Three-Dimensional Craniofacial Landmark Detection in Series of CT Slices Using Multi-Phased Regression Networks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Personal Computer
2.2. Datasets
2.2.1. CT Images
2.2.2. 3D Reconstruction (STL File Creation)
2.2.3. Plotting Anatomical Landmarks
2.3. Neural Networks and Learning Datasets
2.3.1. First-Phase Deep Learning
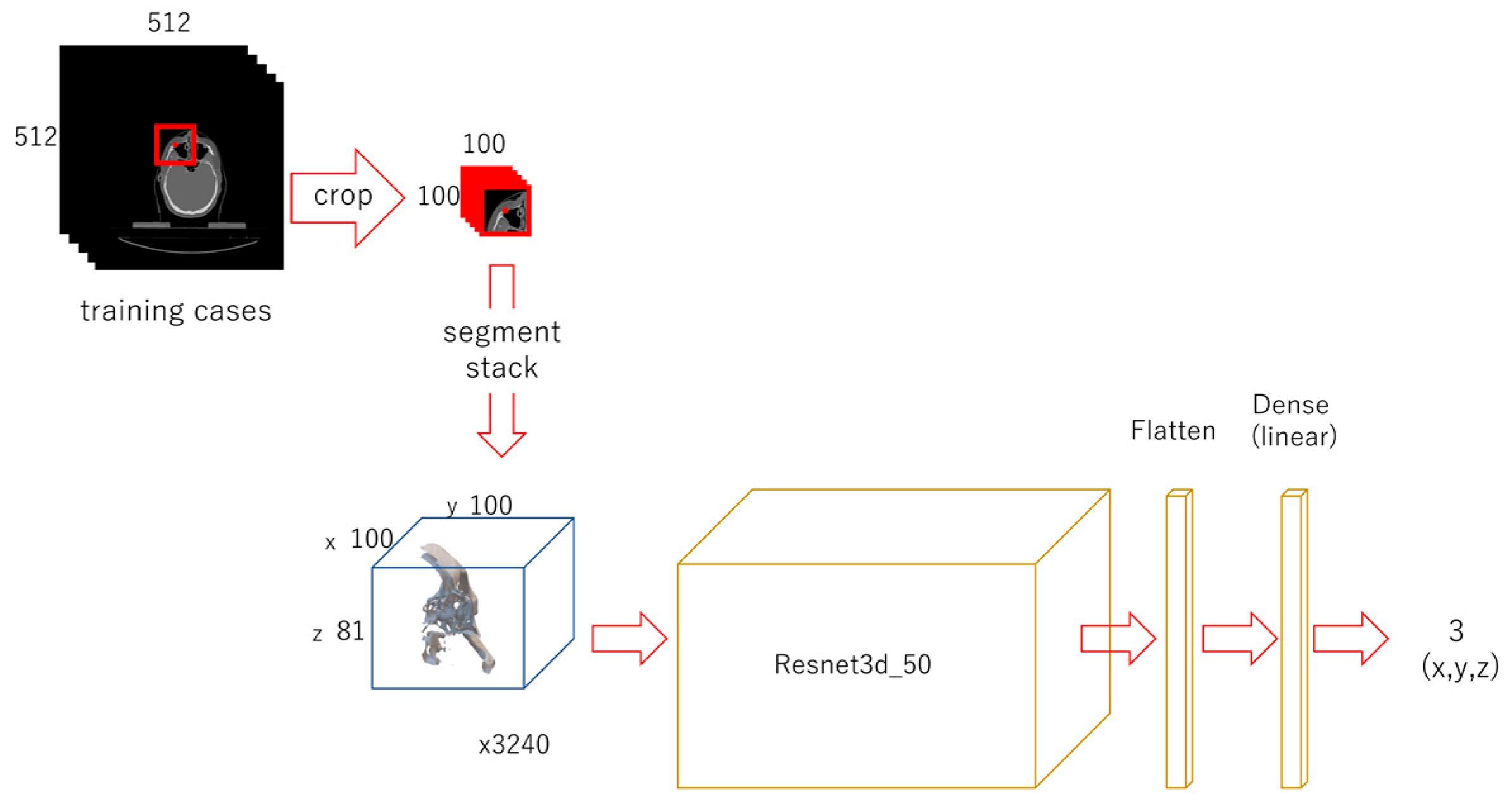
2.3.2. Second-Phase Deep Learning
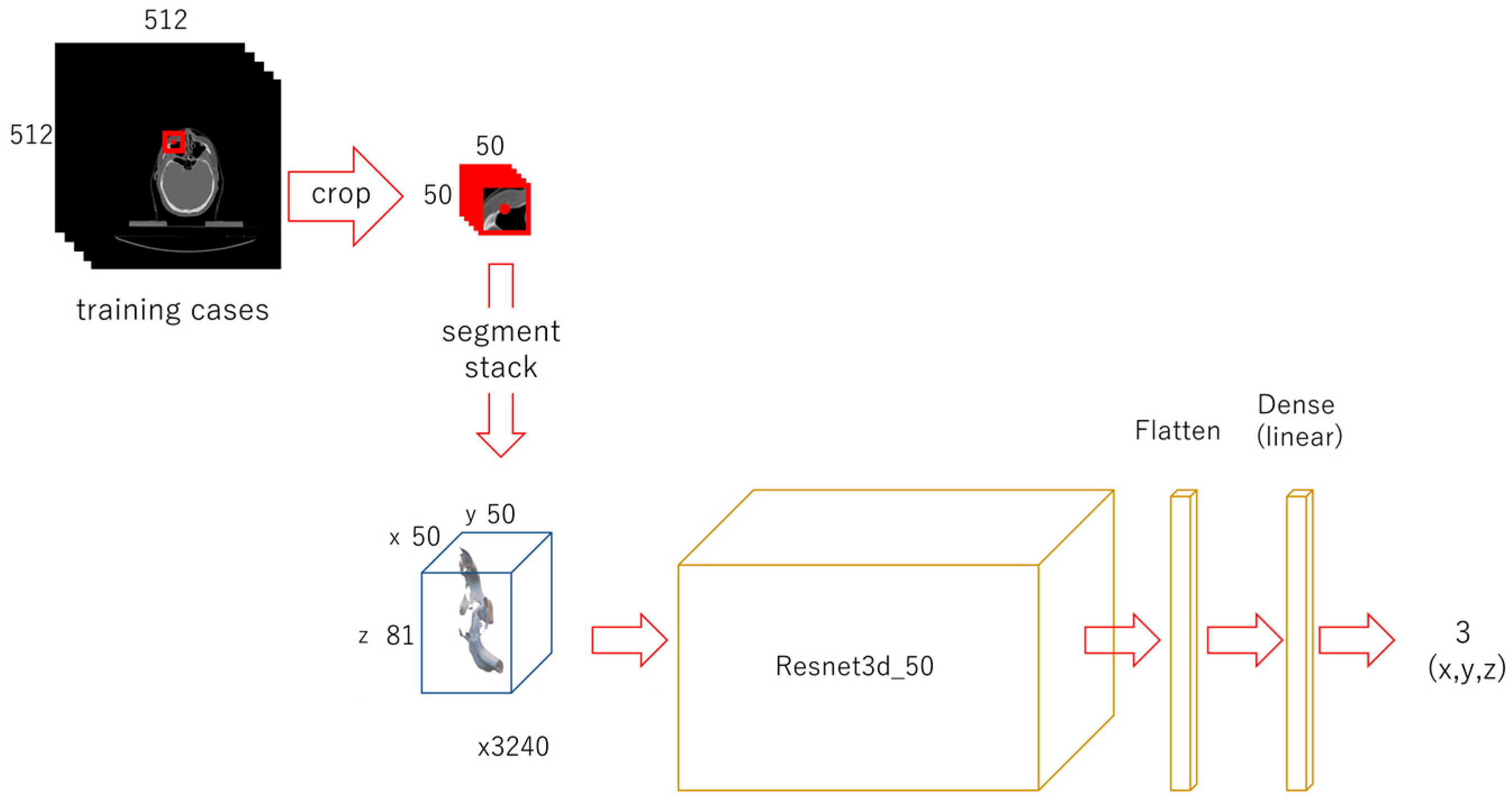
2.3.3. Third-Phase Deep Learning
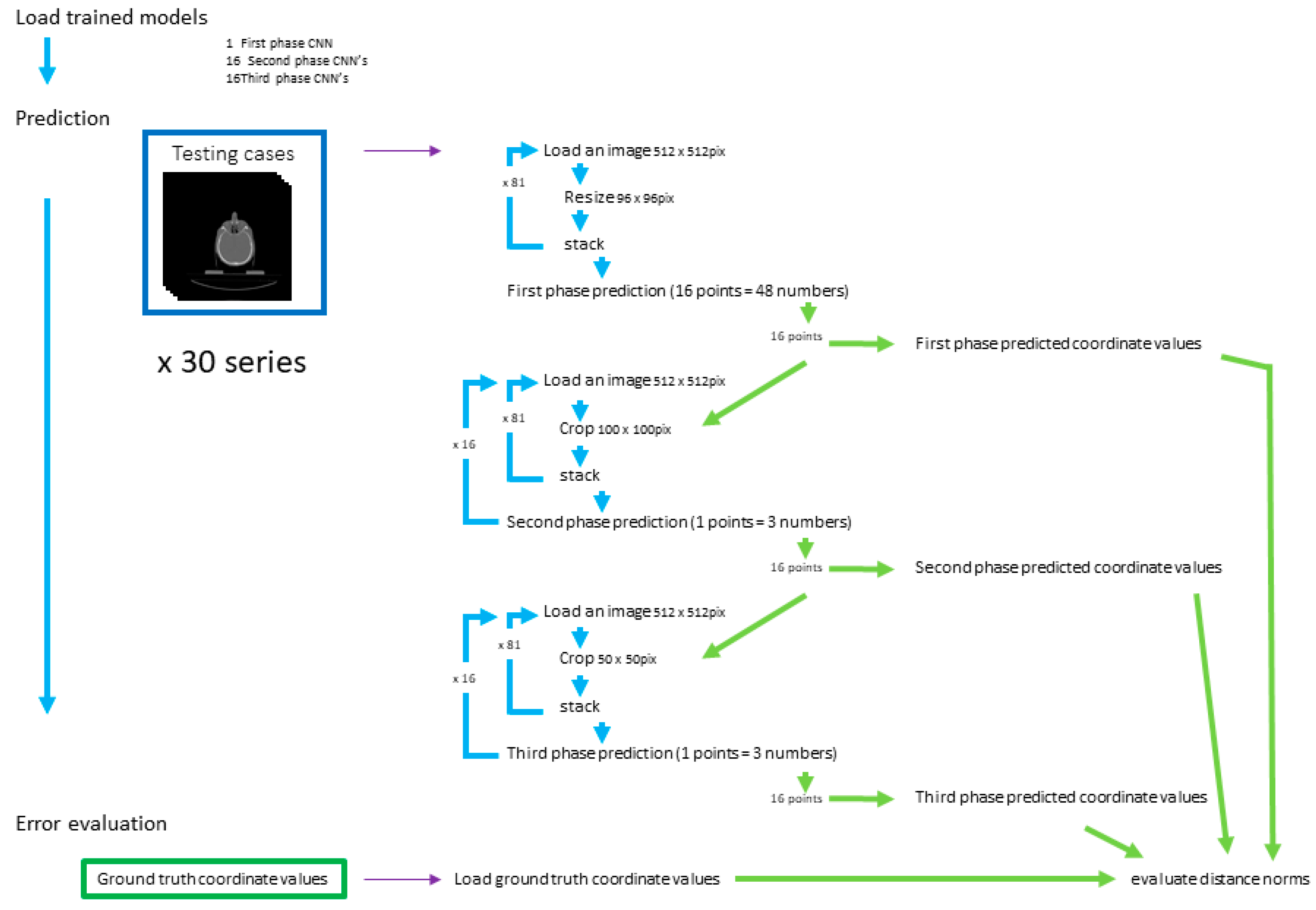
2.4. Evaluation
2.4.1. First-Phase Prediction
2.4.2. Second-Phase Prediction
2.4.3. Third-Phase Prediction
2.4.4. Prediction Error Evaluation
2.4.5. Statistical Analysis
3. Results
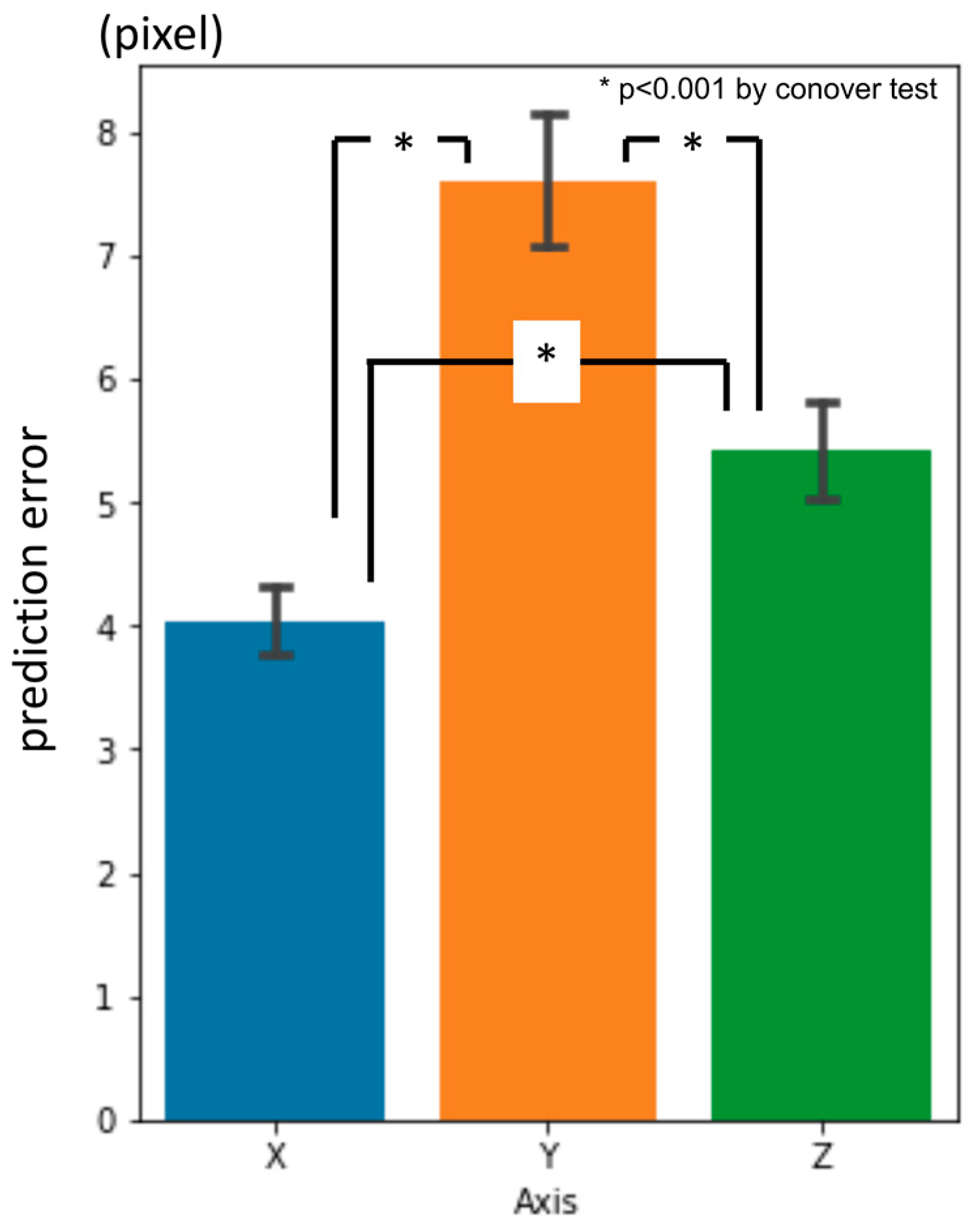
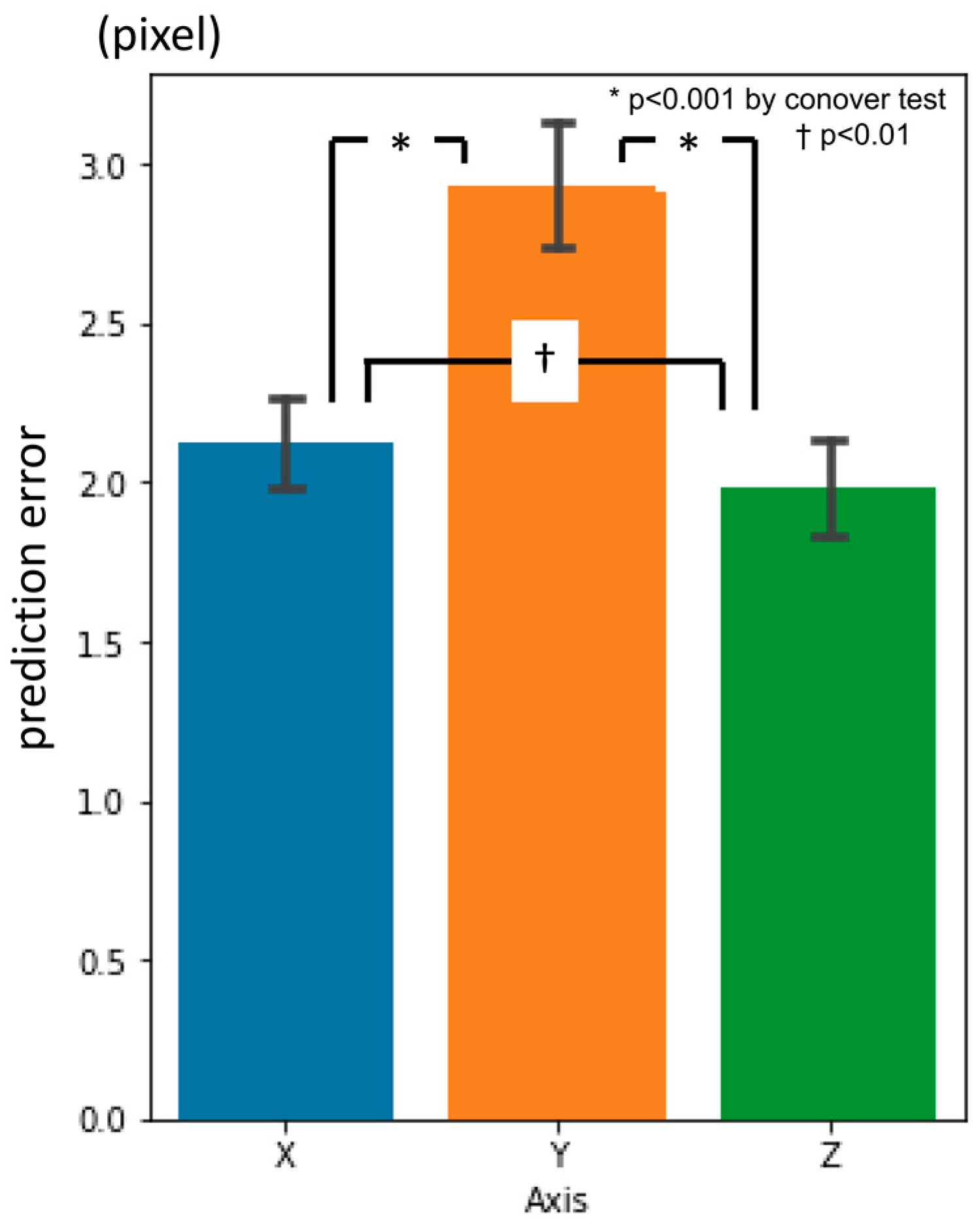
3.1. First-Phase Prediction Error
3.2. Second-Phase Prediction Error
3.3. Third-Phase Prediction Error
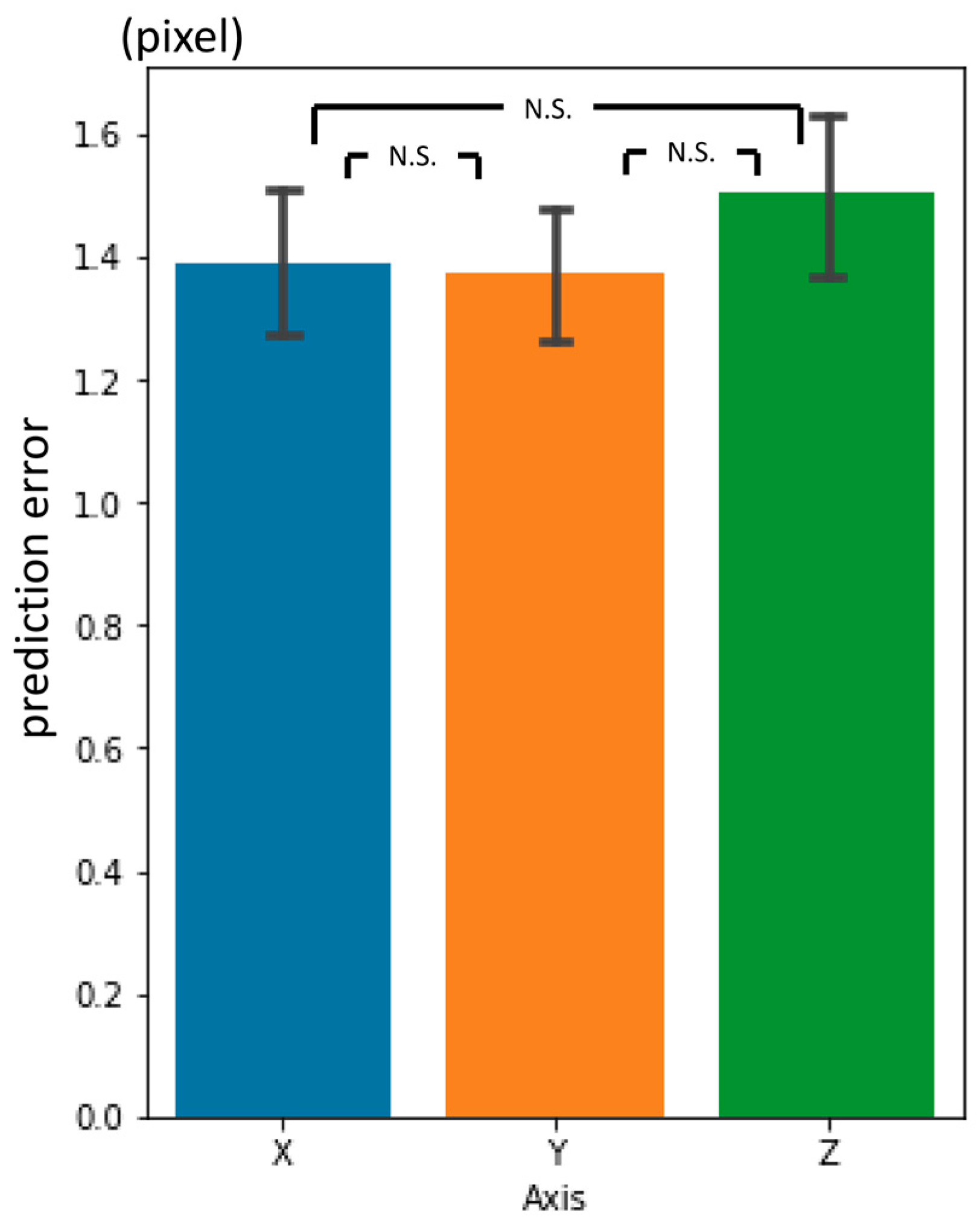
3.4. Inter-Phase and Inter-Observer Plotting Gaps
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Finlay, L.M. Craniometry and Cephalometry: A History Prior to the Advent of Radiography. Angle Orthod. 1980, 50, 312–321. [Google Scholar]
- Birmingham, A.E. The Topographical Anatomy of the Mastoid Region of the Skull; with Special Reference to Operation in This Region. Br. Med. J. 1890, 2, 683–684. [Google Scholar] [CrossRef]
- Scott, S.R.; Lond, M.B.; Eng, F.R.C.S.; Lond, L.R.C.P. A New Method of Demonstrating the Topographical Anatomy of the Adult Human Skull. J. Anat. Physiol. 1906, 40, 171–185. [Google Scholar] [PubMed]
- Ford, E.H.R. The Growth of the Foetal Skull. J. Anat. 1956, 90, 63–72. [Google Scholar] [PubMed]
- Hans, M.G.; Martin Palomo, J.; Valiathan, M. History of Imaging in Orthodontics from Broadbent to Cone-Beam Computed Tomography. Am. J. Orthod. Dentofac. Orthop. 2015, 148, 914–921. [Google Scholar] [CrossRef]
- Broadbent, B.H. A New X-Ray Technique and Its Application to Orthodontia. Angle Orthod. 1931, 1, 45–66. [Google Scholar]
- Hofrath, H. Die Bedeutung der Roentgenfern der Kiefer Anomalien. Fortschr Orthod. 1931, 1, 232–248. [Google Scholar]
- Steiner, C.C. Cephalometrics for You and Me. Am. J. Orthod. 1953, 39, 729–755. [Google Scholar] [CrossRef]
- Jacobson, A. The “Wits” Appraisal of Jaw Disharmony. Am. J. Orthod. 1975, 67, 125–138. [Google Scholar] [CrossRef]
