Ensemble Deep Learning Derived from Transfer Learning for Classification of COVID-19 Patients on Hybrid Deep-Learning-Based Lung Segmentation: A Data Augmentation and Balancing Framework
Abstract
:1. Introduction
2. Background Literature
3. Methodology
3.1. Image Acquisition and Data Demographics
3.2. Overall Pipeline of the System
3.3. Hybrid Deep Learning Architecture of CT Lung Segmentation
3.4. Transfer-Learning-Based Architecture for Classification
3.5. Ensemble Deep Learning Architectures for Classification
| Algorithm 1: EDL generator |
| Result: Combination of TL models score generates five EDLs |
| Input: TL Model predicted score |
| EDL = [ ]; |
| While1 len(EDL) < 6 do: |
| While2 i in range (2,8) do: |
| While3 k in combinations, i: do: |
| NewEDL = GenerateEDL(TLk,i combinations); |
| //GenerateEDL function to generate on predicted score of TLs |
| If ACC of NewEDL >ACC of contituents TLs ACC then |
| EDL.append(NewEDL); |
| Else |
| Print(“Try new combination”) |
| End //End of If |
| End //End of while3 |
| End //End of while2 |
| End //End of while1 |
3.6. Loss Function
3.7. Performance Metric
3.8. Experimental Protocol
3.8.1. Five Data Combinations
- DC1: Training validation and testing using both CroMED (COVID) and NovMED (control).
- DC2: Training validation and testing on both NovMED (COVID) and NovMED (control).
- DC3: Training validation using CroMED (COVID) and NovMED (control) and testing on NovMED (COVID) and NovMED (control).
- DC4: Training validation using NovMED (COVID) and NovMED (control) and testing on CroMED (COVID) and NovMED (control).
- DC5: Training validation and testing on mixed data in which COVID CT scans from Croatia and Italy are mixed; the control of Italy was used.
3.8.2. Experiment 1: Transfer Learning Models using Lung Segmented Data
3.8.3. Experiment 2: Ensemble Deep Learning for Classification
3.8.4. Experiment 3: Effect of EDL Classification over TL Classification with Augmentation
3.8.5. Experiment 4: Unseen Data Analysis
3.9. Experimental Setup
3.10. Power Analysis
4. Results and Performance Evaluation
4.1. PE for HDL Lung Segmentation
4.2. Results of Experiment 1: Transfer Learning Models using Lung Segmented Data
- DC1 results: Table 2 and Figure 12 show that the best accuracy of 97.93% without augmentation and 99.93% with augmentation is shown by MobileNetV2. The mean accuracy of all seven TLs without augmentation is 93.91% and is 97.03% with augmentation. For TL6 (VGG16), the accuracy improves from 90.20% (before augmentation) to 95.61% (after augmentation), so the improvement was 5.41% using DC1 data combination. TL2 (Inception V3) had an accuracies of 93.60% (before augmentation) and 93.97% (after augmentation), so the improvement was 0.37%. Therefore, we see that augmentation has different effects on TL-based classifiers. It is more pronounced in TL6, unlike in TL2. Table 3 shows the COVID precision are significantly increased or comparable after balancing data.
- DC2 results: Table 4 and Figure 13 show that the best accuracy of 90.84% is achieved by InceptionV3 without augmentation, and the best with augmentation of 93.92% is achieved by EfficientNetV2M. The mean accuracy of all seven TLs without augmentation is 84.41% and is 89.85% with augmentation. TL4 (ResNet152), the accuracy improves from 78.16% (before augmentation) to 87.40% (after augmentation) when using DC2 data combination, so the improvement was 11.82%. TL6 (VGG16) had accuracies of 85.6% (before augmentation) and 84.05% (after augmentation), so there was no improvement. Therefore, we see that augmentation has different effects on TL-based classifiers. It is more pronounced in TL4, unlike in TL6. Table 5 shows the effect of augmentation in COVID precision, recall and F1-score. These are significantly increased or comparable after balancing data.
- DC3 results: Table 6 and Figure 14 show that the best accuracies of 85.40% without augmentation and 91.41% with augmentation are achieved by EfficientNetV2M. The mean accuracy of all seven TLs without augmentation is 72.90% and is 82.355% with augmentation. For TL5 (ResNet50), the accuracy improves from 67.17% (before augmentation) to 80% (after augmentation) when using DC3 data combination, so the improvement was 19.10%. TL2 (InceptionV3) had accuracies of 67.58% (before augmentation) and 76.43% (after augmentation), so the improvement was 13.09%. Therefore, we see that augmentation has different effects on TL-based classifiers. It is more pronounced in TL5, unlike in TL2. Augmentation and balancing effects are visible in Table 7. It shows that better results can be achieved after balancing the data.
- DC4 results: Table 8 and Figure 15 show that the best accuracies of 69.40% without augmentation and 81.05% with augmentation are shown by VGG19. The mean accuracy of all seven TLs without augmentation is 47.85% and is 70.39% with augmentation. The augmentation effect was also visible with TL3 (MobileNetV2), which had a lowest accuracy of 27.97% before augmentation and 52.68% after the augmentation, so the improvement is 92.5%. Table 9 has been presented to show augmentation effect for precision, recall and F1-score.
- DC5 results: Table 10 and Figure 16 show that the best accuracy of 95.10% is achieved by InceptionV3 without augmentation, and 95.28% is achieved by VGG16 with augmentation. The mean accuracy of all seven TLs without augmentation is 91.22% and is 93.76% with augmentation. TL6 (VGG16), the accuracy improves from 86.81% (before augmentation) to 95.28% (after augmentation) when using DC5 data combination, so the improvement is 9.75%. TL3 (MobileNetV2) has an accuracies of 92.95% (before augmentation) and 89.07% (after augmentation), so there is no improvement. Therefore, we see that augmentation has different effects on TL-based classifiers. It is more pronounced in TL6, unlike in TL3. In the most of TL models, improvement of precision, recall and F1-score can be seen Table 11, after the balancing and augmenting the data.
- Table 3, Table 5, Table 7, Table 9 and Table 11 also show the precision and a comparison to Experiment 1, which presents the verification of Hypothesis 2 that data augmentation helps in improvement in the performance of TL model. p-value based on the Mann–Whitney test was used for all data combinations.
4.3. Results of Experiment 2: Ensemble Deep Learning for Classification
4.4. Results of Experiment 3: EDL vs. TL Classification with Augmentation
4.5. Results of Experiment 4: Unseen Data Analysis
4.6. Receiver Operating Charaterstics
5. System Reliability
Statistical Test
6. Discussion
6.1. Principal Findings
6.2. Benchmarking
6.3. A Special Note on EDL
6.4. Strengths, Weaknesses, and Extensions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A



Appendix B







Appendix C





References
- Congiu, T.; Demontis, R.; Cau, F.; Piras, M.; Fanni, D.; Gerosa, C.; Botta, C.; Scano, A.; Chighine, A.; Faedda, E. Scanning electron microscopy of lung disease due to COVID-19—A case report and a review of the literature. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 7997–8003. [Google Scholar]
- Suri, J.S.; Agarwal, S.; Elavarthi, P.; Pathak, R.; Ketireddy, V.; Columbu, M.; Saba, L.; Gupta, S.K.; Faa, G.; Singh, I.M.; et al. Inter-Variability Study of COVLIAS 1.0: Hybrid Deep Learning Models for COVID-19 Lung Segmentation in Computed Tomography. Diagnostics 2021, 11, 2025. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S.; Agarwal, S.; Gupta, S.; Puvvula, A.; Viskovic, K.; Suri, N.; Alizad, A.; El-Baz, A.; Saba, L.; Fatemi, M.; et al. Systematic Review of Artificial Intelligence in Acute Respiratory Distress Syndrome for COVID-19 Lung Patients: A Biomedical Imaging Perspective. IEEE J. Biomed. Health Inf. 2021, 25, 4128–4139. [Google Scholar] [CrossRef] [PubMed]
