The Application of Deep Learning on CBCT in Dentistry
Abstract
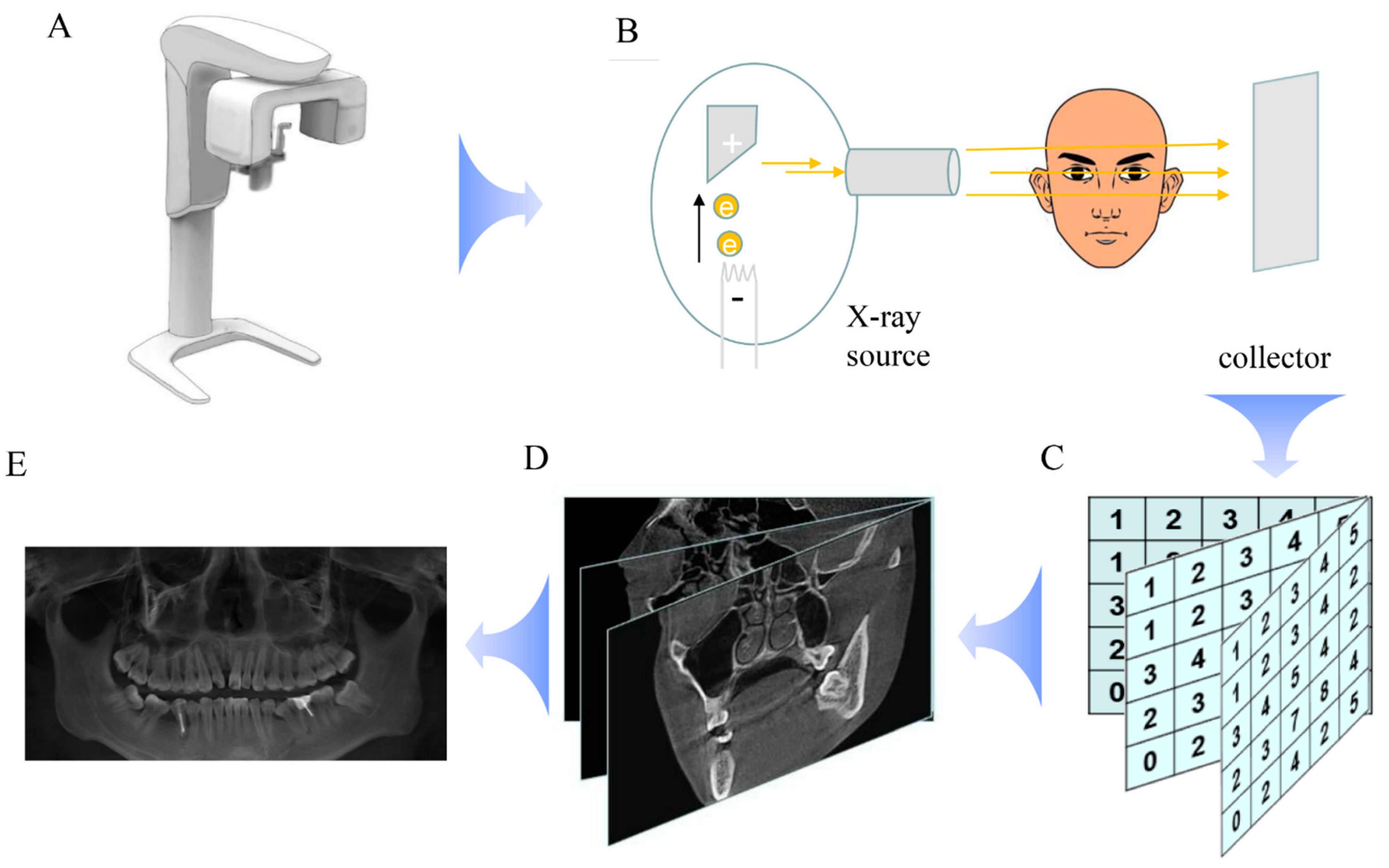
1. Introduction
2. Deep Learning
3. The Application of Deep Learning in CBCT
3.1. The Application of Deep Learning in CBCT in Segmentation of the Upper Airway
3.2. The Application of Deep Learning in CBCT in Segmentation of the Inferior Alveolar Nerve
3.3. The Application of Deep Learning in CBCT in Bone-Related Disease
3.4. The Application of Deep Learning in CBCT in Tooth Segmentation and Endodontics
3.5. The Application of Deep Learning in CBCT in TMJ and Sinus Disease
3.6. The Application of Deep Learning in CBCT in Dental Implant
3.7. The Application of Deep Learning in CBCT in Landmark Localization
4. Conclusions
5. Recommendations for Future Research
Funding
Data Availability Statement
Conflicts of Interest
References
- Kaasalainen, T.; Ekholm, M.; Siiskonen, T.; Kortesniemi, M. Dental cone beam CT: An updated review. Phys. Med. 2021, 88, 193–217. [Google Scholar] [CrossRef] [PubMed]
- Mozzo, P.; Procacci, C.; Tacconi, A.; Martini, P.T.; Andreis, I.A.B. A new volumetric CT machine for dental imaging based on the cone-beam technique: Preliminary results. Eur. Radiol. 1998, 8, 1558–1564. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, R.; Araki, K.; Siewerdsen, J.H.; Thongvigitmanee, S.S. Technical aspects of dental CBCT: State of the art. Dentomaxillofac. Radiol. 2015, 44, 20140224. [Google Scholar] [CrossRef]
- Miracle, A.C.; Mukherji, S.K. Conebeam CT of the head and neck, part 1: Physical principles. AJNR Am. J. Neuroradiol. 2009, 30, 1088–1095. [Google Scholar] [CrossRef]
- Quinto, E.T. An introduction to X-ray tomography and radon transforms. In Proceedings of the American-Mathematical-Society Short Course on the Radon Transform and Applications to Inverse Problems, Atlanta, GA, USA, 3–4 January 2005; pp. 1–23. [Google Scholar]
- Marchant, T.E.; Price, G.J.; Matuszewski, B.J.; Moore, C.J. Reduction of motion artefacts in on-board cone beam CT by warping of projection images. Br. J. Radiol. 2011, 84, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Eshraghi, V.T.; Malloy, K.A.; Tahmasbi, M. Role of Cone-Beam Computed Tomography in the Management of Periodontal Disease. Dent. J. 2019, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Castonguay-Henri, A.; Matenine, D.; Schmittbuhl, M.; de Guise, J.A. Image Quality Optimization and Soft Tissue Visualization in Cone-Beam CT Imaging. In Proceedings of the IUPESM World Congress on Medical Physics and Biomedical Engineering, Prague, Czech Republic, 3–8 June 2018; pp. 283–288. [Google Scholar]
- Muthukrishnan, N.; Maleki, F.; Ovens, K.; Reinhold, C.; Forghani, B.; Forghani, R. Brief History of Artificial Intelligence. Neuroimaging Clin. N. Am. 2020, 30, 393–399. [Google Scholar] [CrossRef]
- Putra, R.H.; Doi, C.; Yoda, N.; Astuti, E.R.; Sasaki, K. Current applications and development of artificial intelligence for digital dental radiography. Dentomaxillofac. Radiol. 2022, 51, 20210197. [Google Scholar] [CrossRef]
- Suzuki, K. Overview of deep learning in medical imaging. Radiol. Phys. Technol. 2017, 10, 257–273. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Carrillo-Perez, F.; Pecho, O.E.; Morales, J.C.; Paravina, R.D.; Della Bona, A.; Ghinea, R.; Pulgar, R.; Perez, M.D.M.; Herrera, L.J. Applications of artificial intelligence in dentistry: A comprehensive review. J. Esthet. Restor. Dent. 2022, 34, 259–280. [Google Scholar] [CrossRef] [PubMed]
- El-Shafai, W.; El-Hag, N.A.; El-Banby, G.M.; Khalaf, A.A.; Soliman, N.F.; Algarni, A.D.; El-Samie, A. An Efficient CNN-Based Automated Diagnosis Framework from COVID-19 CT Images. Comput. Mater. Contin. 2021, 69, 1323–1341. [Google Scholar] [CrossRef]
- Abdou, M.A. Literature review: Efficient deep neural networks techniques for medical image analysis. Neural Comput. Appl. 2022, 34, 5791–5812. [Google Scholar] [CrossRef]
