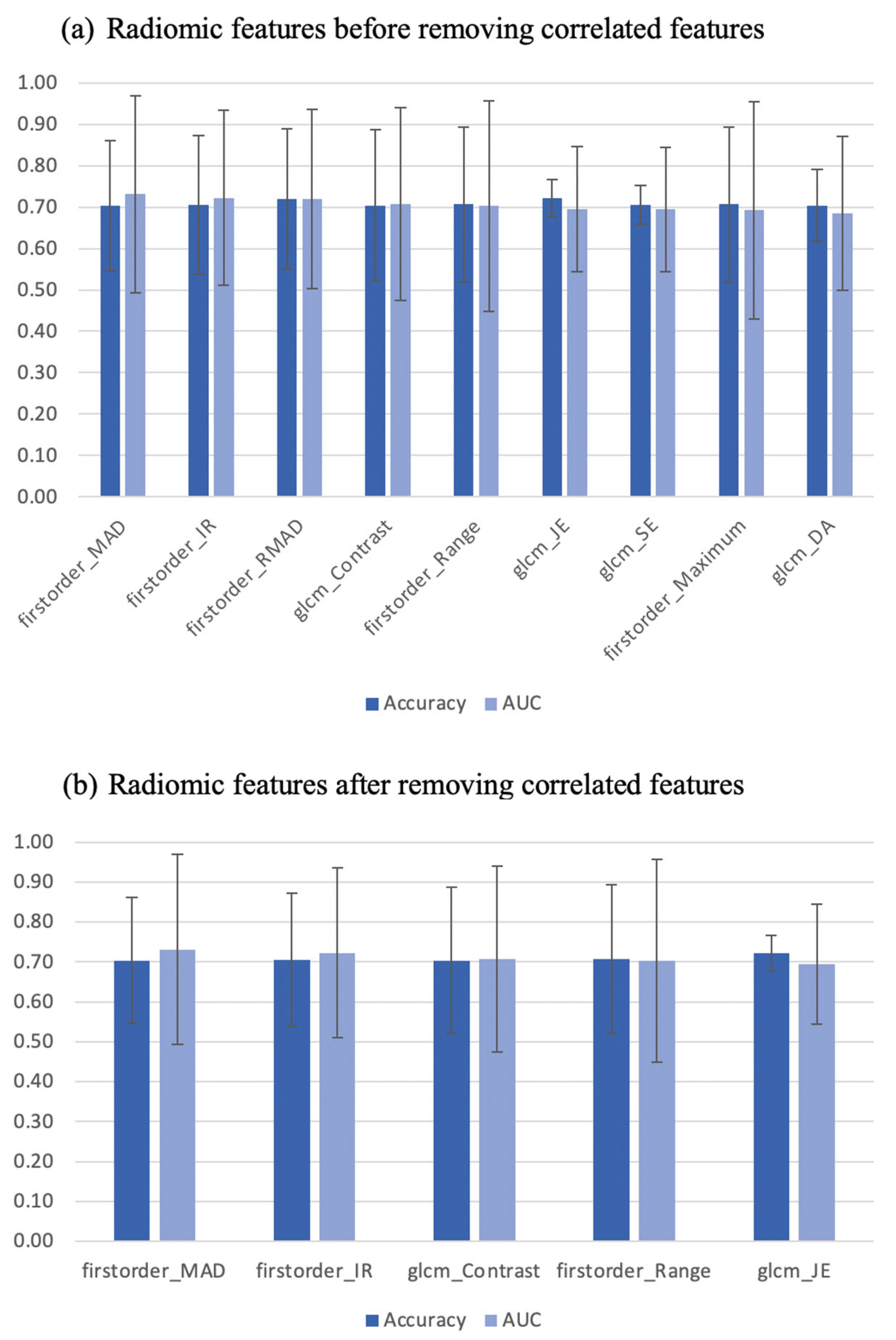
3.1. Individual Features—Diagnostic Utility
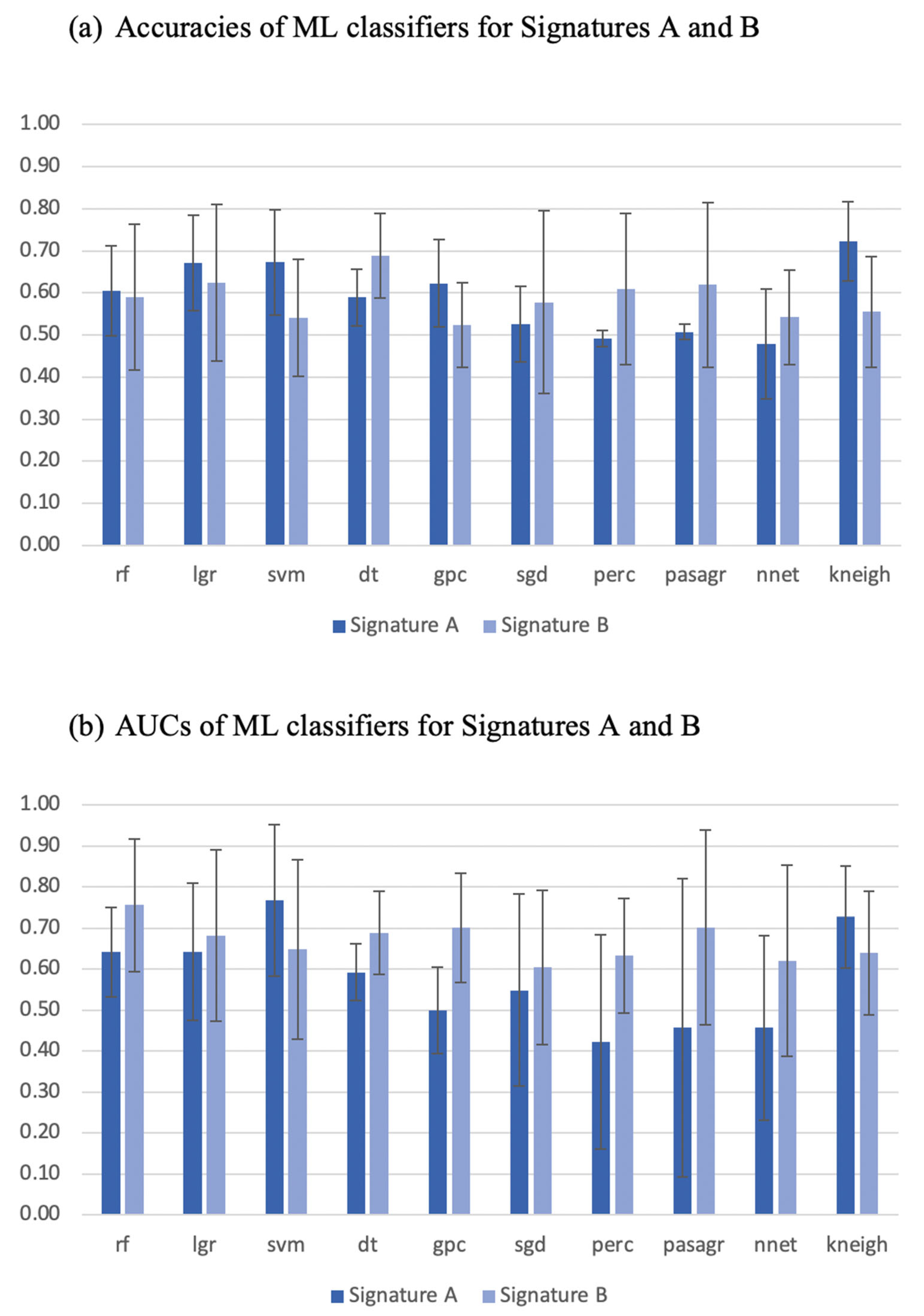
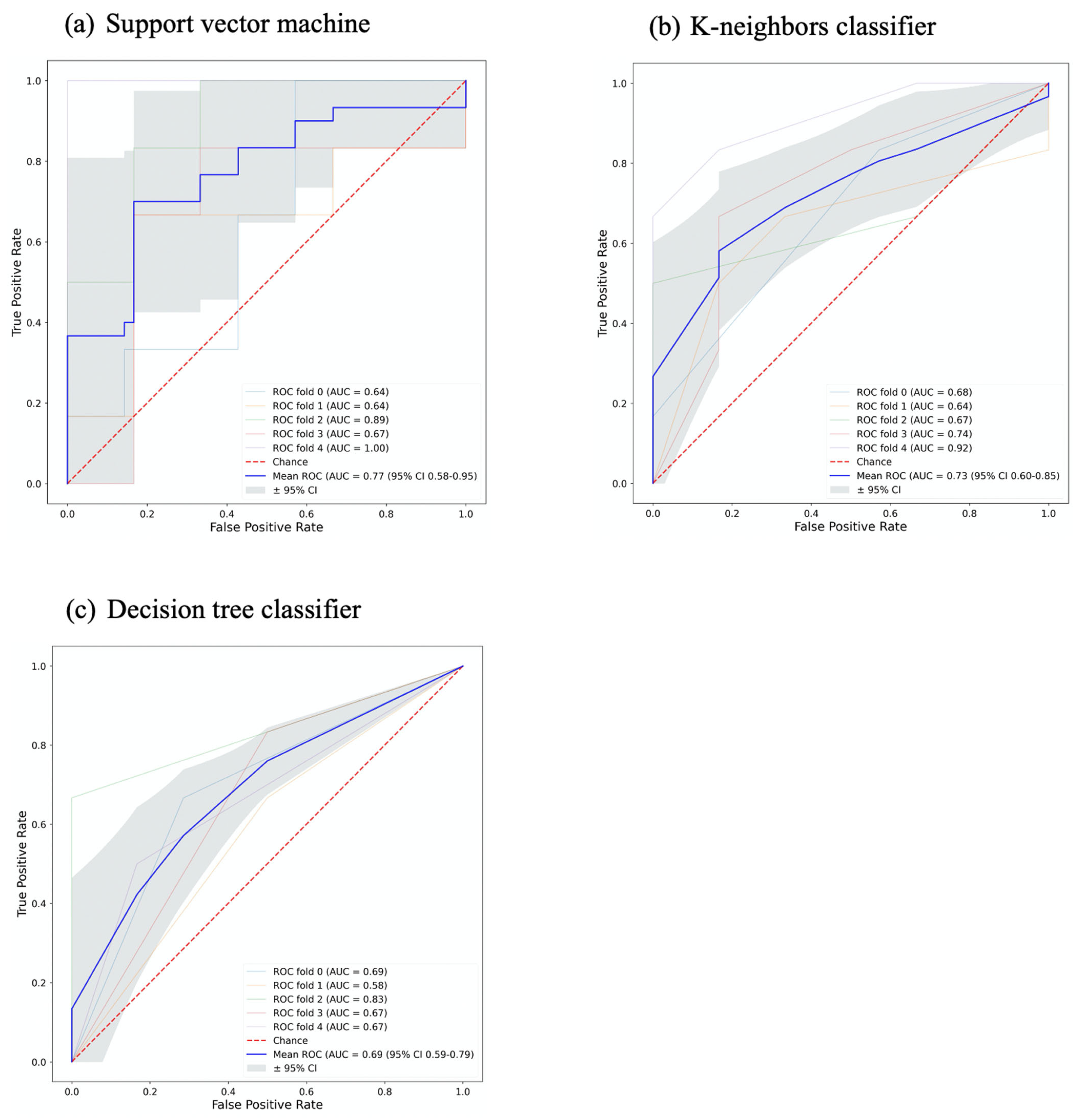
3.2. Signature Building and Machine Learning Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Birnie, D.H.; Nery, P.B.; Ha, A.C.; Beanlands, R.S. Cardiac sarcoidosis. J. Am. Coll. Cardiol. 2016, 68, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Ginelliová, A.; Farkaš, D.; Iannaccone, S.F.; Vyhnálková, V. Sudden unexpected death due to severe pulmonary and cardiac sarcoidosis. Forensic Sci. Med. Pathol. 2016, 12, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Nappi, C.; Ponsiglione, A.; Imbriaco, M.; Cuocolo, A. 18F-FDG PET/CMR in Cardiac Sarcoidosis: A Wild Card in the Deck? Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–3. [Google Scholar]
- Bennett, M.K.; Gilotra, N.A.; Harrington, C.; Rao, S.; Dunn, J.M.; Freitag, T.B.; Halushka, M.K.; Russell, S.D. Evaluation of the role of endomyocardial biopsy in 851 patients with unexplained heart failure from 2000–2009. Circ. Heart Fail. 2013, 6, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Chareonthaitawee, P.; Beanlands, R.S.; Chen, W.; Dorbala, S.; Miller, E.J.; Murthy, V.L.; Birnie, D.H.; Chen, E.S.; Cooper, L.T.; Tung, R.H. Joint SNMMI–ASNC Expert Consensus Document on the Role of 18F-FDG PET/CT in Cardiac Sarcoid Detection and Therapy Monitoring; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Osborne, M.T.; Hulten, E.A.; Murthy, V.L.; Skali, H.; Taqueti, V.R.; Dorbala, S.; DiCarli, M.F.; Blankstein, R. Patient preparation for cardiac fluorine-18 fluorodeoxyglucose positron emission tomography imaging of inflammation. J. Nucl. Cardiol. 