- Munjral, S.; Ahluwalia, P.; Jamthikar, A.D.; Puvvula, A.; Saba, L.; Faa, G.; Singh, I.M.; Chadha, P.S.; Turk, M.; Johri, A.M. Nutrition, atherosclerosis, arterial imaging, cardiovascular risk stratification, and manifestations in COVID-19 framework: A narrative review. Front. Biosci. 2021, 26, 1312–1339. [Google Scholar]
- Sanagala, S.S.; Nicolaides, A.; Gupta, S.K.; Koppula, V.K.; Saba, L.; Agarwal, S.; Johri, A.M.; Kalra, M.S.; Suri, J.S. Ten Fast Transfer Learning Models for Carotid Ultrasound Plaque Tissue Characterization in Augmentation Framework Embedded with Heatmaps for Stroke Risk Stratification. Diagnostics 2021, 11, 2109. [Google Scholar] [CrossRef] [PubMed]
- Jena, B.; Saxena, S.; Nayak, G.K.; Saba, L.; Sharma, N.; Suri, J.S. Artificial intelligence-based hybrid deep learning models for image classification: The first narrative review. Comput. Biol. Med. 2021, 137, 104803. [Google Scholar] [CrossRef]
- Gerosa, C.; Faa, G.; Fanni, D.; Manchia, M.; Suri, J.; Ravarino, A.; Barcellona, D.; Pichiri, G.; Coni, P.; Congiu, T. Fetal programming of COVID-19: May the barker hypothesis explain the susceptibility of a subset of young adults to develop severe disease. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 5876–5884. [Google Scholar]
- Suri, J.S.; Agarwal, S.; Saba, L.; Chabert, G.L.; Carriero, A.; Paschè, A.; Danna, P.; Mehmedović, A.; Faa, G.; Jujaray, T. Multicenter Study on COVID-19 Lung Computed Tomography Segmentation with varying Glass Ground Opacities using Unseen Deep Learning Artificial Intelligence Paradigms: COVLIAS 1.0 Validation. J. Med. Syst. 2022, 46, 62. [Google Scholar] [CrossRef]
- Suri, J.S.; Puvvula, A.; Biswas, M.; Majhail, M.; Saba, L.; Faa, G.; Singh, I.M.; Oberleitner, R.; Turk, M.; Chadha, P.S. COVID-19 pathways for brain and heart injury in comorbidity patients: A role of medical imaging and artificial intelligence-based COVID severity classification: A review. Comput. Biol. Med. 2020, 124, 103960. [Google Scholar] [CrossRef]
- Shalbaf, A.; Vafaeezadeh, M. Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 115–123. [Google Scholar]
- Suri, J.S.; Maindarkar, M.A.; Paul, S.; Ahluwalia, P.; Bhagawati, M.; Saba, L.; Faa, G.; Saxena, S.; Singh, I.M.; Chadha, P.S. Deep Learning Paradigm for Cardiovascular Disease/Stroke Risk Stratification in Parkinson’s Disease Affected by COVID-19: A Narrative Review. Diagnostics 2022, 12, 1543. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Sanagala, S.S.; Gupta, S.K.; Koppula, V.K.; Laird, J.R.; Viswanathan, V.; Sanches, M.J.; Kitas, G.D.; Johri, A.M.; Sharma, N. A Multicenter Study on Carotid Ultrasound Plaque Tissue Characterization and Classification Using Six Deep Artificial Intelligence Models: A Stroke Application. IEEE Trans. Instrum. Meas. 2021, 70, 1–12. [Google Scholar] [CrossRef]
- Skandha, S.S.; Nicolaides, A.; Gupta, S.K.; Koppula, V.K.; Saba, L.; Johri, A.M.; Kalra, M.S.; Suri, J.S. A hybrid deep learning paradigm for carotid plaque tissue characterization and its validation in multicenter cohorts using a supercomputer framework. Comput. Biol. Med. 2022, 141, 105131. [Google Scholar] [CrossRef]
- Saba, L.; Biswas, M.; Suri, H.S.; Viskovic, K.; Laird, J.R.; Cuadrado-Godia, E.; Nicolaides, A.; Khanna, N.N.; Viswanathan, V.; Suri, J.S. Ultrasound-based carotid stenosis measurement and risk stratification in diabetic cohort: A deep learning paradigm. Cardiovasc. Diagn. Ther. 2019, 9, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Saba, L.; Gupta, S.K.; Carriero, A.; Falaschi, Z.; Paschè, A.; Danna, P.; El-Baz, A.; Naidu, S.; Suri, J.S. A novel block imaging technique using nine artificial intelligence models for COVID-19 disease classification, characterization and severity measurement in lung computed tomography scans on an Italian cohort. J. Med. Syst. 2021, 45, 28. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.; Kuppili, V.; Saba, L.; Edla, D.R.; Suri, H.S.; Cuadrado-Godia, E.; Laird, J.R.; Marinhoe, R.T.; Sanches, J.M.; Nicolaides, A.; et al. State-of-the-art review on deep learning in medical imaging. Front. Biosci. 2019, 24, 392–426. [Google Scholar]
- LeCun, Y.; Denker, J.; Solla, S. Optimal brain damage. In Advances in Neural Information Processing Systems, Proceedings of the Neural Information Processing Systems Conference, Denver, CO, USA, 27–30 November 1989; Massachusetts Institute of Technology Press: Cambridge, MA, USA, 1989; Volume 2. [Google Scholar]
- Taylor, R. Interpretation of the correlation coefficient: A basic review. J. Diagn. Med. Sonogr. 1990, 6, 35–39. [Google Scholar] [CrossRef]
- Kozek, T.; Roska, T.; Chua, L.O. Genetic algorithm for CNN template learning. IEEE Trans. Circuits Syst. I Fundam. Theory Appl. 1993, 40, 392–402. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95-International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December 1995; IEEE: Washington, DC, USA, 1995; pp. 1942–1948. [Google Scholar]
- Acharya, U.R.; Kannathal, N.; Ng, E.; Min, L.C.; Suri, J.S. Computer-based classification of eye diseases. In Proceedings of the 2006 International Conference of the IEEE Engineering in Medicine and Biology Society, New York, NY, USA, 30 August–3 September 2006; IEEE: Washington, DC, USA, 2006; pp. 6121–6124. [Google Scholar]
- Molinari, F.; Liboni, W.; Pavanelli, E.; Giustetto, P.; Badalamenti, S.; Suri, J.S. Accurate and automatic carotid plaque characterization in contrast enhanced 2-D ultrasound images. In Proceedings of the 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Lyon, France, 22–26 August 2007; IEEE: Washington, DC, USA, 2007; pp. 335–338. [Google Scholar]
- Acharya, U.R.; Faust, O.; Sree, S.V.; Alvin, A.P.C.; Krishnamurthi, G.; Sanches, J.; Suri, J.S. Atheromatic™: Symptomatic vs. asymptomatic classification of carotid ultrasound plaque using a combination of HOS, DWT & texture. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; IEEE: Washington, DC, USA, 2011; pp. 4489–4492. [Google Scholar]
- El-Baz, A.; Gimel’farb, G.; Suri, J.S. Stochastic Modeling for Medical Image Analysis, 1st ed.; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Murgia, A.; Erta, M.; Suri, J.S.; Gupta, A.; Wintermark, M.; Saba, L. CT imaging features of carotid artery plaque vulnerability. Ann. Transl. Med. 2020, 8, 1261. [Google Scholar] [CrossRef]
- Gozes, O.; Frid-Adar, M.; Greenspan, H.; Browning, P.D.; Zhang, H.; Ji, W.; Bernheim, A.; Siegel, E. Rapid ai development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv 2020, arXiv:2003.05037. [Google Scholar]
- Li, C.; Yang, Y.; Liang, H.; Wu, B. Transfer learning for establishment of recognition of COVID-19 on CT imaging using small-sized training datasets. Knowl.-Based Syst. 2021, 218, 106849. [Google Scholar] [CrossRef]
- Alshazly, H.; Linse, C.; Barth, E.; Martinetz, T.J.S. Explainable COVID-19 detection using chest CT scans and deep learning. Sensors 2021, 21, 455. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.F.H.S. An ensemble approach for multi-stage transfer learning models for COVID-19 detection from chest CT scans. Intell.-Based Med. 2021, 5, 100027. [Google Scholar] [CrossRef] [PubMed]
- Shaik, N.S.; Cherukuri, T.K. Transfer learning based novel ensemble classifier for COVID-19 detection from chest CT-scans. Comput. Biol. Med. 2022, 141, 105127. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.-L.; Liao, Y.-C. Stacking Ensemble and ECA-EfficientNetV2 Convolutional Neural Networks on Classification of Multiple Chest Diseases Including COVID-19. Acad. Radiol. 2022, in press. [CrossRef] [PubMed]