- Du, G.; Cao, X.; Liang, J.; Chen, X.; Zhan, Y. Medical Image Segmentation based on U-Net: A Review. J. Imaging Sci. Technol. 2020, 64, art00009. [Google Scholar] [CrossRef]
- Wu, X.; Wang, S.; Zhang, Y. Survey on theory and application of k-Nearest-Neighbors algorithm. Computer Engineering and Applications. Comput. Eng. Appl. 2017, 53, 1–7. [Google Scholar]
- Zhang, C.; Guo, Y.; Li, M. Review of Development and Application of Artificial Neural Network Models. Comput. Eng. Appl. 2021, 57, 57–69. [Google Scholar]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.W.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. Npj Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019, 17, 9. [Google Scholar] [CrossRef]
- Moraru, A.D.; Costin, D.; Moraru, R.L.; Branisteanu, D.C. Artificial intelligence and deep learning in ophthalmology—Present and future (Review). Exp. Ther. Med. 2020, 20, 3469–3473. [Google Scholar] [CrossRef]
- Ismael, A.M.; Sengur, A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021, 164, 11. [Google Scholar] [CrossRef]
- Lee, J.; Chung, S.W. Deep Learning for Orthopedic Disease Based on Medical Image Analysis: Present and Future. Appl. Sci. 2022, 12, 681. [Google Scholar] [CrossRef]
- Hung, K.F.; Ai, Q.Y.H.; Wong, L.M.; Yeung, A.W.K.; Li, D.T.S.; Leung, Y.Y. Current Applications of Deep Learning and Radiomics on CT and CBCT for Maxillofacial Diseases. Diagnostics 2022, 13, 110. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Bhadauria, H.S.; Singh, A. Descriptive analysis of dental X-ray images using various practical methods: A review. PeerJ Comput. Sci. 2021, 7, e620. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, I.; Rundo, L.; Codari, M.; Di Leo, G.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI applications to medical images: From machine learning to deep learning. Phys. Med. 2021, 83, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, Y.I.; Schuman, J.S.; Shabsigh, R.; Caplan, A.; Al-Aswad, L.A. Ethics of Artificial Intelligence in Medicine and Ophthalmology. Asia-Pac. J. Ophthalmol. 2021, 10, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Shujaat, S.; Jazil, O.; Willems, H.; Van Gerven, A.; Shaheen, E.; Politis, C.; Jacobs, R. Automatic segmentation of the pharyngeal airway space with convolutional neural network. J. Dent. 2021, 111, 103705. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Kim, J.H.; Yu, H.; Jung, H.D.; Chang, S.W.; Park, J.J.; Hong, S.; Cho, H.J.; Choi, Y.J.; Choi, J.; et al. Diagnosis of obstructive sleep apnea with prediction of flow characteristics according to airway morphology automatically extracted from medical images: Computational fluid dynamics and artificial intelligence approach. Comput. Methods Programs Biomed. 2021, 208, 106243. [Google Scholar] [CrossRef]
- Wu, W.; Yu, Y.; Wang, Q.; Liu, D.; Yuan, X. Upper Airway Segmentation Based on the Attention Mechanism of Weak Feature Regions. IEEE Access 2021, 9, 95372–95381. [Google Scholar] [CrossRef]
- Leonardi, R.; Lo Giudice, A.; Farronato, M.; Ronsivalle, V.; Allegrini, S.; Musumeci, G.; Spampinato, C. Fully automatic segmentation of sinonasal cavity and pharyngeal airway based on convolutional neural networks. Am. J. Orthod. Dentofac. Orthop. 2021, 159, 824–835.e821. [Google Scholar] [CrossRef]
- Sin, C.; Akkaya, N.; Aksoy, S.; Orhan, K.; Oz, U. A deep learning algorithm proposal to automatic pharyngeal airway detection and segmentation on CBCT images. Orthod. Craniofac. Res. 2021, 24 (Suppl. S2), 117–123. [Google Scholar] [CrossRef]
- Park, J.; Hwang, J.; Ryu, J.; Nam, I.; Kim, S.-A.; Cho, B.-H.; Shin, S.-H.; Lee, J.-Y. Deep Learning Based Airway Segmentation Using Key Point Prediction. Appl. Sci. 2021, 11, 3501. [Google Scholar] [CrossRef]