2017, 24, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Mushari, N.A.; Soultanidis, G.; Duff, L.; Trivieri, M.G.; Fayad, Z.A.; Robson, P.; Tsoumpas, C. Exploring the utility of radiomic feature extraction to improve the diagnostic accuracy of cardiac sarcoidosis using FDG PET. Front. Med. 2022, 9, 840261. [Google Scholar] [CrossRef]
- Rainey, C.; O’Regan, T.; Matthew, J.; Skelton, E.; Woznitza, N.; Chu, K.-Y.; Goodman, S.; McConnell, J.; Hughes, C.; Bond, R. An insight into the current perceptions of UK radiographers on the future impact of AI on the profession: A cross-sectional survey. J. Med. Imaging Radiat. Sci. 2022, 53, 347–361. [Google Scholar] [CrossRef]
- Duff, L.; Scarsbrook, A.F.; Mackie, S.L.; Frood, R.; Bailey, M.; Morgan, A.W.; Tsoumpas, C. A methodological framework for AI-assisted diagnosis of active aortitis using radiomic analysis of FDG PET–CT images: Initial analysis. J. Nucl. Cardiol. 2022, 29, 3315–3331. [Google Scholar] [CrossRef]
- Dweck, M.R.; Abgral, R.; Trivieri, M.G.; Robson, P.M.; Karakatsanis, N.; Mani, V.; Palmisano, A.; Miller, M.A.; Lala, A.; Chang, H.L. Hybrid magnetic resonance imaging and positron emission tomography with fluorodeoxyglucose to diagnose active cardiac sarcoidosis. JACC Cardiovasc. Imaging 2018, 11, 94–107. [Google Scholar] [CrossRef]