- Xu, Y.; Lam, H.-K.; Jia, G.; Jiang, J.; Liao, J.; Bao, X. Improving COVID-19 CT classification of CNNs by learning parameter-efficient representation. Comput. Biol. Med. 2023, 152, 106417. [Google Scholar] [CrossRef] [PubMed]
- Pathan, S.; Siddalingaswamy, P.; Kumar, P.; MM, M.P.; Ali, T.; Acharya, U.R. Novel ensemble of optimized CNN and dynamic selection techniques for accurate COVID-19 screening using chest CT images. Comput. Biol. Med. 2021, 137, 104835. [Google Scholar] [CrossRef]
- Kundu, R.; Singh, P.K.; Mirjalili, S.; Sarkar, R. COVID-19 detection from lung CT-Scans using a fuzzy integral-based CNN ensemble. Comput. Biol. Med. 2021, 138, 104895. [Google Scholar] [CrossRef]
- Zhou, T.; Lu, H.; Yang, Z.; Qiu, S.; Huo, B.; Dong, Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021, 98, 106885. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Wang, C.; Nie, J.; Kumar, N.; Zhang, Y.; Xiong, Z.; Barnawi, A. EDL-COVID: Ensemble deep learning for COVID-19 case detection from chest X-ray images. IEEE Trans. Ind. Inform. 2021, 17, 6539–6549. [Google Scholar] [CrossRef]
- Ray, E.L.; Wattanachit, N.; Niemi, J.; Kanji, A.H.; House, K.; Cramer, E.Y.; Bracher, J.; Zheng, A.; Yamana, T.K.; Xiong, X. Ensemble forecasts of coronavirus disease 2019 (COVID-19) in the US. MedRXiv 2008. [Google Scholar] [CrossRef]
- Batra, R.; Chan, H.; Kamath, G.; Ramprasad, R.; Cherukara, M.J.; Sankaranarayanan, S.K. Screening of therapeutic agents for COVID-19 using machine learning and ensemble docking studies. J. Phys. Chem. Lett. 2020, 11, 7058–7065. [Google Scholar] [CrossRef] [PubMed]
- Aversano, L.; Bernardi, M.L.; Cimitile, M.; Pecori, R. Deep neural networks ensemble to detect COVID-19 from CT scans. Pattern Recognit. Lett. 2021, 120, 108135. [Google Scholar] [CrossRef] [PubMed]
- Al, A.; Kabir, M.R.; Ar, A.M.; Nishat, M.M.; Faisal, F. COVID-EnsembleNet: An ensemble based approach for detecting COVID-19 by utilising chest X-Ray images. In Proceedings of the 2022 IEEE World AI IoT Congress (AIIoT), Seattle, WA, USA, 6–9 June 2022; IEEE: Washington, DC, USA, 2022; pp. 351–356. [Google Scholar]
- Hosni, M.; Abnane, I.; Idri, A.; de Gea, J.M.C.; Alemán, J.L.F. Reviewing ensemble classification methods in breast cancer. Comput. Methods Programs Biomed. 2019, 177, 89–112. [Google Scholar] [CrossRef]
- Lavanya, D.; Rani, K.U. Ensemble decision tree classifier for breast cancer data. Int. J. Inf. Technol. Converg. Serv. 2012, 2, 17–24. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, B.; Yoon, S.W.; Ko, H.S. A support vector machine-based ensemble algorithm for breast cancer diagnosis. Eur. J. Oper. Res. 2018, 267, 687–699. [Google Scholar] [CrossRef]
- Abdar, M.; Zomorodi-Moghadam, M.; Zhou, X.; Gururajan, R.; Tao, X.; Barua, P.D.; Gururajan, R. A new nested ensemble technique for automated diagnosis of breast cancer. Pattern Recognit. Lett. 2020, 132, 123–131. [Google Scholar] [CrossRef]
- Hsieh, S.-L.; Hsieh, S.-H.; Cheng, P.-H.; Chen, C.-H.; Hsu, K.-P.; Lee, I.-S.; Wang, Z.; Lai, F. Design ensemble machine learning model for breast cancer diagnosis. J. Med. Syst. 2012, 36, 2841–2847. [Google Scholar] [CrossRef]
- Hu, J.; Gu, X.; Gu, X. Mutual ensemble learning for brain tumor segmentation. Neurocomputing 2022, 504, 68–81. [Google Scholar] [CrossRef]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Alassafi, M.O. Brain tumor classification based on fine-tuned models and the ensemble method. Comput. Mater. Contin. 2021, 67, 3967–3982. [Google Scholar] [CrossRef]
- Kang, J.; Ullah, Z.; Gwak, J. MRI-based brain tumor classification using ensemble of deep features and machine learning classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef]
- Younis, A.; Qiang, L.; Nyatega, C.O.; Adamu, M.J.; Kawuwa, H.B. Brain tumor analysis using deep learning and VGG-16 ensembling learning approaches. Appl. Sci. 2022, 12, 7282. [Google Scholar] [CrossRef]
- Almulihi, A.; Saleh, H.; Hussien, A.M.; Mostafa, S.; El-Sappagh, S.; Alnowaiser, K.; Ali, A.A.; Hassan, M.J.D.R. Ensemble Learning Based on Hybrid Deep Learning Model for Heart Disease Early Prediction. Diagnostics 2022, 12, 3215. [Google Scholar] [CrossRef] [PubMed]
- Karadeniz, T.; Maraş, H.H.; Tokdemir, G.; Ergezer, H. Two Majority Voting Classifiers Applied to Heart Disease Prediction. Appl. Sci. 2023, 13, 3767. [Google Scholar] [CrossRef]
- Fitriyani, N.L.; Syafrudin, M.; Alfian, G.; Rhee, J. Development of disease prediction model based on ensemble learning approach for diabetes and hypertension. IEEE Access 2019, 7, 144777–144789. [Google Scholar] [CrossRef]
- Reddy, G.T.; Bhattacharya, S.; Ramakrishnan, S.S.; Chowdhary, C.L.; Hakak, S.; Kaluri, R.; Reddy, M.P.K. An ensemble based machine learning model for diabetic retinopathy classification. In Proceedings of the 2020 International Conference on Emerging Trends in Information Technology and Engineering (IC-ETITE), Vellore, India, 24–25 February 2020; IEEE: Washington, DC, USA, 2020; pp. 1–6. [Google Scholar]
- Jiang, H.; Yang, K.; Gao, M.; Zhang, D.; Ma, H.; Qian, W. An interpretable ensemble deep learning model for diabetic retinopathy disease classification. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: Washington, DC, USA, 2019; pp. 2045–2048. [Google Scholar]
- Mahesh, T.; Kumar, D.; Kumar, V.V.; Asghar, J.; Bazezew, B.M.; Natarajan, R.; Vivek, V. Blended ensemble learning prediction model for strengthening diagnosis and treatment of chronic diabetes disease. Comput. Intell. Neurosci. 2022, 2022, 4451792. [Google Scholar] [CrossRef] [PubMed]
- Antal, B.; Hajdu, A. An ensemble-based system for automatic screening of diabetic retinopathy. Knowl.-Based Syst. 2014, 60, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Thirion, J.-P.; Prima, S.; Subsol, G.; Roberts, N. Statistical analysis of normal and abnormal dissymmetry in volumetric medical images. Med. Image Anal. 2000, 4, 111–121. [Google Scholar] [CrossRef]
- Cootes, T.F.; Taylor, C.J. Statistical models of appearance for medical image analysis and computer vision. In Medical Imaging 2001: Image Processing; SPIE: Philadelphia, PA, USA, 2001; pp. 236–248. [Google Scholar]
- Heimann, T.; Meinzer, H.-P. Statistical shape models for 3D medical image segmentation: A review. Med. Image Anal. 2009, 13, 543–563. [Google Scholar] [CrossRef]
- Duncan, J.S.; Ayache, N. Medical image analysis: Progress over two decades and the challenges ahead. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 85–106. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.L.; Balakrishnan, N. A random-sum Wilcoxon statistic and its application to analysis of ROC and LROC data. J. Stat. Plan. Inference 2011, 141, 335–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez, N.P.; López, M.A.G.; Silva, A.; Ramos, I. Improving the Mann–Whitney statistical test for feature selection: An approach in breast cancer diagnosis on mammography. Artif. Intell. Med. 2015, 63, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Murcia, F.J.; Górriz, J.M.; Ramirez, J.; Puntonet, C.G.; Salas-Gonzalez, D.; Alzheimer’s Disease Neuroimaging Initiative. Computer aided diagnosis tool for Alzheimer’s disease based on Mann–Whitney–Wilcoxon U-test. Expert Syst. Appl. 2012, 39, 9676–9685. [Google Scholar] [CrossRef]
- Lin, Y.; Su, J.; Li, Y.; Wei, Y.; Yan, H.; Zhang, S.; Luo, J.; Ai, D.; Song, H.; Fan, J. High-Resolution Boundary Detection for Medical Image Segmentation with Piece-Wise Two-Sample T-Test Augmented Loss. arXiv 2022, arXiv:2211.02419. [Google Scholar]