- Su, N.C.; van Wijk, A.; Berkhout, E.; Sanderink, G.; De Lange, J.; Wang, H.; van der Heijden, G. Predictive Value of Panoramic Radiography for Injury of Inferior Alveolar Nerve After Mandibular Third Molar Surgery. J. Oral Maxillofac. Surg. 2017, 75, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Cipriano, M.; Allegretti, S.; Bolelli, F.; Di Bartolomeo, M.; Pollastri, F.; Pellacani, A.; Minafra, P.; Anesi, A.; Grana, C. Deep Segmentation of the Mandibular Canal: A New 3D Annotated Dataset of CBCT Volumes. IEEE Access 2022, 10, 11500–11510. [Google Scholar] [CrossRef]
- Jaskari, J.; Sahlsten, J.; Jarnstedt, J.; Mehtonen, H.; Karhu, K.; Sundqvist, O.; Hietanen, A.; Varjonen, V.; Mattila, V.; Kaski, K. Deep Learning Method for Mandibular Canal Segmentation in Dental Cone Beam Computed Tomography Volumes. Sci. Rep. 2020, 10, 5842. [Google Scholar] [CrossRef]
- Lim, H.K.; Jung, S.K.; Kim, S.H.; Cho, Y.; Song, I.S. Deep semi-supervised learning for automatic segmentation of inferior alveolar nerve using a convolutional neural network. BMC Oral. Health 2021, 21, 630. [Google Scholar] [CrossRef]
- Kwak, G.H.; Kwak, E.J.; Song, J.M.; Park, H.R.; Jung, Y.H.; Cho, B.H.; Hui, P.; Hwang, J.J. Automatic mandibular canal detection using a deep convolutional neural network. Sci. Rep. 2020, 10, 5711. [Google Scholar] [CrossRef]
- Jarnstedt, J.; Sahlsten, J.; Jaskari, J.; Kaski, K.; Mehtonen, H.; Lin, Z.Y.; Hietanen, A.; Sundqvist, O.; Varjonen, V.; Mattila, V.; et al. Comparison of deep learning segmentation and multigrader-annotated mandibular canals of multicenter CBCT scans. Sci. Rep. 2022, 12, 18598. [Google Scholar] [CrossRef]
- Lahoud, P.; Diels, S.; Niclaes, L.; Van Aelst, S.; Willems, H.; Van Gerven, A.; Quirynen, M.; Jacobs, R. Development and validation of a novel artificial intelligence driven tool for accurate mandibular canal segmentation on CBCT. J. Dent. 2022, 116, 103891. [Google Scholar] [CrossRef]
- Liu, M.Q.; Xu, Z.N.; Mao, W.Y.; Li, Y.; Zhang, X.H.; Bai, H.L.; Ding, P.; Fu, K.Y. Deep learning-based evaluation of the relationship between mandibular third molar and mandibular canal on CBCT. Clin. Oral. Investig. 2022, 26, 981–991. [Google Scholar] [CrossRef]
- Jeoun, B.S.; Yang, S.; Lee, S.J.; Kim, T.I.; Kim, J.M.; Kim, J.E.; Huh, K.H.; Lee, S.S.; Heo, M.S.; Yi, W.J. Canal-Net for automatic and robust 3D segmentation of mandibular canals in CBCT images using a continuity-aware contextual network. Sci. Rep. 2022, 12, 11. [Google Scholar] [CrossRef]
- Usman, M.; Rehman, A.; Saleem, A.M.; Jawaid, R.; Byon, S.S.; Kim, S.H.; Lee, B.D.; Heo, M.S.; Shin, Y.G. Dual-Stage Deeply Supervised Attention-Based Convolutional Neural Networks for Mandibular Canal Segmentation in CBCT Scans. Sensors 2022, 22, 9877. [Google Scholar] [CrossRef] [PubMed]
- Son, D.M.; Yoon, Y.A.; Kwon, H.J.; An, C.H.; Lee, S.H. Automatic Detection of Mandibular Fractures in Panoramic Radiographs Using Deep Learning. Diagnostics 2021, 11, 933. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.M.; Xia, T.; Kim, J.M.; Zhang, L.F.; Li, B. Combining CNN with Pathological Information for the Detection of Transmissive Lesions of Jawbones from CBCT Images. In Proceedings of the 43rd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE EMBC), Electr Network, Mexico, 1–5 November 2021; pp. 2972–2975. [Google Scholar]
- Yilmaz, E.; Kayikcioglu, T.; Kayipmaz, S. Computer-aided diagnosis of periapical cyst and keratocystic odontogenic tumor on cone beam computed tomography. Comput. Methods Programs Biomed. 2017, 146, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, H.; Feng, C.; Xu, R.; He, Y.; Li, L.; Tu, C. Emerging Applications of Deep Learning in Bone Tumors: Current Advances and Challenges. Front. Oncol. 2022, 12, 908873. [Google Scholar] [CrossRef] [PubMed]
- Jang, T.J.; Kim, K.C.; Cho, H.C.; Seo, J.K. A Fully Automated Method for 3D Individual Tooth Identification and Segmentation in Dental CBCT. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 6562–6568. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, Z.; He, B. Semantic Segmentation of 3D Tooth Model Based on GCNN for CBCT Simulated Mouth Scan Point Cloud Data. J. Comput.-Aided Des. Comput. Graph. 2020, 32, 1162–1170. [Google Scholar]