- Ishida, Y.; Yoshinaga, K.; Miyagawa, M.; Moroi, M.; Kondoh, C.; Kiso, K.; Kumita, S. Recommendations for 18F-fluorodeoxyglucose positron emission tomography imaging for cardiac sarcoidosis: Japanese Society of Nuclear Cardiology recommendations. Ann. Nucl. Med. 2014, 28, 393–403. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Kikinis, R.; Pieper, S.D.; Vosburgh, K.G. 3D Slicer: A platform for subject-specific image analysis, visualization, and clinical support. In Intraoperative Imaging and Image-Guided Therapy; Springer: Berlin/Heidelberg, Germany, 2014; pp. 277–289. [Google Scholar]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef]
- Balagurunathan, Y.; Kumar, V.; Gu, Y.; Kim, J.; Wang, H.; Liu, Y.; Goldgof, D.B.; Hall, L.O.; Korn, R.; Zhao, B. Test–retest reproducibility analysis of lung CT image features. J. Digit. Imaging 2014, 27, 805–823. [Google Scholar] [CrossRef]
- Ergen, B.; Baykara, M. Texture based feature extraction methods for content based medical image retrieval systems. Bio-Med. Mater. Eng. 2014, 24, 3055–3062. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563. [Google Scholar] [CrossRef]
- Lovinfosse, P.; Hatt, M.; Visvikis, D.; Hustinx, R. Heterogeneity analysis of 18F-FDG PET imaging in oncology: Clinical indications and perspectives. Clin. Transl. Imaging 2018, 6, 393–410. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Xu, X.; Zhang, X.; Tian, Q.; Zhang, G.; Liu, Y.; Cui, G.; Meng, J.; Wu, Y.; Liu, T.; Yang, Z. Three-dimensional texture features from intensity and high-order derivative maps for the discrimination between bladder tumors and wall tissues via MRI. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 645–656. [Google Scholar] [CrossRef]