- Chin, M.H.; Muramatsu, N. What is the quality of quality of medical care measures?: Rashomon-like relativism and real-world applications. Perspect. Biol. Med. 2003, 46, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, P.; Yan, J.; Wang, Y.; Li, S.; Jiang, J.; Sun, Z.; Tang, B.; Chang, T.-H.; Wang, S. Real-world data medical knowledge graph: Construction and applications. Artif. Intell. Med. 2020, 103, 101817. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, C.; Nakahira, Y. Medical application on internet of things. In Proceedings of the IET international conference on communication technology and application (ICCTA 2011), Beijing, China, 14–16 October 2011; IET: London, UK, 2011; pp. 660–665. [Google Scholar]
- Endrei, D.; Molics, B.; Ágoston, I. Multicriteria decision analysis in the reimbursement of new medical technologies: Real-world experiences from Hungary. Value Health 2014, 17, 487–489. [Google Scholar] [CrossRef] [Green Version]
- Hudson, J.N. Computer-aided learning in the real world of medical education: Does the quality of interaction with the computer affect student learning? Med. Educ. 2004, 38, 887–895. [Google Scholar] [CrossRef]
- Attallah, O. RADIC: A tool for diagnosing COVID-19 from chest CT and X-ray scans using deep learning and quad-radiomics. Chemom. Intell. Lab. Syst. 2023, 233, 104750. [Google Scholar] [CrossRef]
- Attallah, O.; Ragab, D.A.; Sharkas, M. MULTI-DEEP: A novel CAD system for coronavirus (COVID-19) diagnosis from CT images using multiple convolution neural networks. PeerJ 2020, 8, e10086. [Google Scholar] [CrossRef]
- Mercaldo, F.; Belfiore, M.P.; Reginelli, A.; Brunese, L.; Santone, A. Coronavirus COVID-19 detection by means of explainable deep learning. Sci. Rep. 2023, 13, 462. [Google Scholar] [CrossRef] [PubMed]
- Shah, V.; Keniya, R.; Shridharani, A.; Punjabi, M.; Shah, J.; Mehendale, N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg. Radiol. 2021, 28, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O.; Samir, A. A wavelet-based deep learning pipeline for efficient COVID-19 diagnosis via CT slices. Appl. Soft Comput. 2022, 128, 109401. [Google Scholar] [CrossRef] [PubMed]
- Kini, A.S.; Reddy, A.N.G.; Kaur, M.; Satheesh, S.; Singh, J.; Martinetz, T.; Alshazly, H. Ensemble deep learning and internet of things-based automated COVID-19 diagnosis framework. Contrast Media Mol. Imaging 2022, 2022, 7377502. [Google Scholar] [CrossRef]
- Li, X.; Tan, W.; Liu, P.; Zhou, Q.; Yang, J. Classification of COVID-19 chest CT images based on ensemble deep learning. J. Healthc. Eng. 2021, 2021, 5528441. [Google Scholar] [CrossRef]
- Yang, L.; Wang, S.-H.; Zhang, Y.-D. EDNC: Ensemble deep neural network for COVID-19 recognition. Tomography 2022, 8, 869–890. [Google Scholar] [CrossRef]
- Ragab, D.A.; Attallah, O. FUSI-CAD: Coronavirus (COVID-19) diagnosis based on the fusion of CNNs and handcrafted features. PeerJ Comput. Sci. 2020, 6, e306. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S.; Agarwal, S.; Pathak, R.; Ketireddy, V.; Columbu, M.; Saba, L.; Gupta, S.K.; Faa, G.; Singh, I.M.; Turk, M. COVLIAS 1.0: Lung Segmentation in COVID-19 Computed Tomography Scans Using Hybrid Deep Learning Artificial Intelligence Models. Diagnostics 2021, 11, 1405. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S.; Agarwal, S.; Chabert, G.L.; Carriero, A.; Paschè, A.; Danna, P.S.; Saba, L.; Mehmedović, A.; Faa, G.; Singh, I.M. COVLIAS 2.0-cXAI: Cloud-based explainable deep learning system for COVID-19 lesion localization in computed tomography scans. Diagnostics 2022, 12, 1482. [Google Scholar] [CrossRef]
- Ibrahim, D.A.; Zebari, D.A.; Mohammed, H.J.; Mohammed, M.A. Effective hybrid deep learning model for COVID-19 patterns identification using CT images. Expert Syst. 2022, 39, e13010. [Google Scholar] [CrossRef] [PubMed]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Rafiee, M.J.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. Hybrid deep learning model for diagnosis of COVID-19 using CT scans and clinical/demographic data. In Proceedings of the 2021 IEEE International Conference on Image Processing (ICIP), Anchorage, AK, USA, 19–22 September 2021; IEEE: Washington, DC, USA, 2021; pp. 180–184. [Google Scholar]
- Chola, C.; Mallikarjuna, P.; Muaad, A.Y.; Benifa, J.B.; Hanumanthappa, J.; Al-antari, M.A. A hybrid deep learning approach for COVID-19 diagnosis via CT and X-ray medical images. Comput. Sci. Math. Forum 2021, 2, 13. [Google Scholar]
- Liang, S.; Zhang, W.; Gu, Y. A hybrid and fast deep learning framework for COVID-19 detection via 3D Chest CT Images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 508–512. [Google Scholar]
- R-Prabha, M.; Prabhu, R.; Suganthi, S.; Sridevi, S.; Senthil, G.; Babu, D.V. Design of hybrid deep learning approach for COVID-19 infected lung image segmentation. J. Phys. Conf. Ser. 2021, 2040, 012016. [Google Scholar] [CrossRef]
- Wu, D.; Gong, K.; Arru, C.D.; Homayounieh, F.; Bizzo, B.; Buch, V.; Ren, H.; Kim, K.; Neumark, N.; Xu, P. Severity and consolidation quantification of COVID-19 from CT images using deep learning based on hybrid weak labels. IEEE J. Biomed. Health Inform. 2020, 24, 3529–3538. [Google Scholar] [CrossRef] [PubMed]
- Loey, M.; Manogaran, G.; Khalifa, N.E.M. A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Neural Comput. Appl. 2020, 1–13, online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.; Panigrahi, B.K.; Dey, N.; Rajinikanth, V.; Gandhi, T.K.J.A.I. Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Appl. Intell. Vol. 2021, 51, 571–585. [Google Scholar] [CrossRef]
- Arora, V.; Ng, E.Y.-K.; Leekha, R.S.; Darshan, M.; Singh, A. Transfer learning-based approach for detecting COVID-19 ailment in lung CT scan. Comput. Biol. Med. 2021, 135, 104575. [Google Scholar] [CrossRef]
- Das, S.; Nayak, G.K.; Saba, L.; Kalra, M.; Suri, J.S.; Saxena, S. An artificial intelligence framework and its bias for brain tumor segmentation: A narrative review. Comput. Biol. Med. 2022, 143, 105273. [Google Scholar] [CrossRef]
- Alakus, T.B.; Turkoglu, I. Comparison of deep learning approaches to predict COVID-19 infection. Chaos Solitons Fractals 2020, 140, 110120. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Park, S.; Ye, J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Liu, H.; Shao, M.; Li, S.; Fu, Y. Infinite ensemble for image clustering. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1745–1754. [Google Scholar]
- Orchard, J.; Mann, R. Registering a multisensor ensemble of images. IEEE Trans. Image Process. 2009, 19, 1236–1247. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhou, Z.-H. SOM ensemble-based image segmentation. Neural Process. Lett. 2004, 20, 171–178. [Google Scholar] [CrossRef]
- Chaeikar, S.S.; Ahmadi, A. Ensemble SW image steganalysis: A low dimension method for LSBR detection. Signal Process. Image Commun. 2019, 70, 233–245. [Google Scholar] [CrossRef]
- Li, B.; Goh, K. Confidence-based dynamic ensemble for image annotation and semantics discovery. In Proceedings of the Eleventh ACM International Conference on Multimedia, Berkeley, CA, USA, 2–8 November 2003; pp. 195–206. [Google Scholar]
- Varol, E.; Gaonkar, B.; Erus, G.; Schultz, R.; Davatzikos, C. Feature ranking based nested support vector machine ensemble for medical image classification. In Proceedings of the 2012 9th IEEE international symposium on biomedical imaging (ISBI), Barcelona, Spain, 2–5 May 2012; IEEE: Washington, DC, USA, 2012; pp. 146–149. [Google Scholar]
- Wu, S.; Zhang, H.; Valiant, G.; Ré, C. On the generalization effects of linear transformations in data augmentation. In Proceedings of the International Conference on Machine Learning, Virtual, 13–18 July 2020; pp. 10410–10420. [Google Scholar]
- Perez, L.; Wang, J. The effectiveness of data augmentation in image classification using deep learning. arXiv 2017, arXiv:1712.04621. [Google Scholar]
- Van Dyk, D.A.; Meng, X.-L. The art of data augmentation. J. Comput. Graph. Stat. 2001, 10, 1–50. [Google Scholar] [CrossRef]