- Lahoud, P.; EzEldeen, M.; Beznik, T.; Willems, H.; Leite, A.; Van Gerven, A.; Jacobs, R. Artificial Intelligence for Fast and Accurate 3-Dimensional Tooth Segmentation on Cone-beam Computed Tomography. J. Endod. 2021, 47, 827–835. [Google Scholar] [CrossRef]
- Shaheen, E.; Leite, A.; Alqahtani, K.A.; Smolders, A.; Van Gerven, A.; Willems, H.; Jacobs, R. A novel deep learning system for multi-class tooth segmentation and classification on cone beam computed tomography. A validation study. J. Dent. 2021, 115, 103865. [Google Scholar] [CrossRef]
- Gao, S.; Li, X.G.; Li, X.; Li, Z.; Deng, Y.Q. Transformer based tooth classification from cone-beam computed tomography for dental charting. Comput. Biol. Med. 2022, 148, 7. [Google Scholar] [CrossRef]
- Gerhardt, M.D.; Fontenele, R.C.; Leite, A.F.; Lahoud, P.; Van Gerven, A.; Willems, H.; Smolders, A.; Beznik, T.; Jacobs, R. Automated detection and labelling of teeth and small edentulous regions on cone-beam computed tomography using convolutional neural networks. J. Dent. 2022, 122, 8. [Google Scholar] [CrossRef]
- Hsu, K.; Yuh, D.Y.; Lin, S.C.; Lyu, P.S.; Pan, G.X.; Zhuang, Y.C.; Chang, C.C.; Peng, H.H.; Lee, T.Y.; Juan, C.H.; et al. Improving performance of deep learning models using 3.5D U-Net via majority voting for tooth segmentation on cone beam computed tomography. Sci. Rep. 2022, 12, 19809. [Google Scholar] [CrossRef] [PubMed]
- Orhan, K.; Bayrakdar, I.S.; Ezhov, M.; Kravtsov, A.; Ozyurek, T. Evaluation of artificial intelligence for detecting periapical pathosis on cone-beam computed tomography scans. Int. Endod. J. 2020, 53, 680–689. [Google Scholar] [CrossRef]
- Setzer, F.C.; Shi, K.J.; Zhang, Z.; Yan, H.; Yoon, H.; Mupparapu, M.; Li, J. Artificial Intelligence for the Computer-aided Detection of Periapical Lesions in Cone-beam Computed Tomographic Images. J. Endod. 2020, 46, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qiao, X.; Qi, S.; Zhang, X.; Li, S. Effect of adenoid hypertrophy on the upper airway and craniomaxillofacial region. Transl. Pediatr. 2021, 10, 2563–2572. [Google Scholar] [CrossRef]
- Sherwood, A.A.; Sherwood, A.I.; Setzer, F.C.; Shamili, J.V.; John, C.; Schwendicke, F. A Deep Learning Approach to Segment and Classify C-Shaped Canal Morphologies in Mandibular Second Molars Using Cone-beam Computed Tomography. J. Endod. 2021, 47, 1907–1916. [Google Scholar] [CrossRef] [PubMed]
- Albitar, L.; Zhao, T.Y.; Huang, C.; Mahdian, M. Artificial Intelligence (AI) for Detection and Localization of Unobturated Second Mesial Buccal (MB2) Canals in Cone-Beam Computed Tomography (CBCT). Diagnostics 2022, 12, 3214. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Xie, Z. Deep learning in cone-beam computed tomography image segmentation for the diagnosis and treatment of acute pulpitis. J. Supercomput. 2021, 78, 11245–11264. [Google Scholar] [CrossRef]
- Duan, W.; Chen, Y.; Zhang, Q.; Lin, X.; Yang, X. Refined tooth and pulp segmentation using U-Net in CBCT image. Dentomaxillofac. Radiol. 2021, 50, 20200251. [Google Scholar] [CrossRef]
- Li, Q.; Chen, K.; Han, L.; Zhuang, Y.; Li, J.; Lin, J. Automatic tooth roots segmentation of cone beam computed tomography image sequences using U-net and RNN. J. Xray Sci. Technol. 2020, 28, 905–922. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Cao, D.T.; Hu, Y.N.; Wang, B.X.; Zhang, Y.F.; Tang, R.; Zhuang, J.; Gao, A.T.; Chen, Y.; Lin, Z.T. Diagnosis of in vivo vertical root fracture using deep learning on cone-beam CT images. BMC Oral. Health 2022, 22, 9. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, W.; Dong, J.; Tang, Z.; Zhao, Q. Root Canal Segmentation in CBCT Images by 3D U-Net with Global and Local Combination Loss. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2021, 2021, 3097–3100. [Google Scholar] [CrossRef] [PubMed]