- Björk, A. Sutural Growth of the Upper Face Studied by The Implant Method. Acta Odontol. Scand. 1966, 24, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Doberschütz, P.H.; Schwahn, C.; Krey, K.F. Cephalometric Analyses for Cleft Patients: A Statistical Approach to Compare the Variables of Delaire’s Craniofacial Analysis to Bergen Analysis. Clin. Oral Investig. 2022, 26, 353–364. [Google Scholar] [CrossRef]
- Lévy-Mandel, A.D.; Venetsanopoulos, A.N.; Tsotsos, J.K. Knowledge-Based Landmarking of Cephalograms. Comput. Biomed. Res. 1986, 19, 282–309. [Google Scholar] [CrossRef]
- Parthasarathy, S.; Nugent, S.T.; Gregson, P.G.; Fay, D.F. Automatic Landmarking of Cephalograms. Comput. Biomed. Res. 1989, 22, 248–269. [Google Scholar] [CrossRef]
- Cardillo, J.; Sid-Ahmed, M.A. An Image Processing System for Locating Craniofacial Landmarks. IEEE Trans. Med. Imaging 1994, 13, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Forsyth, D.B.; Davis, D.N. Assessment of an Automated Cephalometric Analysis System. Eur. J. Orthod. 1996, 18, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Giordano, D.; Leonardi, R.; Maiorana, F.; Cristaldi, G.; Distefano, M.L. Automatic Landmarking of Cephalograms by Cellular Neural Networks. In Proceedings of the 10th Conference on Artificial Intelligence in Medicine (AIME 2005), Aberdeen, UK, 23–27 July 2005; Lecture Notes in Computer Science. Springer: Berlin/Heidelberg, Germany, 2005; pp. 333–342. [Google Scholar]
- Yue, W.; Yin, D.; Li, C.; Wang, G.; Xu, T. Automated 2-D Cephalometric Analysis on X-Ray Images by a Model-Based Approach. IEEE Trans. Biomed. Eng. 2006, 53, 1615–1623. [Google Scholar]
- Rueda, S.; Alcañiz, M. An Approach for the Automatic Cephalometric Landmark Detection Using Mathematical Morphology and Active Appearance Models. In Proceedings of the 9th International Conference on Medical Image Computing and Computer-Assisted Intervention—MICCAI 2006, Copenhagen, Denmark, 1–6 October 2006; Lecture Notes in Computer Science. Springer: Berlin/Heidelberg, Germany, 2006; Volume 4190, pp. 159–166. [Google Scholar]
- Kafieh, R.; Sadri, S.; Mehri, A.; Raji, H. Discrimination of Bony Structures in Cephalograms for Automatic Landmark Detection. In Advances in Computer Science and Engineering; Communications in Computer and Information Science; Springer: Berlin/Heidelberg, Germany, 2008; Volume 6, pp. 609–620. [Google Scholar]