- Aquino, N.R.; Gutoski, M.; Hattori, L.T.; Lopes, H.S. The effect of data augmentation on the performance of convolutional neural networks. J. Braz. Comput. Soc. 2017. [Google Scholar]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.-M.; Ciompi, F.; Van Der Laak, J. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parmar, C.; Barry, J.D.; Hosny, A.; Quackenbush, J.; Aerts, H.J. Data Analysis Strategies in Medical ImagingData Science Designs in Medical Imaging. Clin. Cancer Res. 2018, 24, 3492–3499. [Google Scholar] [CrossRef] [Green Version]
- Lai, M. Deep learning for medical image segmentation. arXiv 2015, arXiv:1505.02000. [Google Scholar]
- Abdollahi, B.; Tomita, N.; Hassanpour, S. Data augmentation in training deep learning models for medical image analysis. In Deep Learners Deep Learner Descriptors for Medical Applications; Springer: Cham, Switzerland, 2020; pp. 167–180. [Google Scholar]
- Chen, H.; Sung, J.J. Potentials of AI in medical image analysis in Gastroenterology and Hepatology. J. Gastroenterol. Hepatol. 2021, 36, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.R.; Oda, H.; Zhou, X.; Shimizu, N.; Yang, Y.; Hayashi, Y.; Oda, M.; Fujiwara, M.; Misawa, K.; Mori, K. Graphics, An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput. Med. Imaging 2018, 66, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Gu, R.; Zhang, J.; Huang, R.; Lei, W.; Wang, G.; Zhang, S. Domain composition and attention for unseen-domain generalizable medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Part III 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 241–250. [Google Scholar]
- Kumar, A.; Kim, J.; Lyndon, D.; Fulham, M.; Feng, D. An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J. Biomed. Health Inform. 2016, 21, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Ghesu, F.C.; Georgescu, B.; Mansoor, A.; Yoo, Y.; Gibson, E.; Vishwanath, R.; Balachandran, A.; Balter, J.M.; Cao, Y.; Singh, R. Quantifying and leveraging predictive uncertainty for medical image assessment. Med. Image Anal. 2021, 68, 101855. [Google Scholar] [CrossRef]
- Adams, M.; Chen, W.; Holcdorf, D.; McCusker, M.W.; Howe, P.D.; Gaillard, F. Computer vs. human: Deep learning versus perceptual training for the detection of neck of femur fractures. J. Med. Imaging Radiat. Oncol. 2019, 63, 27–32. [Google Scholar] [CrossRef] [Green Version]
- González, G.; Washko, G.R.; Estépar, R.S.J. Deep learning for biomarker regression: Application to osteoporosis and emphysema on chest CT scans. In Medical Imaging 2018: Image Processing; SPIE: Philadelphia, PA, USA, 2018; pp. 372–378. [Google Scholar]
- Li, P.; Liu, Q.; Tang, D.; Zhu, Y.; Xu, L.; Sun, X.; Song, S. Lesion based diagnostic performance of dual phase 99m Tc-MIBI SPECT/CT imaging and ultrasonography in patients with secondary hyperparathyroidism. BMC Med. Imaging 2017, 17, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Ma, C.; Wang, Z.; Liu, J.-L.; Rui, Y.-P.; Li, Y.-H.; Peng, Y.-F. Effect of region of interest on ADC and interobserver variability in thyroid nodules. BMC Med. Imaging 2019, 19, 55. [Google Scholar] [CrossRef] [Green Version]
- Shia, W.-C.; Chen, D.-R. Classification of malignant tumors in breast ultrasound using a pretrained deep residual network model and support vector machine. Comput. Med. Imaging Graph. 2021, 87, 101829. [Google Scholar] [CrossRef] [PubMed]
- Lahoud, P.; EzEldeen, M.; Beznik, T.; Willems, H.; Leite, A.; Van Gerven, A.; Jacobs, R. Artificial intelligence for fast and accurate 3-dimensional tooth segmentation on cone-beam computed tomography. J. Endod. 2021, 47, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Skandha, S.S.; Gupta, S.K.; Saba, L.; Koppula, V.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; et al. 3-D optimized classification and characterization artificial intelligence paradigm for cardiovascular/stroke risk stratification using carotid ultrasound-based delineated plaque: Atheromatic™ 2.0. Comput. Biol. Med. 2020, 125, 103958. [Google Scholar] [CrossRef]
- Jamthikar, A.; Gupta, D.; Khanna, N.N.; Saba, L.; Laird, J.R.; Suri, J.S. Cardiovascular/stroke risk prevention: A new machine learning framework integrating carotid ultrasound image-based phenotypes and its harmonics with conventional risk factors. Indian Heart J. 2020, 72, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Jamthikar, A.; Gupta, D.; Khanna, N.N.; Saba, L.; Araki, T.; Viskovic, K.; Suri, H.S.; Gupta, A.; Mavrogeni, S.; Turk, M. A low-cost machine learning-based cardiovascular/stroke risk assessment system: Integration of conventional factors with image phenotypes. Cardiovasc. Diagn. Ther. 2019, 9, 420. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, M.; Agarwal, S.; Saba, L.; Chabert, G.L.; Gupta, S.; Carriero, A.; Pasche, A.; Danna, P.; Mehmedovic, A.; Faa, G. Eight pruning deep learning models for low storage and high-speed COVID-19 computed tomography lung segmentation and heatmap-based lesion localization: A multicenter study using COVLIAS 2.0. Comput. Biol. Med. 2022, 146, 105571. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S.; Agarwal, S.; Chabert, G.L.; Carriero, A.; Paschè, A.; Danna, P.S.; Saba, L.; Mehmedović, A.; Faa, G.; Singh, I.M. COVLIAS 1.0 Lesion vs. MedSeg: An Artificial Intelligence Framework for Automated Lesion Segmentation in COVID-19 Lung Computed Tomography Scans. Diagnostics 2022, 12, 1283. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Sharma, N.; Saba, L.; Paraskevas, K.I.; Kalra, M.K.; Johri, A.; Laird, J.R.; Nicolaides, A.N.; Suri, J.S. Unseen artificial intelligence—Deep learning paradigm for segmentation of low atherosclerotic plaque in carotid ultrasound: A multicenter cardiovascular study. Diagnostics 2021, 11, 2257. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Gupta, S.K.; Puvvula, A.; Biswas, M.; Saba, L.; Bit, A.; Tandel, G.S.; Agarwal, M.; Patrick, A. A narrative review on characterization of acute respiratory distress syndrome in COVID-19-infected lungs using artificial intelligence. Comput. Biol. Med. 2021, 130, 104210. [Google Scholar] [CrossRef]
- Verma, A.K.; Kuppili, V.; Srivastava, S.K.; Suri, J.S. An AI-based approach in determining the effect of meteorological factors on incidence of malaria. Front. Biosci.-Landmark 2020, 25, 1202–1229. [Google Scholar]
- Verma, A.K.; Kuppili, V.; Srivastava, S.K.; Suri, J.S. A new backpropagation neural network classification model for prediction of incidence of malaria. Front. Biosci.-Landmark 2020, 25, 299–334. [Google Scholar]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Hasöksüz, M.; Kilic, S.; Sarac, F. Coronaviruses and SARS-CoV-2. Turk. J. Med. Sci. 2020, 50, 549–556. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The architecture of SARS-CoV-2 transcriptome. Cell 2020, 181, 914–921.e910. [Google Scholar] [CrossRef]
- Amanat, F.; Krammer, F. SARS-CoV-2 vaccines: Status report. Immunity 2020, 52, 583–589. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Du, H.; Zhang, J.; Li, Y.Y.; Qu, J.; Zhang, W.; Wang, Y.; Bao, S.; Li, Y. SARS-CoV-2 infection in children. N. Engl. J. Med. 2020, 382, 1663–1665. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, S.; Zarbock, A. Coronaviruses and SARS-CoV-2: A brief overview. Anesth. Analg. 2020, 131, 93–96. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Pedersen, S.F.; Ho, Y.-C. SARS-CoV-2: A storm is raging. J. Clin. Investig. 2020, 130, 2202–2205. [Google Scholar] [CrossRef]
- Yang, X.; He, X.; Zhao, J.; Zhang, Y.; Zhang, S.; Xie, P. COVID-CT-dataset: A CT scan dataset about COVID-19. arXiv 2020, arXiv:2003.13865. [Google Scholar]
- Ter-Sarkisov, A.J.A.I. COVID-ct-mask-net: Prediction of COVID-19 from ct scans using regional features. Appl. Intell. 2022, 52, 9664–9675. [Google Scholar] [CrossRef] [PubMed]