- Le, C.; Deleat-Besson, R.; Prieto, J.; Brosset, S.; Dumont, M.; Zhang, W.; Cevidanes, L.; Bianchi, J.; Ruellas, A.; Gomes, L.; et al. Automatic Segmentation of Mandibular Ramus and Condyles. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2021, 2021, 2952–2955. [Google Scholar] [CrossRef] [PubMed]
- de Dumast, P.; Mirabel, C.; Cevidanes, L.; Ruellas, A.; Yatabe, M.; Ioshida, M.; Ribera, N.T.; Michoud, L.; Gomes, L.; Huang, C.; et al. A web-based system for neural network based classification in temporomandibular joint osteoarthritis. Comput. Med. Imaging Graph. 2018, 67, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Ribera, N.T.; de Dumast, P.; Yatabe, M.; Ruellas, A.; Ioshida, M.; Paniagua, B.; Styner, M.; Goncalves, J.R.; Bianchi, J.; Cevidanes, L.; et al. Shape variation analyzer: A classifier for temporomandibular joint damaged by osteoarthritis. Proc. SPIE Int. Soc. Opt. Eng. 2019, 10950, 517–523. [Google Scholar] [CrossRef]
- Serindere, G.; Bilgili, E.; Yesil, C.; Ozveren, N. Evaluation of maxillary sinusitis from panoramic radiographs and cone-beam computed tomographic images using a convolutional neural network. Imaging Sci. Dent. 2022, 52, 187. [Google Scholar] [CrossRef]
- Jung, S.K.; Lim, H.K.; Lee, S.; Cho, Y.; Song, I.S. Deep Active Learning for Automatic Segmentation of Maxillary Sinus Lesions Using a Convolutional Neural Network. Diagnostics 2021, 11, 688. [Google Scholar] [CrossRef]
- Sorkhabi, M.M.; Saadat Khajeh, M. Classification of alveolar bone density using 3-D deep convolutional neural network in the cone-beam CT images: A 6-month clinical study. Measurement 2019, 148, 106945. [Google Scholar] [CrossRef]
- Xiao, Y.J.; Liang, Q.H.; Zhou, L.; He, X.Z.; Lv, L.F.; Chen, J.; Su, E.D.; Guo, J.B.; Wu, D.; Lin, L. Construction of a new automatic grading system for jaw bone mineral density level based on deep learning using cone beam computed tomography. Sci. Rep. 2022, 12, 7. [Google Scholar] [CrossRef]
- Yong, T.H.; Yang, S.; Lee, S.J.; Park, C.; Kim, J.E.; Huh, K.H.; Lee, S.S.; Heo, M.S.; Yi, W.J. QCBCT-NET for direct measurement of bone mineral density from quantitative cone-beam CT: A human skull phantom study. Sci. Rep. 2021, 11, 15083. [Google Scholar] [CrossRef]
- Al-Sarem, M.; Al-Asali, M.; Alqutaibi, A.Y.; Saeed, F. Enhanced Tooth Region Detection Using Pretrained Deep Learning Models. Int. J. Environ. Res. Public Health 2022, 19, 15414. [Google Scholar] [CrossRef]
- Kurt Bayrakdar, S.; Orhan, K.; Bayrakdar, I.S.; Bilgir, E.; Ezhov, M.; Gusarev, M.; Shumilov, E. A deep learning approach for dental implant planning in cone-beam computed tomography images. BMC Med. Imaging 2021, 21, 86. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Shi, M.; Xiang, D.; Zeng, P.; Gong, Z.; Liu, H.; Liu, Q.; Chen, Z.; Xia, J.; Chen, Z. Construction of an end-to-end regression neural network for the determination of a quantitative index sagittal root inclination. J. Periodontol. 2022, 93, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.L.; Zheng, H.R.; Huang, J.Q.; Yang, Y.; Wu, Y.P.; Ge, L.H.; Wang, L.P. The Construction and Evaluation of a Multi-Task Convolutional Neural Network for a Cone-Beam Computed-Tomography-Based Assessment of Implant Stability. Diagnostics 2022, 12, 2673. [Google Scholar] [CrossRef] [PubMed]
- Torosdagli, N.; Liberton, D.K.; Verma, P.; Sincan, M.; Lee, J.S.; Bagci, U. Deep Geodesic Learning for Segmentation and Anatomical Landmarking. IEEE Trans. Med. Imaging 2019, 38, 919–931. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.; Wang, F.; Deng, H.H.; Wang, L.; Xiao, D.; Kuang, T.; Lin, H.-Y.; Gateno, J.; Shen, S.G.F.; Yap, P.-T.; et al. Multi-task Dynamic Transformer Network for Concurrent Bone Segmentation and Large-Scale Landmark Localization with Dental CBCT. Med. Image Comput. Comput.-Assist. Interv. 2020, 12264, 807–816. [Google Scholar] [CrossRef]
