Artificial Intelligence in Renal Cell Carcinoma Histopathology: Current Applications and Future Perspectives
Abstract
1. Introduction
2. Evidence Acquisition
3. Basics of Artificial Intelligence and Its Application in Histopathology
| Machine learning: Machine learning is a specific branch of artificial intelligence, based on algorithms that enable computer systems to learn, make predictions, and decisions based on data, without the need for explicit programming instructions to do so. | Whole-slide images: Digital representations of entire microscope slides created by scanning glass slides with high-resolution scanners. |
| Deep learning: A subfield of machine learning where algorithms are trained for a task or set of tasks by subjecting a multi-layered artificial neural network to a training data. It eliminates the need for manual feature engineering by allowing the networks to learn directly from raw input data during the training process. The acquired algorithm is subsequently utilized for tasks such as classification, detection, or segmentation. The term "deep" refers to the use of artificial neural networks comprising numerous layers, thus referred to as deep neural networks. | Convolutional neural network: In deep learning, a class of artificial nural network consisting of convolutional of a sequence of convolutional layers to process an input data and produce an output. Each layer implements the convolution operation between the input data and a set of filters. These filter values are learned automatically during training, allowing the network to extract relevant features from the data in an end-to-end fashion (learning the optimal value of all parameters of the model simultaneously rather than sequentially) |
| Digital pathology: The process of digitizing the conventional diagnostic approach. It is accomplished through the utilization of whole-slide scanners and computer screens | Pathomics: The analysis by computational algorithms of digital pathology data, to extract meaningful features. These features are then used to build models for diagnostics, prognostics, and therapeutics purposes |
| Computational pathology: Computational analysis of digital images acquired by scanning pathology slides | Image segmentation: The process of dividing a digital pathology image into distinct regions or objects of interest (for example nuclei or tumor region) to enable analysis and extraction of specific features. |
4. Artificial Intelligence Aided Diagnosis of RCC Subtypes
4.1. RCC Diagnosis and Subtyping in Biopsy Specimens
4.2. RCC Diagnosis and Subtyping in Surgical Resection Specimens
| Group | Aim | Number of Patients | Training Process | Accuracy on the Test Set | External Validation (N of Patients) | Accuracy on the External Validation Cohort | Algorithm |
|---|---|---|---|---|---|---|---|
| Fenstermaker et al. [55] | (1) RCC diagnosis, (2) subtyping, (3) grading | (1) 15 ccRCC; (2) 15 pRCC; (3) 12 chRCC. | No significant error decrease in 25 epochs in training was recorded. Next, a validation dataset was used. Training was halted when the performance on the validation set ceased to improve. | (1) 99.1%; (2) 97.5%; (3) 98.4% | N.A. | N.A. | CNN: 6 different convolutional layers, 2 layers of 32 filters, 2 layers of 64 filters, and 2 layers of 128 filters. |
| Zhu et al. [59] | RCC subtyping | (1) 486 SR (30 NT, 27 RO, 38 chRCC, 310 ccRCC, 81 pRCC); (2) 79 RMB (24 RO, 34 ccRCC, 21 pRCC). | The models were trained for 40 epochs. The trained model assigned a confidence score for each patch. Finally, a comparison of the trained models was completed. | (1) 97% on SRS, (2) 97% on RMB | 0 RO 109 ChRCC 505 ccRCC 294 pRCC: | 95% accuracy (only SRs) | DNN: we tested four versions of ResNet: ResNet-18, ResNet-34, ResNet-50, and ResNet-101. ResNet-18 was selected for the highest average F1-score on the developement set (0.96) |