- Shakouri, S.; Bakhshali, M.A.; Layegh, P.; Kiani, B.; Masoumi, F.; Nakhaei, S.A.; Mostafavi, S.M. COVID19-CT-dataset: An open-access chest CT image repository of 1000+ patients with confirmed COVID-19 diagnosis. BMC Res. Notes 2021, 14, 178. [Google Scholar] [CrossRef] [PubMed]
- Khaniabadi, P.M.; Bouchareb, Y.; Al-Dhuhli, H.; Shiri, I.; Al-Kindi, F.; Khaniabadi, B.M.; Zaidi, H.; Rahmim, A. Two-step machine learning to diagnose and predict involvement of lungs in COVID-19 and pneumonia using CT radiomics. Comput. Biol. Med. 2022, 150, 106165. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Dai, Q. A self-supervised COVID-19 CT recognition system with multiple regularizations. Comput. Biol. Med. 2022, 150, 106–149. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the Computer Vision–ECCV 2022 Workshops, Tel Aviv, Israel, 23–27 October 2022; Part III. Springer: Berlin/Heidelberg, Germany, 2023; pp. 205–218. [Google Scholar]
- Sha, Y.; Zhang, Y.; Ji, X.; Hu, L. Transformer-unet: Raw image processing with unet. arXiv 2021, arXiv:2109.08417. [Google Scholar]
- Torbunov, D.; Huang, Y.; Yu, H.; Huang, J.; Yoo, S.; Lin, M.; Viren, B.; Ren, Y. Uvcgan: Unet vision transformer cycle-consistent gan for unpaired image-to-image translation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 2–7 January 2023; pp. 702–712. [Google Scholar]
- Yan, X.; Tang, H.; Sun, S.; Ma, H.; Kong, D.; Xie, X. After-unet: Axial fusion transformer unet for medical image segmentation. In Proceedings of the IEEE/CVF winter conference on applications of computer vision, Waikoloa, HI, USA, 3–8 January 2022; p. 39713981. [Google Scholar]
- Xie, Y.; Zhang, J.; Shen, C.; Xia, Y. Cotr: Efficiently bridging cnn and transformer for 3d medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Part III 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 171–180. [Google Scholar]
- Johri, A.M.; Singh, K.V.; Mantella, L.E.; Saba, L.; Sharma, A.; Laird, J.R.; Utkarsh, K.; Singh, I.M.; Gupta, S.; Kalra, M.S. Deep learning artificial intelligence framework for multiclass coronary artery disease prediction using combination of conventional risk factors, carotid ultrasound, and intraplaque neovascularization. Comput. Biol. Med. 2022, 150, 106018. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S.; Bhagawati, M.; Paul, S.; Protogerou, A.D.; Sfikakis, P.P.; Kitas, G.D.; Khanna, N.N.; Ruzsa, Z.; Sharma, A.M.; Saxena, S. A powerful paradigm for cardiovascular risk stratification using multiclass, multi-label, and ensemble-based machine learning paradigms: A narrative review. Diagnostics 2022, 12, 722. [Google Scholar] [CrossRef] [PubMed]
- Jamthikar, A.D.; Gupta, D.; Mantella, L.E.; Saba, L.; Laird, J.R.; Johri, A.M.; Suri, J.S. Multiclass machine learning vs.conventional calculators for stroke/CVD risk assessment using carotid plaque predictors with coronary angiography scores as gold standard: A 500 participants study. Int. J. Cardiovasc. Imaging 2021, 37, 1171–1187. [Google Scholar] [CrossRef]
- Tandel, G.S.; Balestrieri, A.; Jujaray, T.; Khanna, N.N.; Saba, L.; Suri, J.S. Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Comput. Biol. Med. 2020, 122, 103804. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Sharma, N.; Kalra, M.K.; Viskovic, K.; Saba, L.; Suri, J.S. Four types of multiclass frameworks for pneumonia classification and its validation in X-ray scans using seven types of deep learning artificial intelligence models. Diagnostics 2022, 12, 652. [Google Scholar]





























| SN | Dataset (Name) | Country of Origin | Patients | Images (Before Augmentation) | Images (After Augmentation) | Image Dimensions * | Training: Testing Split |
|---|---|---|---|---|---|---|---|
| 1 | CroMED (COVID) | Croatia | 80 | 5396 | 10,792 | 512 × 512 | K5 |
| 2 | NovMED (COVID) | Novara, Italy | 60 | 5797 | 11,594 | 768 × 768 | K5 |
| 3 | NovMED (Control) | Novara, Italy | 12 | 1855 | 11,130 | 768 × 768 | K5 |
| TL Statistics on DC1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Without Augmentation | Balance + With Augmentation | ||||||||||
| TL Type | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | |
| TL1 | EfficientNetV2M | 93.93 | 0.44 | 0.26 | 0.92 | <0.0001 | 98.44 | 0.49 | 0.48 | 0.98 | <0.0001 |
| TL2 | InceptionV3 | 93.65 | 0.39 | 0.19 | 0.87 | <0.0001 | 93.97 | 0.49 | 0.47 | 0.95 | <0.0001 |
| TL3 | MobileNetV2 | 97.93 | 0.42 | 0.23 | 0.95 | <0.0001 | 99.99 | 0.49 | 0.5 | 1.00 | <0.0001 |
| TL4 | ResNet152 | 92.9 | 0.38 | 0.18 | 0.86 | <0.0001 | 96.98 | 0.49 | 0.47 | 0.97 | <0.0001 |
| TL5 | ResNet50 | 97.1 | 0.41 | 0.22 | 0.94 | <0.0001 | 97.44 | 0.49 | 0.48 | 0.97 | <0.0001 |
| TL6 | VGG16 | 90.2 | 0.42 | 0.23 | 0.85 | <0.0001 | 95.61 | 0.49 | 0.47 | 0.95 | <0.0001 |
| TL7 | VGG19 | 91.72 | 0.39 | 0.19 | 0.85 | <0.0001 | 96.8 | 0.49 | 0.48 | 0.96 | <0.0001 |
| Mean ACC of all TLs: 93.91% | Mean ACC of all TLs: 97.03% | ||||||||||
| TL Statistics on DC1 | |||||||
|---|---|---|---|---|---|---|---|
| Without Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.97 | 0.86 | 0.95 | 0.91 | 0.96 | 0.88 |
| TL2 | InceptionV3 | 0.93 | 1.00 | 1.00 | 0.78 | 0.96 | 0.88 |
| TL3 | MobileNetV2 | 0.97 | 1.00 | 1.00 | 0.92 | 0.99 | 0.96 |
| TL4 | ResNet152 | 0.91 | 1.00 | 1.00 | 0.72 | 0.95 | 0.84 |
| TL5 | ResNet50 | 0.94 | 0.98 | 0.99 | 0.81 | 0.97 | 0.89 |
| TL6 | VGG16 | 0.92 | 0.84 | 0.95 | 0.77 | 0.94 | 0.8 |
| TL7 | VGG19 | 0.97 | 0.81 | 0.93 | 0.92 | 0.95 | 0.86 |
| Balance + With Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.97 | 0.93 | 0.92 | 0.98 | 0.95 | 0.95 |
| TL2 | InceptionV3 | 0.99 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 |
| TL3 | MobileNetV2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| TL4 | ResNet152 | 0.99 | 1.00 | 1.00 | 0.99 | 0.99 | 0.99 |
| TL5 | ResNet50 | 0.99 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 |
| TL6 | VGG16 | 0.94 | 0.92 | 0.92 | 0.95 | 0.93 | 0.93 |
| TL7 | VGG19 | 0.92 | 0.95 | 0.95 | 0.92 | 0.93 | 0.93 |
| TL Statistics on DC2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Without Augmentation | Balance + With Augmentation | ||||||||||
| TL Type | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | |
| TL1 | EfficientNetV2M | 84.96 | 0.39 | 0.19 | 0.75 | <0.0001 | 93.92 | 0.49 | 0.45 | 0.93 | <0.0001 |
| TL2 | InceptionV3 | 90.84 | 0.35 | 0.15 | 0.81 | <0.0001 | 90.04 | 0.49 | 0.44 | 0.81 | <0.0001 |
| TL3 | MobileNetV2 | 85.22 | 0.37 | 0.16 | 0.74 | <0.0001 | 89.77 | 0.49 | 0.49 | 0.89 | <0.0001 |
| TL4 | ResNet152 | 78.16 | 0.2 | 0.04 | 0.56 | <0.0001 | 87.4 | 0.49 | 0.52 | 0.87 | <0.0001 |
| TL5 | ResNet50 | 79.47 | 0.44 | 0.27 | 0.74 | <0.0001 | 93.53 | 0.49 | 0.46 | 0.89 | <0.0001 |
| TL6 | VGG16 | 85.62 | 0.43 | 0.24 | 0.8 | <0.0001 | 84.05 | 0.49 | 0.45 | 0.83 | <0.0001 |
| TL7 | VGG19 | 86.66 | 0.42 | 0.22 | 0.8 | <0.0001 | 90.3 | 0.49 | 0.43 | 0.9 | <0.0001 |
| Mean ACC of all TLs: 84.41% | Mean ACC of all TLs: 89.85% | ||||||||||
| TL Statistics on DC2 | |||||||
|---|---|---|---|---|---|---|---|
| Without Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.9 | 0.69 | 0.9 | 0.68 | 0.9 | 0.68 |
| TL2 | InceptionV3 | 0.87 | 0.38 | 0.64 | 0.71 | 0.74 | 0.5 |
| TL3 | MobileNetV2 | 0.87 | 0.8 | 0.96 | 0.54 | 0.81 | 0.65 |
| TL4 | ResNet152 | 0.88 | 0.48 | 0.76 | 0.68 | 0.82 | 0.56 |
| TL5 | ResNet50 | 0.89 | 0.53 | 0.81 | 0.69 | 0.85 | 0.6 |
| TL6 | VGG16 | 0.93 | 0.62 | 0.85 | 0.79 | 0.89 | 0.7 |
| TL7 | VGG19 | 0.9 | 0.6 | 0.85 | 0.7 | 0.87 | 0.65 |
| Balance + With Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.86 | 0.94 | 0.95 | 0.84 | 0.9 | 0.89 |
| TL2 | InceptionV3 | 0.85 | 0.83 | 0.83 | 0.84 | 0.84 | 0.84 |