- Lang, Y.; Lian, C.; Xiao, D.; Deng, H.; Yuan, P.; Gateno, J.; Shen, S.G.F.; Alfi, D.M.; Yap, P.-T.; Xia, J.J.; et al. Automatic Localization of Landmarks in Craniomaxillofacial CBCT Images Using a Local Attention-Based Graph Convolution Network. Med. Image Comput. Comput.-Assist. Interv. 2020, 12264, 817–826. [Google Scholar] [CrossRef]
- Chen, X.; Lian, C.; Deng, H.H.; Kuang, T.; Lin, H.Y.; Xiao, D.; Gateno, J.; Shen, D.; Xia, J.J.; Yap, P.T. Fast and Accurate Craniomaxillofacial Landmark Detection via 3D Faster R-CNN. IEEE Trans. Med. Imaging 2021, 40, 3867–3878. [Google Scholar] [CrossRef]
- Lang, Y.K.; Lian, C.F.; Xiao, D.Q.; Deng, H.N.; Thung, K.H.; Yuan, P.; Gateno, J.; Kuang, T.S.; Alfi, D.M.; Wang, L.; et al. Localization of Craniomaxillofacial Landmarks on CBCT Images Using 3D Mask R-CNN and Local Dependency Learning. IEEE Trans. Med. Imaging 2022, 41, 2856–2866. [Google Scholar] [CrossRef]
- Ahn, J.; Nguyen, T.P.; Kim, Y.J.; Kim, T.; Yoon, J. Automated analysis of three-dimensional CBCT images taken in natural head position that combines facial profile processing and multiple deep-learning models. Comput. Methods Programs Biomed. 2022, 226, 107123. [Google Scholar] [CrossRef]
- Alberts, I.L.; Mercolli, L.; Pyka, T.; Prenosil, G.; Shi, K.Y.; Rominger, A.; Afshar-Oromieh, A. Large language models (LLM) and ChatGPT: What will the impact on nuclear medicine be? Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 1549–1552. [Google Scholar] [CrossRef]


| Authors | DL Models | Year | Training Dataset | Validation/Test Dataset | Functions | Best Performance of DL | Time- Consuming |
|---|---|---|---|---|---|---|---|
| Jacobs et al. [28] | 3D U-Net | 2021 | 48 | 25 | Segmentation of pharyngeal airway space | Precision: 0.97 ± 0.02 Recall: 0.98 ± 0.01 Accuracy: 1.00 ± 0.00 DSC: 0.98 ± 0.01 IoU: 0.96 ± 0.02 95HD: 0.82 ± 0.41 mm | No |
| Choi et al. [29] | CNN | 2021 | 73 for segmentation 121 for OSAHS diagnose | 15 for segmentation 52 for OSAHS diagnose | Segmentation of upper airway, computational fluid dynamics and OSAHS assessment |
Sensitivity: 0.893 ± 0.048 Specificity: 0.593 ± 0.053 F1 score: 0.74 ± 0.033 DSC: 0.76 ± 0.041
Sensitivity: 0.893 ± 0.048 Specificity: 0.862 ± 0.047 F1 score: 0.0876 ± 0.033 | 6 min |
| Yuan et al. [30] | CNN | 2021 | 102 | 21 for validation 31 for test | Segmentation of upper airway | Precision: 0.914 Recall: 0.864 DSC: 0.927 95HD: 8.3 | No |
| Spampinato et al. [31] | CNN | 2021 | 20 | 20 | Segmentation of sinonasal cavity and pharyngeal airway | DSC: 0.8387 Matching percentage: 0.8535 for tolerance 0.5 mm 0.9344 for tolerance 1.0 mm | No |
| Oz et al. [32] | CNN | 2021 | 214 | 46 for validation 46 for test | Segmentation of upper airway | DSC: 0.919 IoU: 0.993 | No |
| Lee et al. [33] | Regres-sion Neural Network | 2021 | 243 | 72 | Segmentation of upper airway | r2 = 0.975, p < 0.001 | No |
| Authors | DL Models | Year | Training Dataset | Validation/Test Dataset | Functions | Best Performance of DL | Time- Consuming |
|---|---|---|---|---|---|---|---|
| Grana et al. [35] | CNN | 2022 | 68 | 8 for validation 15 for test | IAN detection | IoU: 0.45 DSC: 0.62 | No |
| Kaski et al. [36] | CNN | 2020 | 128 | IAN detection | Precision: 0.85 Recall: 0.64 DSC: 0.6 (roughly) | No | |
| Song et al. [37] | CNN | 2021 | 83 | 50 | IAN detection | 0.58 ± 0.08 | 86.4 ± 61.8 s |
| Hwang et al. [38] | 3D U-Net | 2020 | 102 | IAN detection | Background accuracy: 0.999 Mandibular canal accuracy: 0.927 Global accuracy: 0.999 IoU: 0.577 | No | |
| Nalampang et al. [39] | CNN | 2022 | 882 | 100 for validation 150 for test | IAN detection | Accuracy: 0.99 | No |
| Jacobs et al. [40] | CNN | 2022 | 166 | 30 for validation 39 for test | IAN detection, relationship between IAN and the third molar | Precision: 0.782 Recall: 0.792 Accuracy: 0.999 DSC: 0.774 IoU: 0.636 HD: 0.705 | 21.2 ± 2.79 s |