| Chen et al. [67] | (1) RCC diagnosis, (2) subtyping, (3) survival prediction | (1) and (2) 362 NT, 362 ccRCC, 128 pRCC, 84 chRCC; (3) 283 ccRCC. | LASSO was used to identify RCC-related digital pathological factors and their coefficients in the training cohort. LASSO–Cox regression was used to identify survival-related digital pathological factors and their coefficients in the training cohort. | (1) 94.5% vs. NT (2) 97% vs. pRCC and chRCC (3) 88.8%, 90.0%, 89.6% in 1–3–5 y DFS | (1) and (2) 150 NP, 150 ccRCC, 52 pRCC, and 84 chRCC; (3) 120ccRCC. | (1) 87.6% vs. NP; (2) 81.4% vs. pRCC and chRCC; (3) 72.0%, 80.9%, 85.9% in 1-, 3-, or 5-year DFS. | Segmentation and feature extraction pipeline via CellProfiler: (1) and (2) LASSO; (2) LASSO–Cox regression analysis |
| Tabibu et al. [66] | (1) RCC diagnosis; (2) subtyping, | (1) 509 NT; (2) 1027 ccRCC; (3) 303 pRCC; (4) 254 chRCC. | Training was terminated when validation accuracy stabilized for 4–5 epochs. Data augmentation included random patches, vertical flip, rotation, and noise addition. Weighted resampling was used to address class imbalance. Training parameters remained unchanged. | (1) 93.9% ccRCC vs. NP 87.34% chRCC vs. NP (2) 92.16% subtyping | N.A. | N.A. | CNN (Resnet 18 and 34 architecture based); DAG-SVM on top of CNN for subtyping. |
| Abdeltawab et al. [73] | RCC subtyping | (1) 27 ccRCC; (2) 14 ccpRCC. | Each image was divided into overlapping patches of different sizes for feature recognition at different sizes. Multiple CNNs outperformed a single CNN for learning features at different scales. Patch overlap of 50% for learning from diverse viewpoints. | 91% in ccpRCC | 10 ccRCC. | 90% in ccRCC | Three CNNs were used for small, medium, and large patch sizes. The CNNs shared the same architecture: a series of convolutional layers intervened by max-pooling layers, followed by two fully connected layers. Finally, there was a soft-max layer |
5. Pathomics in Disease Prognosis
5.1. Cancer Grading
5.2. Molecular-Morphological Connections and AI-Based Therapy Response Prediction
5.3. Prognosis Prediction Models Based on Computational Pathology
6. Future Perspectives
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | artificial intelligence |
| AUC | area under curve |
| BFPS | block filtering post-pruning search |
| ccpRCC | clear cell papillary renal cell carcinoma |
| ccRCC | clear cell renal cell carcinoma |
| chRCC | chromophobe renal cell carcinoma |
| CNA | copy number alteration |
| CNN | convolutional neural network |
| CT | computed tomography |
| DAG-SVM | Directed Acyclic Graph Support Vector Machine |
| DCNN | deep convoluted neural network |
| DFS | disease free survival |
| DL | deep learning |
| DNN | deep neural network |
| EGFR | Epidermal growth factor receptor |
| FCNN | fully-connected neural network |
| grad-CAM | gradient-weighted class activation mapping |
| IMDC | International Metastatic Renal Cell Carcinoma Database Consortium |
| KRAS | V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| lmQCM | local maximum quasi-clique merging |
| ML | machine learning |
| mRCC | metastatic renal cell carcinoma |
| MRI | magnetic resonance imaging |
| MSKCC | Memorial Sloan Kettering Cancer Center |
| N.A. | not applicable |
| NP | normal parenchyma |
| NT | normal tissue |
| OS | overall survival |
| PFS | Progression-free survival |
| pRCC | papillary renal cell carcinoma |
| RCC | renal cell carcinoma |
| ResNet | residual neural network architecture |
| RMB | renal mass biopsy |
| RO | renal oncocytoma |
| SVM | support vector machine |
| TCGA | The Cancer Genome Atlas |
| TKI | Tyrosine kinase inhibitors |
| UISS | UCLA Integrated Staging System for renal cell carcinoma |