- Tanikawa, C.; Yagi, M.; Takada, K. Automated Cephalometry: System Performance Reliability Using Landmark-Dependent Criteria. Angle Orthod. 2009, 79, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, S.; Sotsuka, Y.; Kawai, K.; Ishise, H.; Kakibuchi, M. Personal Computer-Based Cephalometric Landmark Detection with Deep Learning, Using Cephalograms on the Internet. J. Craniofac. Surg. 2019, 30, 91–95. [Google Scholar] [CrossRef]
- Leonardi, R.; Giordano, D.; Maiorana, F. Automatic Cephalometric Analysis a Systematic Review. Angle Orthod. 2008, 78, 145–151. [Google Scholar] [CrossRef]
- Wang, C.W.; Huang, C.T.; Hsieh, M.C.; Li, C.H.; Chang, S.W.; Li, W.C.; Vandaele, R.; Marée, R.; Jodogne, S.; Geurts, P.; et al. Evaluation and Comparison of Anatomical Landmark Detection Methods for Cephalometric X-Ray Images: A Grand Challenge. IEEE Trans. Med. Imaging 2015, 34, 1890–1900. [Google Scholar] [CrossRef]
- Wang, C.W.; Huang, C.T.; Lee, J.H.; Li, C.H.; Chang, S.W.; Siao, M.J.; Lai, T.M.; Ibragimov, B.; Vrtovec, T.; Ronneberger, O.; et al. A Benchmark for Comparison of Dental Radiography Analysis Algorithms. Med. Image Anal. 2016, 31, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Lindner, C.; Wang, C.-W.; Huang, C.-T.; Li, C.-H.; Chang, S.-W.; Cootes, T.F. Fully Automatic System for Accurate Localisation and Analysis of Cephalometric Landmarks in Lateral Cephalograms. Sci. Rep. 2016, 6, 33581. [Google Scholar] [CrossRef] [PubMed]
- Arik, S.Ö.; Ibragimov, B.; Xing, L. Fully Automated Quantitative Cephalometry Using Convolutional Neural Networks. J. Med. Imaging 2017, 4, 014501. [Google Scholar] [CrossRef]
- Nishimoto, S. Cephalometric Landmark Location with Multi-Phase Deep Learning. In Proceedings of the 34th Annual Conference of the Japanese Society for Artificial Intelligence, Online, 9–12 June 2020; p. 2Q6GS1001. [Google Scholar]
- Nishimoto, S.; Kawai, K.; Fujiwara, T.; Ishise, H.; Kakibuchi, M. Locating Cephalometric Landmarks with Multi-Phase Deep Learning. medRxiv 2020. [Google Scholar] [CrossRef]
- Nishimoto, S.; Kawai, K.; Fujiwara, T.; Ishise, H.; Kakibuchi, M. Locating Cephalometric Landmarks with Multi-Phase Deep Learning. J. Dent. Health Oral Res. 2023, 4, 1–13. [Google Scholar] [CrossRef]