| Fu et al. [41] | CNN | 2022 | 154 | 30 for validation 45 for test | IAN detection, relationship between IAN and the third molar |
DSC: 0.9730 IoU: 0.9606
DSC: 0.9248 IoU: 0.9003 | 6.1 ± 1.0 s for segmentation 7.4 ± 1.0 s for classifying relation |
| Yi et al. [42] | Canal-Net | 2022 | 30 | 20 for validation 20 for test | IAN detection | Precision: 0.89 ± 0.06 Recall: 0.88 ± 0.06 DSC: 0.87 ± 0.05 Jaccard index: 0.80 ± 0.06 Mean curve distance: 0.62 ± 0.10 Volume of error: 0.10 ± 0.04 Relative volume difference: 0.14 ± 0.04 | No |
| Shin et al. [43] | CNN | 2022 | 400 | 500 | IAN detection | Precision: 0.69 Recall: 0.832 DSC: 0.751 F1 score: 0.759 IoU: 0.795 | No |
| Authors | DL Models | Year | Training Dataset | Validation/Test Dataset | Functions | Best Performance of DL | Time- Consuming |
|---|---|---|---|---|---|---|---|
| Li et al. [45] | CNN | 2021 | 282 | 71 | Jaw bone lesions detection | Overall accuracy: 0.8049 | No |
| Kayipmaz et al. [46] | CNN | 2017 | 50 | Periapical cyst and KCOT lesions classification | Accuracy: 1 F1 score: 1 | No | |
| Authors | DL Models | Year | Training Dataset | Validation/Test Dataset | Functions | Best Performance of DL | Time- Consuming |
|---|---|---|---|---|---|---|---|
| Jin et al. [48] | Unknown | 2022 | 216 | 223 | Tooth identification and segmentation |
Recall: 0.9013 ± 0.0530 F1 score: 0.9335 ± 0.0254
Recall: 0.9371 ± 0.0208 DSC: 0.9479 ± 0.0134 HD: 1.66 ± 0.72 mm | No |
| He et al. [49] | cGAN | 2020 | 15,750 teeth | 4200 teeth | Tooth identification and segmentation |
Lateral incisor: 0.92 ± 0.068 Canine: 0.90 ± 0.053 First premolar: 0.91 ± 0.032 Second premolar: 0.93 ± 0.026 First molar: 0.92 ± 0.112 Second molar: 0.90 ± 0.035 | No |
| Jacobs et al. [50] | CNN | 2021 | 2095 slice | 328 for validation 501 for optimization | Tooth segmentation |
DSC: 0.937 ± 0.02
DSC: 0.940 ± 0.018 | R-AI 72 ± 33.02 s F-AI 30 ± 8.64 s |
| Jacobs et al. [51] | 3D U-Net | 2021 | 140 | 35 for validation 11 for test | Tooth identification and segmentation | Precision: 0.98 ± 0.02 IoU: 0.82 ± 0.05 Recall: 0.83 ± 0.05 DSC: 0.90 ± 0.03 95HD: 0.56 ± 0.38 mm | 7 ± 1.2 h for experts 13.7 ± 1.2 s for DL |
| Deng et al. [52] | CNN | 2022 | 450 | 104 | Tooth identification and segmentation | Accuracy: 0.913 AUC: 0.997 | No |
| Jacobs et al. [53] | CNN | 2022 | 140 | 35 | Tooth identification and segmentation | Accuracy of teeth detection: 0.997 Accuracy of missing teeth detection: 0.99 IoU: 0.96 95HD: 0.33 | 1.5 s |
| Ozyurek et al. [55] | CNN | 2020 | 2800 | 153 | Periapical pathosis detection and their volumes calculation | Detection rate: 0.928 | No |
| Li et al. [56] | U-Net | 2020 | 61 | 12 | Periapical lesion, tooth, bone, material segmentation | Accuracy: 0.93 Specificity: 0.88 DSC: 0.78 | No |
| Schwendicke et al. [58] | Xception U-Net | 2021 | 100 | 35 | Detect the C-shaped root canal of the second molar | DSC: 0.768 ± 0.0349 Sensitivity: 0.786 ± 0.0378 | No |
| Mahdian et al. [59] | U-Net | 2022 | 90 | 10 | Unobturated mesial buccal 2 (MB2) canals on endodontically obturated maxillary molars | Accuracy: 0.9 DSC: 0.768 Sensitivity: 0.8 Specificity: 1 | No |
| Xie et al [60] | cGAN | 2021 | Improved group 40 Traditional group 40 | Different tooth parts segmentation | Omit, Precision, TRP, FRP, and DSC | No | |
| Yang et al. [61] | RPN, FRN, U-Net | 2021 | 20 | Tooth and pulp segmentation |
ASD: 0.104 ± 0.019 mm RVD: 0.049 ± 0.017
ASD: 0.137 ± 0.019 mm RVD: 0.053 ± 0.010 | No | |
| Lin et al. [62] | U-Net, AGs, RNN | 2020 | 1160 | 361 | Root segmentation | IoU: 0.914 DSC: 0.955 Precision: 0.958 Recall: 0.953 | No |
| Lin et al. [63] | ResNet50, VGG19, DenseNet169 | 2022 | 839 | 279 | Vertical root fracture diagnosis |
Sensitivity: 0.970 Specificity: 0.985
Sensitivity: 0.927 Specificity: 0.970
Sensitivity: 0.941 Specificity: 0.985 | No |
| Zhao et al. [64] | 3D U-Net | 2021 | 51 | 17 | Root canal system detection | DSC: 0.952 | 350 ms |