| VEGFR-TKI | VEGF receptor-tyrosine kinase inhibitors |
| VHL | Von-Hippel-Lindau tumor suppressor |
| WSI | whole slide imaging |
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Garfield, K.; LaGrange, C.A. Renal Cell Cancer; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Bukavina, L.; Bensalah, K.; Bray, F.; Carlo, M.; Challacombe, B.; Karam, J.A.; Kassouf, W.; Mitchell, T.; Montironi, R.; O’Brien, T.; et al. Epidemiology of Renal Cell Carcinoma: 2022 Update. Eur. Urol. 2022, 82, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs—Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Cimadamore, A.; Caliò, A.; Marandino, L.; Marletta, S.; Franzese, C.; Schips, L.; Amparore, D.; Bertolo, R.; Muselaers, S.; Erdem, S.; et al. Hot topics in renal cancer pathology: Implications for clinical management. Expert Rev. Anticancer. Ther. 2022, 22, 1275–1287. [Google Scholar] [CrossRef]
- Fuhrman, S.A.; Lasky, L.C.; Limas, C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am. J. Surg. Pathol. 1982, 6, 655–664. [Google Scholar] [CrossRef]
- Zhang, L.; Zha, Z.; Qu, W.; Zhao, H.; Yuan, J.; Feng, Y.; Wu, B. Tumor necrosis as a prognostic variable for the clinical outcome in patients with renal cell carcinoma: A systematic review and meta-analysis. BMC Cancer 2018, 18, 870. [Google Scholar] [CrossRef]
- Sun, M.; Shariat, S.F.; Cheng, C.; Ficarra, V.; Murai, M.; Oudard, S.; Pantuck, A.J.; Zigeuner, R.; Karakiewicz, P.I. Prognostic factors and predictive models in renal cell carcinoma: A contemporary review. Eur. Urol. 2011, 60, 644–661. [Google Scholar] [CrossRef]
- Hora, M.; Albiges, L.; Bedke, J.; Campi, R.; Capitanio, U.; Giles, R.H.; Ljungberg, B.; Marconi, L.; Klatte, T.; Volpe, A.; et al. European Association of Urology Guidelines Panel on Renal Cell Carcinoma Update on the New World Health Organization Classification of Kidney Tumours 2022: The Urologist’s Point of View. Eur. Urol. 2023, 83, 97–100. [Google Scholar] [CrossRef]
- Mimma, R.; Anna, C.; Matteo, B.; Gaetano, P.; Carlo, G.; Guido, M.; Camillo, P. Clinico-pathological implications of the 2022 WHO Renal Cell Carcinoma classification. Cancer Treat. Rev. 2023, 116, 102558. [Google Scholar] [CrossRef]
- Baidoshvili, A.; Bucur, A.; Van Leeuwen, J.; Van Der Laak, J.; Kluin, P.; Van Diest, P.J. Evaluating the benefits of digital pathology implementation: Time savings in laboratory logistics. Histopathology 2018, 73, 784–794. [Google Scholar] [CrossRef]
- Shmatko, A.; Ghaffari Laleh, N.; Gerstung, M.; Kather, J.N. Artificial intelligence in histopathology: Enhancing cancer research and clinical oncology. Nat. Cancer 2022, 3, 1026–1038. [Google Scholar] [CrossRef]
- Roussel, E.; Capitanio, U.; Kutikov, A.; Oosterwijk, E.; Pedrosa, I.; Rowe, S.P.; Gorin, M.A. Novel Imaging Methods for Renal Mass Characterization: A Collaborative Review. Eur. Urol. 2022, 81, 476–488. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef] [PubMed]
- Colling, R.; Pitman, H.; Oien, K.; Rajpoot, N.; Macklin, P.; CM-Path AI in Histopathology Working Group; Snead, D.; Sackville, T.; Verrill, C. Artificial intelligence in digital pathology: A roadmap to routine use in clinical practice. J. Pathol. 2019, 249, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Glembin, M.; Obuchowski, A.; Klaudel, B.; Rydzinski, B.; Karski, R.; Syty, P.; Jasik, P.; Narożański, W.J. Enhancing Renal Tumor Detection: Leveraging Artificial Neural Networks in Computed Tomography Analysis. Med. Sci. Monit. 2023, 29, e939462. [Google Scholar] [CrossRef] [PubMed]
- Volpe, A.; Patard, J.J. Prognostic factors in renal cell carcinoma. World J. Urol. 2010, 28, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.D.; Rini, B.I. Predicting Response to Immunotherapy in Metastatic Renal Cell Carcinoma. Cancers 2020, 12, 2662. [Google Scholar] [CrossRef]