| TL3 | MobileNetV2 | 0.73 | 1.00 | 1.00 | 0.61 | 0.84 | 0.76 |
| TL4 | ResNet152 | 0.8 | 0.78 | 0.78 | 0.8 | 0.79 | 0.79 |
| TL5 | ResNet50 | 0.83 | 0.83 | 0.84 | 0.83 | 0.84 | 0.83 |
| TL6 | VGG16 | 0.83 | 0.78 | 0.78 | 0.83 | 0.8 | 0.81 |
| TL7 | VGG19 | 0.84 | 0.88 | 0.89 | 0.83 | 0.86 | 0.85 |
| TL Statistics on DC3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Without Augmentation | Balance + With Augmentation | ||||||||||
| TL Type | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | |
| TL1 | EfficientNetV2M | 85.4 | 0.39 | 0.12 | 0.71 | <0.0001 | 91.41 | 0.49 | 0.49 | 0.91 | <0.0001 |
| TL2 | InceptionV3 | 67.6 | 0.48 | 0.38 | 0.65 | <0.0001 | 76.43 | 0.48 | 0.63 | 0.76 | <0.0001 |
| TL3 | MobileNetV2 | 67.3 | 0.47 | 0.35 | 0.63 | <0.0001 | 86.75 | 0.48 | 0.38 | 0.86 | <0.0001 |
| TL4 | ResNet152 | 78.1 | 0.18 | 0.03 | 0.57 | <0.0001 | 82.28 | 0.49 | 0.59 | 0.82 | <0.0001 |
| TL5 | ResNet50 | 67.2 | 0.49 | 0.4 | 0.66 | <0.0001 | 80 | 0.48 | 0.61 | 0.8 | <0.0001 |
| TL6 | VGG16 | 73.7 | 0.48 | 0.37 | 0.73 | <0.0001 | 81 | 0.48 | 0.61 | 0.8 | <0.0001 |
| TL7 | VGG19 | 71.2 | 0.48 | 0.38 | 0.7 | <0.0001 | 78.63 | 0.49 | 0.56 | 0.78 | <0.0001 |
| Mean ACC of all TLs: 72.90% | Mean ACC of all TLs: 82.35% | ||||||||||
| TL Statistics on DC3 | |||||||
|---|---|---|---|---|---|---|---|
| Without Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.87 | 0.79 | 0.95 | 0.58 | 0.91 | 0.67 |
| TL2 | InceptionV3 | 0.84 | 0.41 | 0.69 | 0.63 | 0.76 | 0.5 |
| TL3 | MobileNetV2 | 0.82 | 0.4 | 0.72 | 0.55 | 0.77 | 0.46 |
| TL4 | ResNet152 | 0.77 | 1.00 | 1.00 | 0.14 | 0.87 | 0.25 |
| TL5 | ResNet50 | 0.85 | 0.41 | 0.68 | 0.65 | 0.75 | 0.5 |
| TL6 | VGG16 | 0.89 | 0.49 | 0.74 | 0.72 | 0.81 | 0.58 |
| TL7 | VGG19 | 0.87 | 0.46 | 0.72 | 0.7 | 0.79 | 0.55 |
| Balance + With Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.91 | 0.92 | 0.92 | 0.91 | 0.91 | 0.91 |
| TL2 | InceptionV3 | 0.86 | 0.71 | 0.63 | 0.9 | 0.72 | 0.79 |
| TL3 | MobileNetV2 | 0.8 | 0.98 | 0.99 | 0.75 | 0.88 | 0.85 |
| TL4 | ResNet152 | 0.89 | 0.78 | 0.74 | 0.91 | 0.8 | 0.84 |
| TL5 | ResNet50 | 0.88 | 0.75 | 0.69 | 0.91 | 0.78 | 0.82 |
| TL6 | VGG16 | 0.89 | 0.76 | 0.7 | 0.92 | 0.78 | 0.83 |
| TL7 | VGG19 | 0.82 | 0.76 | 0.72 | 0.85 | 0.77 | 0.8 |
| TL Statistics on DC4 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Without Augmentation | Balance + With Augmentation | ||||||||||
| TL Type | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | |
| TL1 | EfficientNetV2M | 49.7 | 0.45 | 0.7 | 0.63 | <0.0001 | 67.57 | 0.41 | 0.77 | 0.68 | <0.0001 |
| TL2 | InceptionV3 | 45.6 | 0.47 | 0.66 | 0.55 | <0.0001 | 60.52 | 0.33 | 0.87 | 0.61 | <0.0001 |
| TL3 | MobileNetV2 | 28 | 0.26 | 0.92 | 0.49 | <0.0001 | 52.68 | 0.21 | 0.95 | 0.53 | <0.0001 |
| TL4 | ResNet152 | 35.2 | 0.42 | 0.75 | 0.47 | <0.0001 | 73.92 | 0.46 | 0.68 | 0.74 | <0.0001 |
| TL5 | ResNet50 | 39.7 | 0.4 | 0.78 | 0.56 | <0.0001 | 78.41 | 0.48 | 0.6 | 0.78 | <0.0001 |
| TL6 | VGG16 | 67.5 | 0.49 | 0.45 | 0.7 | <0.0001 | 78.59 | 0.47 | 0.66 | 0.78 | <0.0001 |
| TL7 | VGG19 | 69.4 | 0.49 | 0.41 | 0.7 | <0.0001 | 81.05 | 0.48 | 0.63 | 0.81 | <0.0001 |
| Mean ACC of all TLs: 47.85% | Mean ACC of all TLs: 70.39% | ||||||||||
| TL Statistics on DC4 | |||||||
|---|---|---|---|---|---|---|---|
| Without Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.93 | 0.31 | 0.37 | 0.91 | 0.52 | 0.47 |
| TL2 | InceptionV3 | 0.82 | 0.27 | 0.36 | 0.75 | 0.5 | 0.4 |
| TL3 | MobileNetV2 | 0.72 | 0.24 | 0.08 | 0.92 | 0.14 | 0.38 |
| TL4 | ResNet152 | 0.73 | 0.23 | 0.23 | 0.72 | 0.35 | 0.35 |
| TL5 | ResNet50 | 0.87 | 0.27 | 0.24 | 0.89 | 0.38 | 0.42 |
| TL6 | VGG16 | 0.9 | 0.41 | 0.64 | 0.77 | 0.75 | 0.53 |
| TL7 | VGG19 | 0.88 | 0.42 | 0.69 | 0.72 | 0.77 | 0.53 |
| Balance + With Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.92 | 0.61 | 0.4 | 0.97 | 0.56 | 0.74 |
| TL2 | InceptionV3 | 0.96 | 0.55 | 0.24 | 0.99 | 0.38 | 0.71 |
| TL3 | MobileNetV2 | 0.78 | 0.42 | 0.69 | 0.72 | 0.77 | 0.53 |
| TL4 | ResNet152 | 0.9 | 0.64 | 0.49 | 0.94 | 0.64 | 0.76 |
| TL5 | ResNet50 | 0.88 | 0.72 | 0.67 | 0.9 | 0.76 | 0.8 |
| TL6 | VGG16 | 0.94 | 0.71 | 0.62 | 0.96 | 0.75 | 0.81 |
| TL7 | VGG19 | 0.94 | 0.74 | 0.68 | 0.95 | 0.78 | 0.83 |
| TL Statistics on DC5 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Without Augmentation | Balance + With Augmentation | ||||||||||
| TL Type | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | |
| TL1 | EfficientNetV2M | 89.9 | 0.32 | 0.12 | 0.76 | <0.0001 | 94.14 | 0.47 | 0.33 | 0.93 | <0.0001 |
| TL2 | InceptionV3 | 95.1 | 0.29 | 0.09 | 0.83 | <0.0001 | 94.86 | 0.45 | 0.29 | 0.92 | <0.0001 |
| TL3 | MobileNetV2 | 93 | 0.32 | 0.12 | 0.82 | <0.0001 | 89.07 | 0.48 | 0.36 | 0.88 | <0.0001 |
| TL4 | ResNet152 | 89.7 | 0.34 | 0.14 | 0.78 | <0.0001 | 95.22 | 0.45 | 0.29 | 0.93 | <0.0001 |
| TL5 | ResNet50 | 91.8 | 0.25 | 0.06 | 0.72 | <0.0001 | 94.62 | 0.45 | 0.28 | 0.92 | <0.0001 |
| TL6 | VGG16 | 86.8 | 0.31 | 0.11 | 0.68 | <0.0001 | 95.28 | 0.45 | 0.28 | 0.93 | <0.0001 |
| TL7 | VGG19 | 92.3 | 0.27 | 0.08 | 0.75 | <0.0001 | 93.19 | 0.45 | 0.29 | 0.9 | <0.0001 |
| Mean ACC of all TLs: 91.22% | Mean ACC of all TLs: 93.76% | ||||||||||
| TL Statistics on DC5 | |||||||
|---|---|---|---|---|---|---|---|
| Without Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.93 | 0.67 | 0.95 | 0.57 | 0.94 | 0.61 |
| TL2 | InceptionV3 | 0.95 | 0.98 | 1.00 | 0.66 | 0.97 | 0.79 |
| TL3 | MobileNetV2 | 0.95 | 0.79 | 0.97 | 0.69 | 0.96 | 0.73 |
| TL4 | ResNet152 | 0.94 | 0.64 | 0.94 | 0.63 | 0.94 | 0.64 |
| TL5 | ResNet50 | 0.92 | 0.93 | 0.99 | 0.45 | 0.95 | 0.61 |
| TL6 | VGG16 | 0.94 | 0.39 | 0.82 | 0.69 | 0.88 | 0.5 |
| TL7 | VGG19 | 0.93 | 0.88 | 0.99 | 0.53 | 0.96 | 0.66 |
| Balance + With Augmentation | |||||||
| TL Type | COVID Precision | Control Precision | COVID Recall | Control Recall | COVID F1 Score | Control F1 Score | |
| TL1 | EfficientNetV2M | 0.96 | 0.91 | 0.95 | 0.92 | 0.96 | 0.91 |
| TL2 | InceptionV3 | 0.94 | 0.98 | 0.99 | 0.86 | 0.96 | 0.92 |
| TL3 | MobileNetV2 | 0.94 | 0.81 | 0.89 | 0.88 | 0.92 | 0.84 |
| TL4 | ResNet152 | 0.94 | 0.99 | 0.99 | 0.88 | 0.97 | 0.93 |
| TL5 | ResNet50 | 0.93 | 0.99 | 1.00 | 0.85 | 0.96 | 0.91 |
| TL6 | VGG16 | 0.94 | 0.99 | 1.00 | 0.86 | 0.97 | 0.92 |
| TL7 | VGG19 | 0.92 | 0.95 | 0.98 | 0.84 | 0.95 | 0.89 |
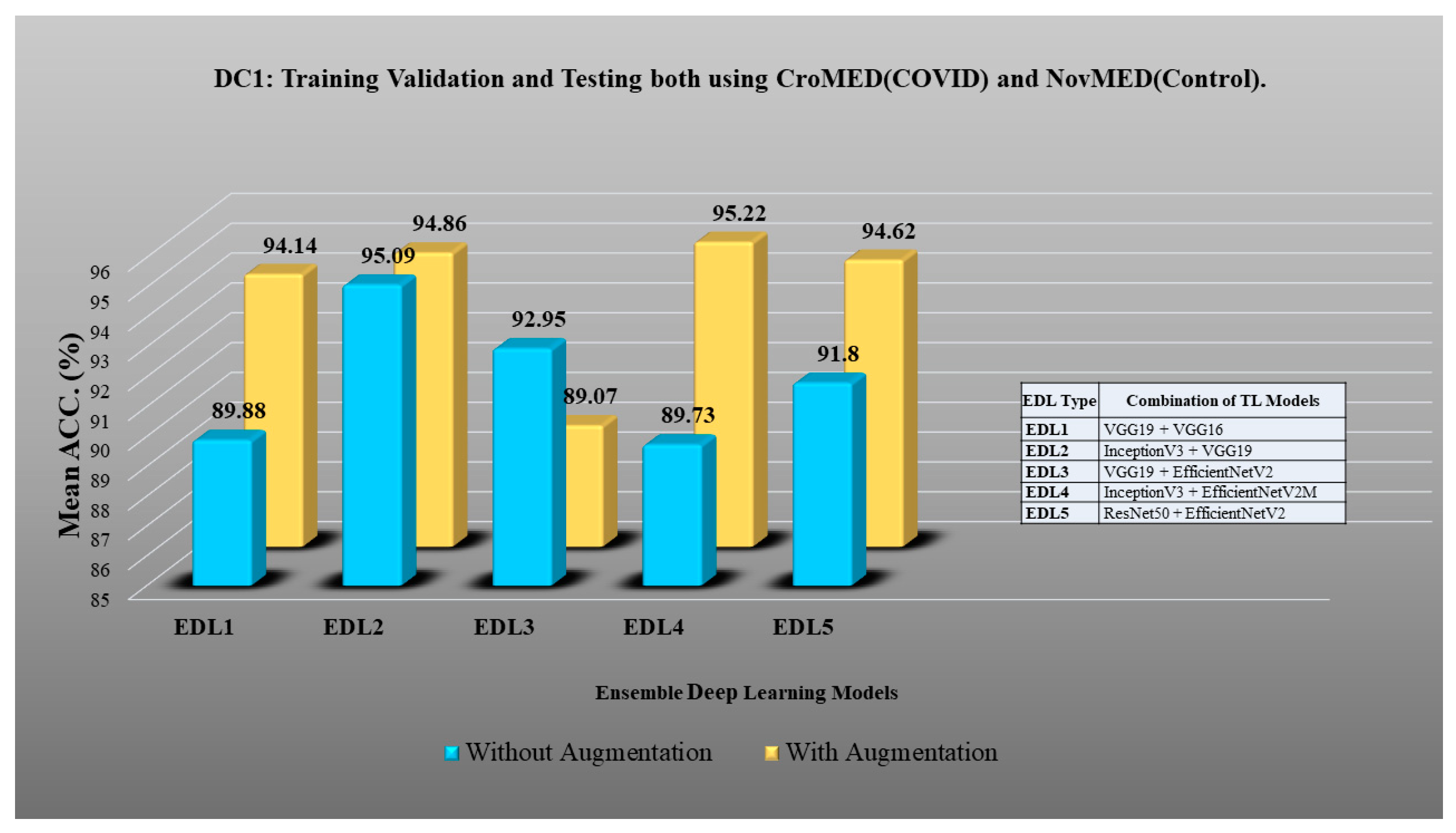
| EDL Statistics on DC1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EDL Type | Without Augmentation | Balance + With Augmentation | ||||||||
| Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | |
| EDL1 | 92.82 | 0.4 | 0.2 | 0.87 | <0.0001 | 96.89 | 0.49 | 0.47 | 0.96 | <0.0001 |
| EDL2 | 94.06 | 0.39 | 0.19 | 0.88 | <0.0001 | 95.43 | 0.49 | 0.46 | 0.95 | <0.0001 |
| EDL3 | 94.48 | 0.41 | 0.21 | 0.9 | <0.0001 | 98.26 | 0.49 | 0.48 | 0.98 | <0.0001 |
| EDL4 | 95.31 | 0.42 | 0.23 | 0.92 | <0.0001 | 96.8 | 0.49 | 0.47 | 0.96 | <0.0001 |
| EDL5 | 98.62 | 0.42 | 0.24 | 0.97 | <0.0001 | 97.99 | 0.49 | 0.48 | 0.98 | <0.0001 |
| Mean ACC of all EDLs: 95.05% | Mean ACC of all EDLs: 97.07% | |||||||||
| EDL Statistics on DC2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Without Augmentation | Balance + With Augmentation | |||||||||
| EDL Type | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value |
| EDL1 | 85.09 | 0.36 | 0.15 | 0.73 | <0.0001 | 93.74 | 0.49 | 0.46 | 0.93 | <0.0001 |
| EDL2 | 87.32 | 0.39 | 0.19 | 0.75 | <0.0001 | 91.89 | 0.49 | 0.46 | 0.91 | <0.0001 |
| EDL3 | 87.58 | 0.39 | 0.19 | 0.8 | <0.0001 | 93.65 | 0.49 | 0.45 | 0.93 | <0.0001 |
| EDL4 | 88.75 | 0.4 | 0.2 | 0.82 | <0.0001 | 91.36 | 0.49 | 0.47 | 0.91 | <0.0001 |
| EDL5 | 89.41 | 0.39 | 0.19 | 0.82 | <0.0001 | 92.86 | 0.49 | 0.46 | 0.92 | <0.0001 |
| Mean ACC of all EDLs: 87.63% | Mean ACC of all EDLs: 92.70% | |||||||||