- Kim, H.; Shim, E.; Park, J.; Kim, Y.J.; Lee, U.; Kim, Y. Web-Based Fully Automated Cephalometric Analysis by Deep Learning. Comput. Methods Programs Biomed. 2020, 194, 105513. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Liu, Y.; Oh, S.H.; Ahn, H.W.; Kim, S.H.; Nelson, G. Automatic Cephalometric Landmark Identification System Based on the Multi-Stage Convolutional Neural Networks with CBCT Combination Images. Sensors 2021, 21, 505. [Google Scholar] [CrossRef]
- Chen, R.; Ma, Y.; Chen, N.; Lee, D.; Wang, W. Cephalometric Landmark Detection by AttentiveFeature Pyramid Fusion and Regression-Voting. In Proceedings of the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, Shenzhen, China, 13–17 October 2019; Lecture Notes in Computer Science. Springer: Cham, Switzerland, 2019; Volume 11766, pp. 873–881. [Google Scholar]
- Junaid, N.; Khan, N.; Ahmed, N.; Abbasi, M.S.; Das, G.; Maqsood, A.; Ahmed, A.R.; Marya, A.; Alam, M.K.; Heboyan, A. Development, Application, and Performance of Artificial Intelligence in Cephalometric Landmark Identification and Diagnosis: A Systematic Review. Healthcare 2022, 10, 2454. [Google Scholar] [CrossRef]
- Khalid, M.A.; Zulfiqar, K.; Bashir, U.; Shaheen, A.; Iqbal, R.; Rizwan, Z.; Rizwan, G.; Fraz, M.M. CEPHA29: Automatic Cephalometric Landmark Detection Challenge 2023. arXiv 2022, arXiv:2212.04808. [Google Scholar]
- Grayson, B.H.; McCarthy, J.G.; Bookstein, F. Analysis of Craniofacial Asymmetry by Multiplane Cephalometry. Am. J. Orthod. 1983, 84, 217–224. [Google Scholar] [CrossRef]
- Bütow, K.W.; van der Walt, P.J. The Use of Triangle Analysis for Cephalometric Analysis in Three Dimensions. J. Maxillofac. Surg. 1984, 12, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Grayson, B.; Cutting, C.; Bookstein, F.L.; Kim, H.; McCarthy, J.G. The Three-Dimensional Cephalogram: Theory, Techniques, and Clinical Application. Am. J. Orthod. Dentofac. Orthop. 1988, 94, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Miyajima, T.; Minami, K.; Sakuda, M. An Accurate Three-Dimensional Cephalometric System: A Solution for the Correction of Cephalic Malpositioning. J. Orthod. 2001, 28, 143–149. [Google Scholar] [CrossRef]
- Matteson, S.R.; Bechtold, W.; Phillips, C.; Staab, E.V. A Method for Three-Dimensional Image Reformation for Quantitative Cephalometric Analysis. J. Oral Maxillofac. Surg. 1989, 47, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Varghese, S.; Kailasam, V.; Padmanabhan, S.; Vikraman, B.; Chithranjan, A. Evaluation of the Accuracy of Linear Measurements on Spiral Computed Tomography-Derived Three-Dimensional Images and Its Comparison with Digital Cephalometric Radiography. Dentomaxillofac. Radiol. 2010, 39, 216–223. [Google Scholar] [CrossRef]