- Duff, L.; Tsoumpas, C. Automated diagnosis and prediction in cardiovascular diseases using tomographic imaging. In Big Data in Multimodal Medical Imaging; Taylor & Francis Group: Abingdon, UK, 2019; pp. 71–102. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Leger, S.; Vallières, M.; Löck, S. Image biomarker standardisation initiative. arXiv 2016, arXiv:1612.07003. [Google Scholar]
- Avard, E.; Shiri, I.; Hajianfar, G.; Abdollahi, H.; Kalantari, K.R.; Houshmand, G.; Kasani, K.; Bitarafan-Rajabi, A.; Deevband, M.R.; Oveisi, M. Non-contrast Cine Cardiac Magnetic Resonance image radiomics features and machine learning algorithms for myocardial infarction detection. Comput. Biol. Med. 2022, 141, 105145. [Google Scholar] [CrossRef]
- Tana, C.; Mantini, C.; Donatiello, I.; Mucci, L.; Tana, M.; Ricci, F.; Cipollone, F.; Giamberardino, M.A. Clinical Features and Diagnosis of Cardiac Sarcoidosis. J. Clin. Med. 2021, 10, 1941. [Google Scholar] [CrossRef] [PubMed]
- Vallières, M.; Freeman, C.R.; Skamene, S.R.; El Naqa, I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys. Med. Biol. 2015, 60, 5471. [Google Scholar] [CrossRef] [PubMed]
| Feature | CSactive Mean | CSinactive Mean | U Statistic | p-Value | Effect Size |
|---|---|---|---|---|---|
| glcm_Cluster Shade | 9.27 | 7.69 | 701 | 0.0007 | 0.09 |
| glcm_Cluster Prominence | 79.37 | 85.63 | 693 | 0.0010 | 0.03 |
| firstorder_Variance | 721.11 | 485.32 | 689 | 0.0013 | 0.50 |
| glrlm_Gray Level Variance | 1.36 | 1.01 | 689 | 0.0013 | 0.38 |
| gldm_Gray Level Variance | 1.21 | 0.83 | 677 | 0.0023 | 0.50 |
| glcm_Maximal Correlation Coefficient | 0.43 | 0.36 | 676 | 0.0024 | 0.60 |