- Wang, D.; Khosla, A.; Gargeya, R.; Irshad, H.; Beck, A.H.; Israel, B. Deep Learning for Identifying Metastatic Breast Cancer. arXiv 2016, arXiv:1606.05718. [Google Scholar] [CrossRef]
- Hayashi, Y. Black Box Nature of Deep Learning for Digital Pathology: Beyond Quantitative to Qualitative Algorithmic Performances. In Artificial Intelligence and Machine Learning for Digital Pathology; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2020; Volume 12090, pp. 95–101. [Google Scholar] [CrossRef]
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Bulten, W.; Pinckaers, H.; van Boven, H.; Vink, R.; de Bel, T.; van Ginneken, B.; van der Laak, J.; Hulsbergen-van de Kaa, C.; Litjens, G. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: A diagnostic study. Lancet Oncol. 2020, 21, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Ström, P.; Kartasalo, K.; Olsson, H.; Solorzano, L.; Delahunt, B.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Grignon, D.J.; Humphrey, P.A.; et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: A population-based, diagnostic study. Lancet Oncol. 2020, 21, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018, 23, 181–193.e7. [Google Scholar] [CrossRef]
- Kapil, A.; Wiestler, T.; Lanzmich, S.; Silva, A.; Steele, K.; Rebelatto, M.; Schmidt, G.; Brieu, N. DASGAN—Joint Domain Adaptation and Segmentation for the Analysis of Epithelial Regions in Histopathology PD-L1 Images. arXiv 2019, arXiv:1906.11118. [Google Scholar]
- Sha, L.; Osinski, B.L.; Ho, I.Y.; Tan, T.L.; Willis, C.; Weiss, H.; Beaubier, N.; Mahon, B.M.; Taxter, T.J.; Yip, S.S.F. Multi-Field-of-View Deep Learning Model Predicts Nonsmall Cell Lung Cancer Programmed Death-Ligand 1 Status from Whole-Slide Hematoxylin and Eosin Images. J. Pathol. Inform. 2019, 10, 24. [Google Scholar] [CrossRef]
- Creighton, C.J.; Morgan, M.; Gunaratne, P.H.; Wheeler, D.A.; Gibbs, R.A.; Gordon Robertson, A.; Chu, A.; Beroukhim, R.; Cibulskis, K.; Signoretti, S.; et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef]
- Krajewski, K.M.; Pedrosa, I. Imaging Advances in the Management of Kidney Cancer. J. Clin. Oncol. 2018, 36, 3582–3590. [Google Scholar] [CrossRef]
- Roussel, E.; Campi, R.; Amparore, D.; Bertolo, R.; Carbonara, U.; Erdem, S.; Ingels, A.; Kara, Ö.; Marandino, L.; Marchioni, M.; et al. Expanding the Role of Ultrasound for the Characterization of Renal Masses. J. Clin. Med. 2022, 11, 1112. [Google Scholar] [CrossRef]
- Shuch, B.; Hofmann, J.N.; Merino, M.J.; Nix, J.W.; Vourganti, S.; Linehan, W.M.; Schwartz, K.; Ruterbusch, J.J.; Colt, J.S.; Purdue, M.P.; et al. Pathologic validation of renal cell carcinoma histology in the Surveillance, Epidemiology, and End Results program. Urol. Oncol. Semin. Orig. Investig. 2013, 32, 23.e9–23.e13. [Google Scholar] [CrossRef]
- Al-Aynati, M.; Chen, V.; Salama, S.; Shuhaibar, H.; Treleaven, D.; Vincic, L. Interobserver and Intraobserver Variability Using the Fuhrman Grading System for Renal Cell Carcinoma. Arch. Pathol. Lab. Med. 2003, 127, 593–596. [Google Scholar] [CrossRef]
- Williamson, S.R.; Rao, P.; Hes, O.; Epstein, J.I.; Smith, S.C.; Picken, M.M.; Zhou, M.; Tretiakova, M.S.; Tickoo, S.K.; Chen, Y.-B.; et al. Challenges in pathologic staging of renal cell carcinoma: A study of interobserver variability among urologic pathologists. Am. J. Surg. Pathol. 2018, 42, 1253–1261. [Google Scholar] [CrossRef]
- Gavrielides, M.A.; Gallas, B.D.; Lenz, P.; Badano, A.; Hewitt, S.M. Observer variability in the interpretation of HER2/neu immunohistochemical expression with unaided and computer-aided digital microscopy. Arch. Pathol. Lab. Med. 2011, 135, 233–242. [Google Scholar] [CrossRef]
- Ficarra, V.; Martignoni, G.; Galfano, A.; Novara, G.; Gobbo, S.; Brunelli, M.; Pea, M.; Zattoni, F.; Artibani, W. Prognostic Role of the Histologic Subtypes of Renal Cell Carcinoma after Slide Revision. Eur. Urol. 2006, 50, 786–794. [Google Scholar] [CrossRef]