- Ghoneima, A.; Albarakati, S.; Baysal, A.; Uysal, T.; Kula, K. Measurements from Conventional, Digital and CT-Derived Cephalograms: A Comparative Study. Aust. Orthod. J. 2012, 28, 232–239. [Google Scholar]
- Dot, G.; Rafflenbeul, F.; Arbotto, M.; Gajny, L.; Rouch, P.; Schouman, T. Accuracy and Reliability of Automatic Three-Dimensional Cephalometric Landmarking. Int. J. Oral Maxillofac. Surg. 2020, 49, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, S.; Bahrampour, E.; Soltanimehr, E.; Zamani, A.; Oshagh, M.; Moattari, M.; Mehdizadeh, A. The Accuracy of a Designed Software for Automated Localization of Craniofacial Landmarks on CBCT Images. BMC Med. Imaging 2014, 14, 32. [Google Scholar] [CrossRef]
- Gupta, A.; Kharbanda, O.P.; Sardana, V.; Balachandran, R.; Sardana, H.K. A Knowledge-Based Algorithm for Automatic Detection of Cephalometric Landmarks on CBCT Images. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, 1737–1752. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, Y.; Wang, L.; Tang, Z.; Xia, J.J.; Shen, D. Automatic Craniomaxillofacial Landmark Digitization via Segmentation-Guided Partially-Joint Regression Forest Model and Multiscale Statistical Features. IEEE Trans. Biomed. Eng. 2016, 63, 1820–1829. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, M.; Wang, L.; Chen, S.; Yuan, P.; Li, J.; Shen, S.G.F.; Tang, Z.; Chen, K.C.; Xia, J.J.; et al. Joint Craniomaxillofacial Bone Segmentation and Landmark Digitization by Context-Guided Fully Convolutional Networks. In Proceedings of the 20th International Conference on Medical Image Computing and Computer-Assisted Intervention—MICCAI 2017, Quebec City, QC, Canada, 11–13 September 2017; Lecture Notes in Computer Science. Springer: Cham, Switzerland, 2017; Volume 10434, pp. 720–728. [Google Scholar]
- De Jong, M.A.; Gül, A.; De Gijt, J.P.; Koudstaal, M.J.; Kayser, M.; Wolvius, E.B.; Böhringer, S. Automated Human Skull Landmarking with 2D Gabor Wavelets. Phys. Med. Biol. 2018, 63, 105011. [Google Scholar] [CrossRef]