| firstorder_Mean Absolute Deviation | 19.05 | 14.50 | 674 | 0.0026 | 0.75 |
| glcm_Correlation | 0.36 | 0.28 | 673 | 0.0028 | 0.61 |
| glszm_Size Zone Non Uniformity | 57.69 | 38.59 | 673 | 0.0028 | 0.81 |
| glrlm_Run Entropy | 2.98 | 2.81 | 672 | 0.0029 | 0.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mushari, N.A.; Soultanidis, G.; Duff, L.; Trivieri, M.G.; Fayad, Z.A.; Robson, P.M.; Tsoumpas, C. Exploring the Utility of Cardiovascular Magnetic Resonance Radiomic Feature Extraction for Evaluation of Cardiac Sarcoidosis. Diagnostics 2023, 13, 1865. https://doi.org/10.3390/diagnostics13111865
Mushari NA, Soultanidis G, Duff L, Trivieri MG, Fayad ZA, Robson PM, Tsoumpas C. Exploring the Utility of Cardiovascular Magnetic Resonance Radiomic Feature Extraction for Evaluation of Cardiac Sarcoidosis. Diagnostics. 2023; 13(11):1865. https://doi.org/10.3390/diagnostics13111865
Chicago/Turabian StyleMushari, Nouf A., Georgios Soultanidis, Lisa Duff, Maria G. Trivieri, Zahi A. Fayad, Philip M. Robson, and Charalampos Tsoumpas. 2023. "Exploring the Utility of Cardiovascular Magnetic Resonance Radiomic Feature Extraction for Evaluation of Cardiac Sarcoidosis" Diagnostics 13, no. 11: 1865. https://doi.org/10.3390/diagnostics13111865
APA StyleMushari, N. A., Soultanidis, G., Duff, L., Trivieri, M. G., Fayad, Z. A., Robson, P. M., & Tsoumpas, C. (2023). Exploring the Utility of Cardiovascular Magnetic Resonance Radiomic Feature Extraction for Evaluation of Cardiac Sarcoidosis. Diagnostics, 13(11), 1865. https://doi.org/10.3390/diagnostics13111865