| EDL Statistics on DC3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Without Augmentation | Balance + With Augmentation | |||||||||
| EDL Type | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value |
| EDL1 | 68.27 | 0.48 | 0.38 | 0.66 | <0.0001 | 82.92 | 0.49 | 0.52 | 0.8 | <0.0001 |
| EDL2 | 68.55 | 0.48 | 0.39 | 0.67 | <0.0001 | 79.63 | 0.48 | 0.61 | 0.79 | <0.0001 |
| EDL3 | 72.41 | 0.48 | 0.37 | 0.71 | <0.0001 | 78.17 | 0.49 | 0.58 | 0.78 | <0.0001 |
| EDL4 | 83.86 | 0.38 | 0.17 | 0.73 | <0.0001 | 81.73 | 0.49 | 0.58 | 0.81 | <0.0001 |
| EDL5 | 86.34 | 0.36 | 0.15 | 0.75 | <0.0001 | 82.46 | 0.49 | 0.59 | 0.82 | <0.0001 |
| Mean ACC of all EDLs: 75.88% | Mean ACC of all EDLs: 80.98% | |||||||||
| EDL Statistics on DC4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Without Augmentation | Balance + With Augmentation | |||||||||
| EDL Type | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value |
| EDL1 | 71.24 | 0.49 | 0.44 | 0.74 | <0.0001 | 80.88 | 0.48 | 0.63 | 0.81 | <0.0001 |
| EDL2 | 70.71 | 0.49 | 0.47 | 0.76 | <0.0001 | 80.61 | 0.47 | 0.65 | 0.8 | <0.0001 |
| EDL3 | 43 | 0.46 | 0.67 | 0.52 | <0.0001 | 75.94 | 0.47 | 0.65 | 0.76 | <0.0001 |
| EDL4 | 44.31 | 0.45 | 0.7 | 0.56 | <0.0001 | 78.32 | 0.48 | 0.61 | 0.78 | <0.0001 |
| EDL5 | 70.71 | 0.48 | 0.47 | 0.76 | <0.0001 | 80.35 | 0.47 | 0.64 | 0.8 | <0.0001 |
| Mean ACC of all EDLs: 59.99% | Mean ACC of all EDLs: 79.22% | |||||||||
| EDL Statistics on DC5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Without Augmentation | Balance + With Augmentation | |||||||||
| EDL Type | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value | Mean ACC (%) | SD (%) | Mean PR | AUC (0–1) | p-Value |
| EDL1 | 93.18 | 0.26 | 0.07 | 0.75 | <0.0001 | 96.59 | 0.46 | 0.3 | 0.95 | <0.0001 |
| EDL2 | 94.17 | 0.29 | 0.09 | 0.81 | <0.0001 | 96.35 | 0.46 | 0.3 | 0.94 | <0.0001 |
| EDL3 | 92.95 | 0.28 | 0.08 | 0.77 | <0.0001 | 95.28 | 0.45 | 0.28 | 0.92 | <0.0001 |
| EDL4 | 92.49 | 0.3 | 0.1 | 0.78 | <0.0001 | 96.65 | 0.46 | 0.3 | 0.95 | <0.0001 |
| EDL5 | 94.17 | 0.31 | 0.1 | 0.83 | <0.0001 | 93.37 | 0.46 | 0.31 | 0.91 | <0.0001 |
| Mean ACC of all EDLs: 93.39% | Mean ACC of all EDLs: 95.64% | |||||||||
| TL Type | Paired t-Test | Mann–Whitney | Wilcoxon | |
|---|---|---|---|---|
| TL1 | EfficientNetV2M | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| TL2 | InceptionV3 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| TL3 | MobileNetV2 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| TL4 | ResNet152 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| TL5 | ResNet50 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| TL6 | VGG16 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| TL7 | VGG19 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| EDL | Paired t-Test | Mann–Whitney | Wilcoxon |
|---|---|---|---|
| EDL1 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| EDL2 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| EDL3 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| EDL4 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| EDL5 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
| SN | Yr | Author | Models | TL | Dataset | DS | Accu (%) | Pre (%) | Re (%) | F1 (%) | AUC (0–1) | p-Value | Cli. Val. | Sci Val. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2021 | Alshazly et al. [28] | DenseNet201 | TL | COVID-CT | 746 | 92.9 | 91.3 | - | 92.5 | 0.93 | - | ✗ | ✗ |
| 2 | 2021 | Alshazly et al. [28] | DenseNet169 | TL | COVID-CT | 746 | 91.2 | 88.1 | - | 90.8 | 0.91 | - | ✗ | ✗ |
| 3 | 2021 | Cruz et al. [29] | DenseNet161 | TL | COVID-CT | 746 | 82.76 | 85.39 | 77.55 | 81.28 | 0.89 | - | ✗ | ✗ |
| 4 | 2021 | Cruz et al. [29] | VGG16 | TL | COVID-CT | 746 | 81.77 | 79.05 | 84.69 | 81.77 | 0.9 | - | ✗ | ✗ |
| 5 | 2022 | Shaik et al. [30] | MobileNetV2 | TL | SARS-CoV-2 | 2482 | 97.38 | 97.41 | 97.35 | 97.38 | 0.97 | - | ✗ | ✗ |
| 6 | 2022 | Shaik et al. [30] | MobileNetV2 | TL | COVID-CT | 746 | 88.67 | 88.5 | 88.61 | 88.55 | 0.88 | - | ✗ | ✗ |
| 7 | 2022 | Huang et al. [31] | EfficientNetV2M | TL | COVID-CT | 7463 | 95.66 | 95.67 | 95.58 | 95.65 | 0.97 | - | ✗ | ✗ |
| 8 | 2023 | Xu et al. [32] | DenseNet121 | TL | COVIDx-CT 2A | 3745 | 99.44 | 99.89 | - | - | - | - | ✗ | ✗ |
| 9 | 2023 | Proposed | MobileNetV2 (Best) | TL | Dataset1 | 5797 1855 | 99.99 | 100 | 100 | 100 | 1.00 | <0.0001 | ✗ | ✓ |
| SN | Yr | Name | Models | EDL | Dataset | DS | Accu (%) | Pre (%) | Re (%) | F1 (%) | AUC (0–1) | p-Value | Cli. Val. | Sci. Val. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2021 | Pathan et al. [33] | ResNet50, AlexNet, VGG19, DenseNet, Inception V3 | EDL | COVID-CT | 746 | 97.00 | 97.00 | 97.00 | 0.97 | - | ✗ | ✗ | |
| 2 | 2021 | Kundu et al. [34] | VGG-11, GoogLeNet, SqueezeNet v1, Wide ResNet-50-2 | EDL | SARS-CoV-2 | 2482 | 98.93 | 98.93 | 98.93 | 98.93 | 0.98 | - | ✗ | ✗ |
| 3 | 2021 | Tao et al. [35] | AlexNet, GoogleNet, ResNet | EDL | COVID-CT | 2933 | 99.05 | - | - | 98.59 | 0 | ✗ | ✗ | |
| 4 | 2021 | Cruz et al. [29] | VGG16, ResNet50, Wide-ResNet50, DenseNet161/169, InceptionV3 | EDL | COVID-CT | 746 | 90.7 | 93.27 | 89.69 | 94.05 | 0.95 | - | ✗ | ✗ |
| 5 | 2022 | Shaik et al. [30] | VGG16, ResNet50, | EDL | COVID-CT | 746 | 91.33 | 91.29 | 91.16 | 91.22 | 0.91 | - | ✗ | ✗ |
| 6 | 2022 | Khanibadi et al. [138] | Naïve Bays, Support Vector Machine | EDL | COVID-CT | 746 | 93.00 | 92.7 | 93.5 | 94.4 | 0.94 | - | ✗ | ✗ |
| 7 | 2022 | Lu et al. [139] | Self-Supervised model with Loss function | EDL | COVID-CT | 746 | 94.3 | 0.94 | 0.93 | 0.94 | 0.98 | <0.0001 | ✗ | ✗ |
| 8 | 2022 | Huang et al. [31] | EfficientNet B0-B4 and V2M | EDL | COVID-CT | 7463 | 98.84 | 98.87 | 98.93 | 98.92 | 0.99 | 0 | ✗ | ✗ |
| 9 | Proposed | ResNet152, MobileNetV2 | EDL | Dataset1 * | 7652 | 99.99 | 100 | 100 | 100 | 100 | <0.0001 | ✗ | ✓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubey, A.K.; Chabert, G.L.; Carriero, A.; Pasche, A.; Danna, P.S.C.; Agarwal, S.; Mohanty, L.; Nillmani; Sharma, N.; Yadav, S.; et al. Ensemble Deep Learning Derived from Transfer Learning for Classification of COVID-19 Patients on Hybrid Deep-Learning-Based Lung Segmentation: A Data Augmentation and Balancing Framework. Diagnostics 2023, 13, 1954. https://doi.org/10.3390/diagnostics13111954
Dubey AK, Chabert GL, Carriero A, Pasche A, Danna PSC, Agarwal S, Mohanty L, Nillmani, Sharma N, Yadav S, et al. Ensemble Deep Learning Derived from Transfer Learning for Classification of COVID-19 Patients on Hybrid Deep-Learning-Based Lung Segmentation: A Data Augmentation and Balancing Framework. Diagnostics. 2023; 13(11):1954. https://doi.org/10.3390/diagnostics13111954
Chicago/Turabian StyleDubey, Arun Kumar, Gian Luca Chabert, Alessandro Carriero, Alessio Pasche, Pietro S. C. Danna, Sushant Agarwal, Lopamudra Mohanty, Nillmani, Neeraj Sharma, Sarita Yadav, and et al. 2023. "Ensemble Deep Learning Derived from Transfer Learning for Classification of COVID-19 Patients on Hybrid Deep-Learning-Based Lung Segmentation: A Data Augmentation and Balancing Framework" Diagnostics 13, no. 11: 1954. https://doi.org/10.3390/diagnostics13111954
APA StyleDubey, A. K., Chabert, G. L., Carriero, A., Pasche, A., Danna, P. S. C., Agarwal, S., Mohanty, L., Nillmani, Sharma, N., Yadav, S., Jain, A., Kumar, A., Kalra, M. K., Sobel, D. W., Laird, J. R., Singh, I. M., Singh, N., Tsoulfas, G., Fouda, M. M., ... Suri, J. S. (2023). Ensemble Deep Learning Derived from Transfer Learning for Classification of COVID-19 Patients on Hybrid Deep-Learning-Based Lung Segmentation: A Data Augmentation and Balancing Framework. Diagnostics, 13(11), 1954. https://doi.org/10.3390/diagnostics13111954