- Montúfar, J.; Romero, M.; Scougall-Vilchis, R.J. Automatic 3-Dimensional Cephalometric Landmarking Based on Active Shape Models in Related Projections. Am. J. Orthod. Dentofac. Orthop. 2018, 153, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Montúfar, J.; Romero, M.; Scougall-Vilchis, R.J. Hybrid Approach for Automatic Cephalometric Landmark Annotation on Cone-Beam Computed Tomography Volumes. Am. J. Orthod. Dentofac. Orthop. 2018, 154, 140–150. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, A.Q.; Kascenas, A.; Henry, J.; Wyeth, D.; Shepherd, M.; Beveridge, E.; Clunie, L.; Sansom, C.; Šeduikytė, E.; Muir, K.; et al. Attaining Human-Level Performance with Atlas Location Autocontext for Anatomical Landmark Detection in 3D CT Data. In Proceedings of the Computer Vision—ECCV 2018 Workshops, Munich, Germany, 8–14 September 2018; Lecture Notes in Computer Science. Springer: Cham, Switzerland, 2018; Volume 11131, pp. 470–484. [Google Scholar]
- Kang, S.H.; Jeon, K.; Kim, H.-J.; Seo, J.K.; Lee, S.-H. Automatic Three-Dimensional Cephalometric Annotation System Using Three-Dimensional Convolutional Neural Networks. arXiv 2018, arXiv:1811.07889. [Google Scholar] [CrossRef]
- Torosdagli, N.; Liberton, D.K.; Verma, P.; Sincan, M.; Lee, J.S.; Bagci, U. Deep Geodesic Learning for Segmentation and Anatomical Landmarking. IEEE Trans. Med. Imaging 2019, 38, 919–931. [Google Scholar] [CrossRef]
- Yun, H.S.; Jang, T.J.; Lee, S.M.; Lee, S.H.; Seo, J.K. Learning-Based Local-to-Global Landmark Annotation for Automatic 3D Cephalometry. Phys. Med. Biol. 2020, 65, 0850181. [Google Scholar] [CrossRef] [PubMed]
- Dot, G.; Schouman, T.; Chang, S.; Rafflenbeul, F.; Kerbrat, A.; Rouch, P.; Gajny, L. Automatic 3-Dimensional Cephalometric Landmarking via Deep Learning. J. Dent. Res. 2022, 101, 1380–1387. [Google Scholar] [CrossRef]
- Yun, H.S.; Hyun, C.M.; Baek, S.H.; Lee, S.H.; Seo, J.K. A Semi-Supervised Learning Approach for Automated 3D Cephalometric Landmark Identification Using Computed Tomography. PLoS ONE 2022, 17, e0275114. [Google Scholar] [CrossRef] [PubMed]
- Blake, G. Head-Neck-Radiomics-HN1—The Cancer Imaging Archive (TCIA) Public Access—Cancer Imaging Archive Wiki. Available online: https://wiki.cancerimagingarchive.net/display/Public/Head-Neck-Radiomics-HN1 (accessed on 1 February 2021).
- Lasso, A. Lassoan/ExtractSkin.Py. Available online: https://gist.github.com/lassoan/1673b25d8e7913cbc245b4f09ed853f9 (accessed on 1 February 2021).
- Jihong, J. GitHub—JihongJu/Keras-Resnet3d: Implementations of ResNets for Volumetric Data, Including a Vanilla Resnet in 3D. Available online: https://github.com/JihongJu/keras-resnet3d (accessed on 1 February 2021).