| Authors | DL Models | Year | Training Dataset | Validation/Test Dataset | Functions | Best Performance of DL | Time- Consuming |
|---|---|---|---|---|---|---|---|
| Soroushmehr et al. [65] | U-Net | 2021 | 90 | 19 | Mandibular condyles and ramus segmentation | Sensitivity: 0.93 ± 0.06 Specificity: 0.9998 ± 0.0001 Accuracy: 0.9996 ± 0.0003 F1 score: 0.91 ± 0.03 | No |
| Prieto et al. [66] | Web-based system based on neural network | 2018 | 259 | 34 | TMJ OA classification | No | No |
| Prieto et al. [67] | SVA | 2019 | 259 | 34 | TMJ OA classification | Accuracy: 0.92 | No |
| Ozveren et al. [68] | CNN | 2022 | 237 | 59 | Maxillary sinusitis evaluation | Accuracy: 0.997 Sensitivity: 1 Specificity: 0.993 | No |
| Song et al. [69] | 3D U-Net | 2021 | 70 | 20 | Sinus lesion segmentation | DSC: 0.75~0.77 Accuracy: 0.91 | 1824 s for manual 855.9 s for DL |
| Authors | DL Models | Year | Training Dataset | Validation/Test Dataset | Functions | Best Performance of DL | Time- Consuming |
|---|---|---|---|---|---|---|---|
| Khajeh et al. [70] | CNN | 2019 | 620 | 54 for validation 43 for test | Bone density classification | Accuracy: 0.991 Precision: 0.952 | 76.8 ms |
| Lin et al. [71] | Nested-U-Net | 2022 | 605 | 68 | Bone density classification | Accuracy: 0.91 DSC: 0.75 | No |
| Yi et al. [72] | QCBCT-NET | 2021 | 200 | Bone mineral density measurement | Pearson correlation coefficients: 0.92 | No | |
| Saeed et al. [73] | CNN | 2022 | 350 | 100 for validation 50 for test | Missing tooth regions detection | Accuracy: 0.933 Recall: 0.91 Precision: 0.96 F1 score: 0.97 | No |
| Shumilov et al. [74] | 3D U-Net | 2021 | 75 | Bone height\thickness\canals, missing tooth, sinus measuring |
Sinuses/fossae: 0.664 Missing tooth: 0.953 | No | |
| Chen et al. [75] | CNN | 2022 | 2920 | 824 for validation 400 for test | Perioperative plan | ICCs: 0.895 | 0.001 s for DL 64~107 s for manual work |
| Wang et al. [76] | CNN | 2022 | 1000 | 150 | Implant stability | Precision: 0.9733 Accuracy: 0.9976 IoU: 0.944 Recall: 0.9687 | No |
| Authors | DL Models | Year | Training Dataset | Validation/Test Dataset | Functions | Best Performance of DL | Time- Consuming |
|---|---|---|---|---|---|---|---|
| Bagci et al. [77] | Long short-term memory network | 2019 | 20,480 | 5120 | Mandible segmentation and 9 automatic landmarks | DSC: 0.9382 95HD: 5.47 IoU: 1 Sensitivity: 0.9342 Specificity: 0.9997 | No |
| Shen et al. [78] | Multi-task dynamic transformer network | 2020 | no | no | 64 CMF landmarks | DSC: 0.9395 ± 0.0130 | No |
| Shen et al. [79] | U-Net, graph convolution network | 2020 | 20 | 5 for validation 10 for test | 60 CMF landmarks | Accuracy: 1.69 mm | 1~3 min for DL |
| Yap et al. [80] | 3D faster R-CNN, 3D MS-UNet | 2021 | 60 | 60 | 18 CMF landmarks | Accuracy: 0.79 ± 0.62 mm | 26.6 s for DL |
| Wang et al. [81] | 3D Mask R-CNN | 2022 | 25 | 25 | 105 CMF landmarks | Accuracy: 1.38 ± 0.95 mm | No |
| Yoon et al. [82] | Mask R-CNN | 2022 | 170 | 30 | 23 CMF landmarks |
length: 1 mm angle: <2° | 25~35 min for manual 17 s for DL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, W.; Zhang, J.; Wang, N.; Li, J.; Hu, L. The Application of Deep Learning on CBCT in Dentistry. Diagnostics 2023, 13, 2056. https://doi.org/10.3390/diagnostics13122056
Fan W, Zhang J, Wang N, Li J, Hu L. The Application of Deep Learning on CBCT in Dentistry. Diagnostics. 2023; 13(12):2056. https://doi.org/10.3390/diagnostics13122056
Chicago/Turabian StyleFan, Wenjie, Jiaqi Zhang, Nan Wang, Jia Li, and Li Hu. 2023. "The Application of Deep Learning on CBCT in Dentistry" Diagnostics 13, no. 12: 2056. https://doi.org/10.3390/diagnostics13122056
APA StyleFan, W., Zhang, J., Wang, N., Li, J., & Hu, L. (2023). The Application of Deep Learning on CBCT in Dentistry. Diagnostics, 13(12), 2056. https://doi.org/10.3390/diagnostics13122056