- Mestiri, M.; Kamel, H. Reeb Graph for Automatic 3D Cephalometry. Int. J. Image Process. 2014, 8, 2014–2031. [Google Scholar]
- Neelapu, B.C.; Kharbanda, O.P.; Sardana, V.; Gupta, A.; Vasamsetti, S.; Balachandran, R.; Sardana, H.K. Automatic Localization of Three-Dimensional Cephalometric Landmarks on CBCT Images by Extracting Symmetry Features of the Skull. Dentomaxillofac. Radiol. 2018, 47, 20170054. [Google Scholar] [CrossRef]
- Codari, M.; Caffini, M.; Tartaglia, G.M.; Sforza, C.; Baselli, G. Computer-Aided Cephalometric Landmark Annotation for CBCT Data. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 113–121. [Google Scholar] [CrossRef]
- Payer, C.; Štern, D.; Bischof, H.; Urschler, M. Integrating Spatial Configuration into Heatmap Regression Based CNNs for Landmark Localization. Med. Image Anal. 2019, 54, 207–219. [Google Scholar] [CrossRef]
- Gonçalves, F.A.; Schiavon, L.; Pereira Neto, J.S.; Nouer, D.F. Comparison of Cephalometric Measurements from Three Radiological Clinics. Braz. Oral Res. 2006, 20, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Paini de Abreu, D.; Salvatore Freitas, K.M.; Nomura, S.; Pinelli Valarelli, F.; Hermont Cançado, R. Comparison among Manual and Computerized Cephalometrics Using the Softwares Dolphin Imaging and Dentofacial Planner. Dent. Oral Craniofacial Res. 2016, 2, 1–5. [Google Scholar] [CrossRef]
- Moon, J.H.; Hwang, H.W.; Yu, Y.; Kim, M.G.; Donatelli, R.E.; Lee, S.J. How Much Deep Learning Is Enough for Automatic Identification to Be Reliable? A Cephalometric Example. Angle Orthod. 2020, 90, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, S. Automatic Landmark Prediction in Craniofacial CT Images. In Proceedings of the 35th Annual Conference of the Japanese Society for Artificial Intelligence, Online, 8–11 June 2021; p. 4I4GS7e04. [Google Scholar]


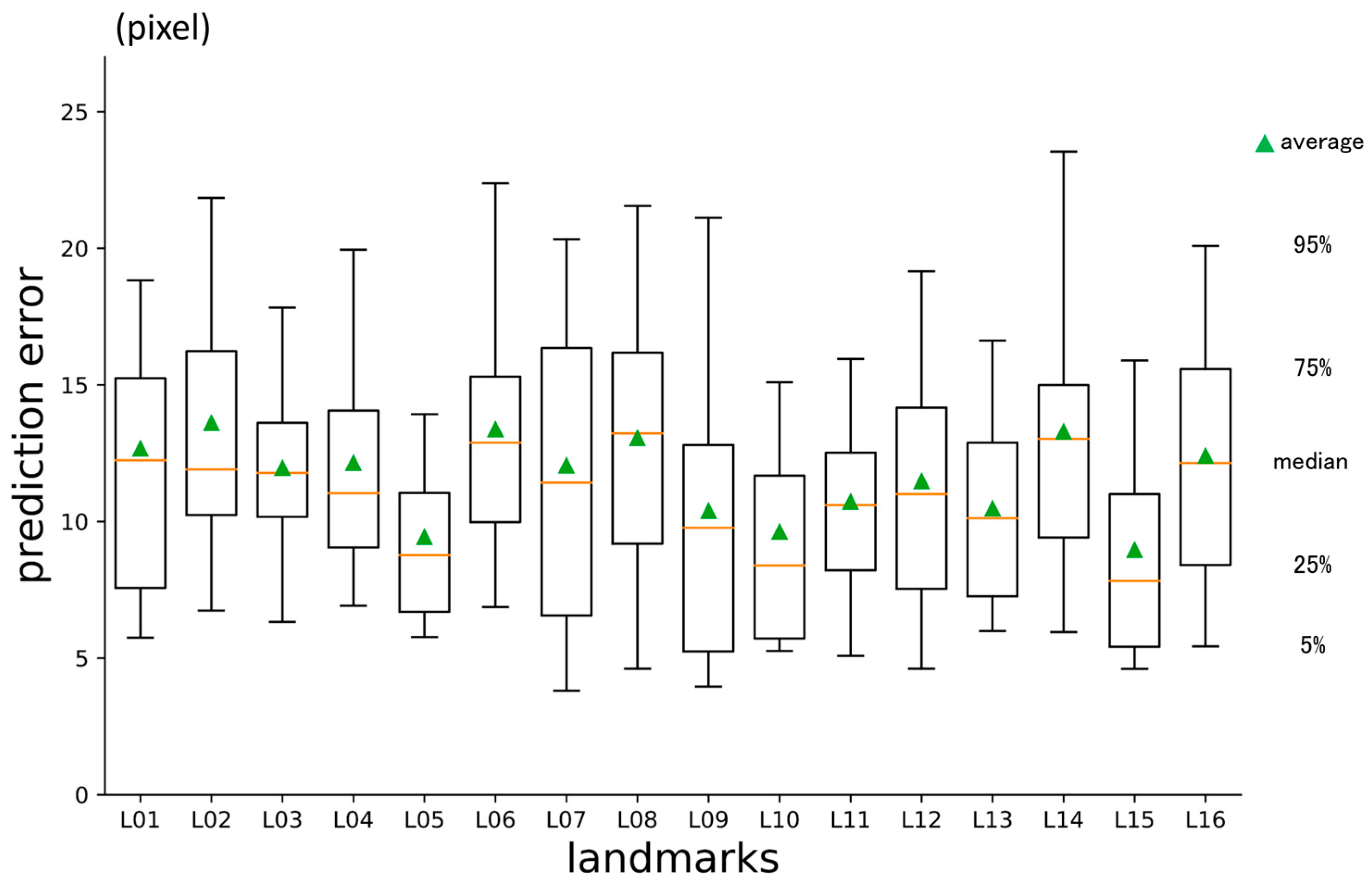
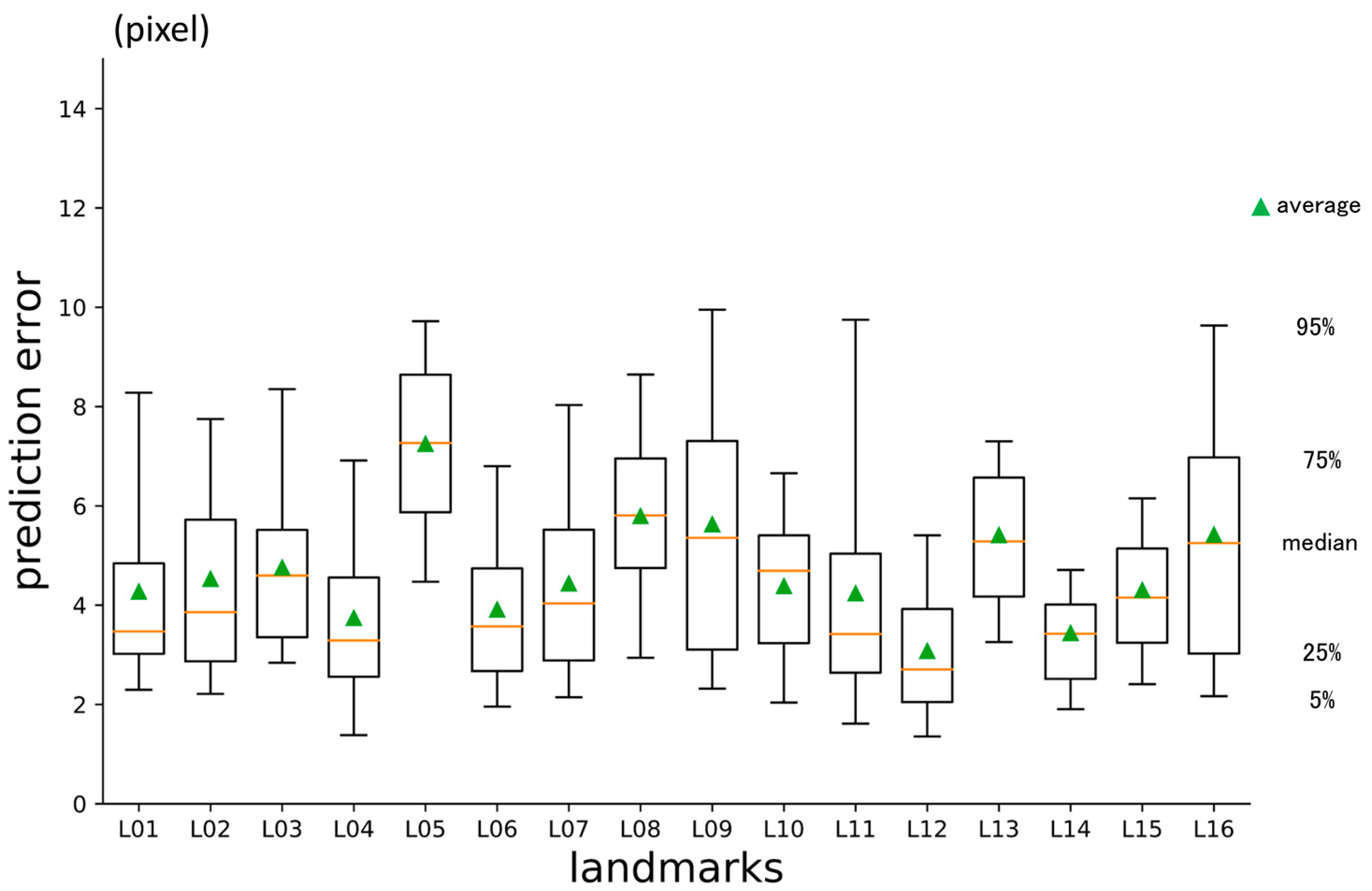
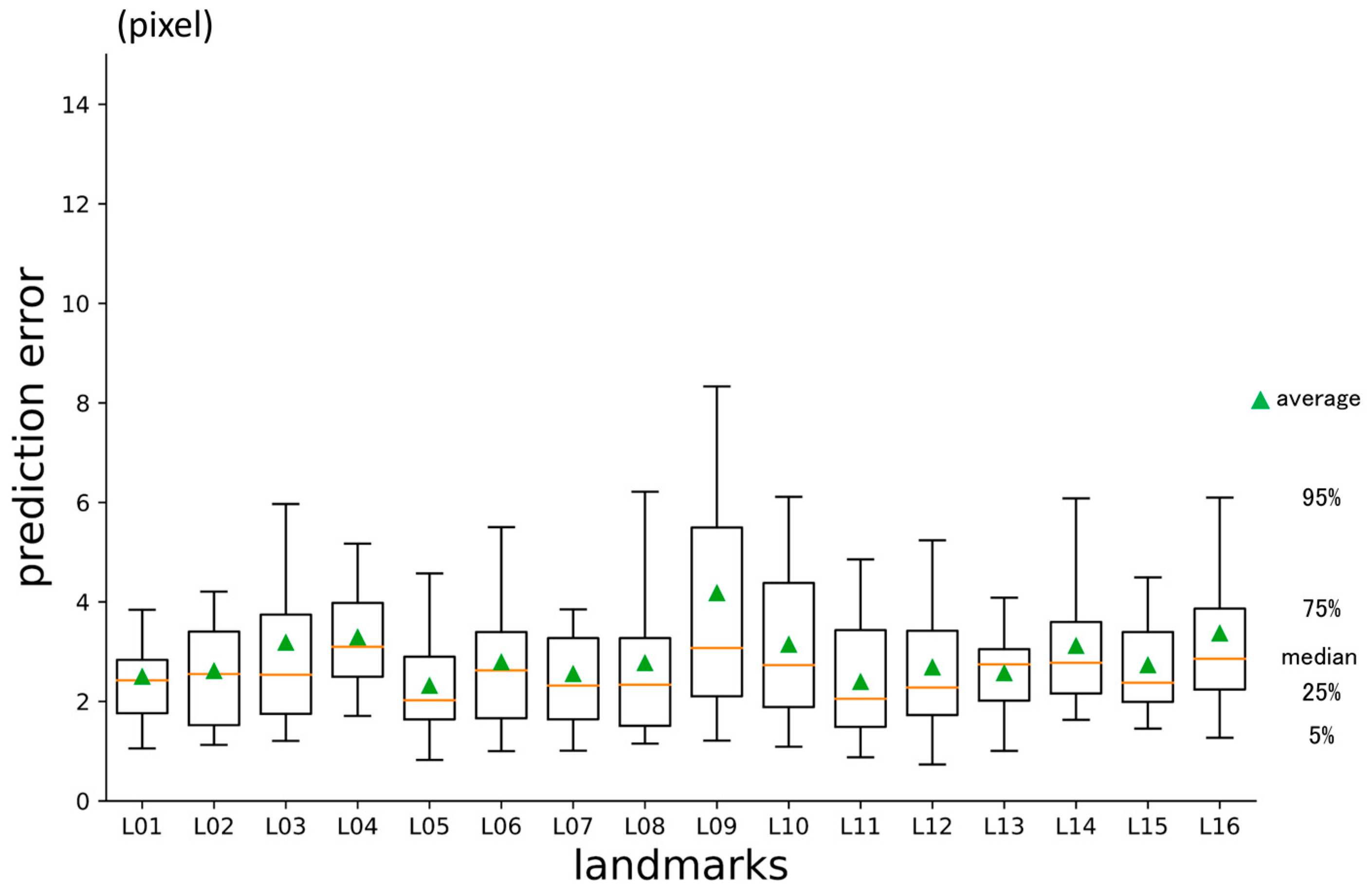
| No | Abbreviation | Description |
|---|---|---|
| L01 | A | Point A |
| L02 | AntNS | Anterior nasal spine |
| L03 | LGoni | Left gonion |
| L04 | LOrbi | Left inferior lateral orbital rim |
| L05 | LPori | Left porion |
| L06 | LsupO | Left supra orbital incisura |
| L07 | Mento | Menton |
| L08 | Nasio | Nasion |
| L09 | PocEx | External occipital protuberance |
| L10 | PosNS | Posterior nasal spine |
| L11 | RGoni | Right gonion |
| L12 | ROrbi | Right inferior lateral orbital rim |
| L13 | RPori | Right porion |
| L14 | RsupO | Right supra orbital incisura |
| L15 | Sella | Center of sella turcica |
| L16 | XstaG | Top of crista galli |
| First Phase | Second Phase | Third Phase | Inter-Observer Gaps | ||||
|---|---|---|---|---|---|---|---|
| Average | 11.6 | _ * _ | 4.66 | _ * _ | 2.88 | _ N.S. _ | 3.08 |
| Median | 10.89 | 4.22 | 2.56 | 2.4 | |||
| Stdev | 5.64 | 2.27 | 1.67 | 2.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishimoto, S.; Saito, T.; Ishise, H.; Fujiwara, T.; Kawai, K.; Kakibuchi, M. Three-Dimensional Craniofacial Landmark Detection in Series of CT Slices Using Multi-Phased Regression Networks. Diagnostics 2023, 13, 1930. https://doi.org/10.3390/diagnostics13111930
Nishimoto S, Saito T, Ishise H, Fujiwara T, Kawai K, Kakibuchi M. Three-Dimensional Craniofacial Landmark Detection in Series of CT Slices Using Multi-Phased Regression Networks. Diagnostics. 2023; 13(11):1930. https://doi.org/10.3390/diagnostics13111930
Chicago/Turabian StyleNishimoto, Soh, Takuya Saito, Hisako Ishise, Toshihiro Fujiwara, Kenichiro Kawai, and Masao Kakibuchi. 2023. "Three-Dimensional Craniofacial Landmark Detection in Series of CT Slices Using Multi-Phased Regression Networks" Diagnostics 13, no. 11: 1930. https://doi.org/10.3390/diagnostics13111930