- Lang, H.; Lindner, V.; de Fromont, M.; Molinié, V.; Letourneux, H.; Meyer, N.; Martin, M.; Jacqmin, D. Multicenter determination of optimal interobserver agreement using the Fuhrman grading system for renal cell carcinoma. Cancer 2004, 103, 625–629. Available online: https://acsjournals.onlinelibrary.wiley.com/doi/full/10.1002/cncr.20812 (accessed on 1 February 2023). [CrossRef] [PubMed]
- Smaldone, M.C.; Egleston, B.; Hollingsworth, J.M.; Hollenbeck, B.K.; Miller, D.C.; Morgan, T.M.; Kim, S.P.; Malhotra, A.; Handorf, E.; Wong, Y.-N.; et al. Understanding Treatment Disconnect and Mortality Trends in Renal Cell Carcinoma Using Tumor Registry Data. Med. Care 2017, 55, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Kutikov, A.; Smaldone, M.C.; Egleston, B.L.; Manley, B.J.; Canter, D.J.; Simhan, J.; Boorjian, S.A.; Viterbo, R.; Chen, D.Y.; Greenberg, R.E.; et al. Anatomic Features of Enhancing Renal Masses Predict Malignant and High-Grade Pathology: A Preoperative Nomogram Using the RENAL Nephrometry Score. Eur. Urol. 2011, 60, 241–248. [Google Scholar] [CrossRef]
- Pierorazio, P.M.; Patel, H.D.; Johnson, M.H.; Sozio, S.; Sharma, R.; Iyoha, E.; Bass, E.; Allaf, M.E. Distinguishing malignant and benign renal masses with composite models and nomograms: A systematic review and meta-analysis of clinically localized renal masses suspicious for malignancy. Cancer 2016, 122, 3267–3276. [Google Scholar] [CrossRef]
- Joshi, S.; Kutikov, A. Understanding Mutational Drivers of Risk: An Important Step Toward Personalized Care for Patients with Renal Cell Carcinoma. Eur. Urol. Focus 2016, 3, 428–429. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.M.; Gill, I.S.; Ellison, L.M. The Evolving Presentation of Renal Carcinoma in the United States: Trends From the Surveillance, Epidemiology, and End Results Program. J. Urol. 2006, 176, 2397–2400. [Google Scholar] [CrossRef] [PubMed]
- Sohlberg, E.M.; Metzner, T.J.; Leppert, J.T. The Harms of Overdiagnosis and Overtreatment in Patients with Small Renal Masses: A Mini-review. Eur. Urol. Focus 2019, 5, 943–945. [Google Scholar] [CrossRef] [PubMed]
- Campi, R.; Stewart, G.D.; Staehler, M.; Dabestani, S.; Kuczyk, M.A.; Shuch, B.M.; Finelli, A.; Bex, A.; Ljungberg, B.; Capitanio, U. Novel Liquid Biomarkers and Innovative Imaging for Kidney Cancer Diagnosis: What Can Be Implemented in Our Practice Today? A Systematic Review of the Literature. Eur. Urol. Oncol. 2021, 4, 22–41. [Google Scholar] [CrossRef]
- Warren, H.; Palumbo, C.; Caliò, A.; Tran, M.G.B.; Campi, R.; European Association of Urology (EAU) Young Academic Urologists (YAU) Renal Cancer Working Group. Oncocytoma on renal mass biopsy: Why is surgery even performed? World J. Urol. 2023, 41, 1709–1710. [Google Scholar] [CrossRef] [PubMed]
- Kutikov, A.; Smaldone, M.C.; Uzzo, R.G.; Haifler, M.; Bratslavsky, G.; Leibovich, B.C. Renal Mass Biopsy: Always, Sometimes, or Never? Eur. Urol. 2016, 70, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Lane, B.R.; Samplaski, M.K.; Herts, B.R.; Zhou, M.; Novick, A.C.; Campbell, S.C. Renal Mass Biopsy—A Renaissance? J. Urol. 2008, 179, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Sinks, A.; Miller, C.; Holck, H.; Zeng, L.; Gaston, K.; Riggs, S.; Matulay, J.; Clark, P.E.; Roy, O. Renal Mass Biopsy Mandate Is Associated With Change in Treatment Decisions. J. Urol. 2023, 210, 72–78. [Google Scholar] [CrossRef]
- Marconi, L.; Dabestani, S.; Lam, T.B.; Hofmann, F.; Stewart, F.; Norrie, J.; Bex, A.; Bensalah, K.; Canfield, S.E.; Hora, M.; et al. Systematic Review and Meta-analysis of Diagnostic Accuracy of Percutaneous Renal Tumour Biopsy. Eur. Urol. 2016, 69, 660–673. [Google Scholar] [CrossRef]
- Evans, A.J.; Delahunt, B.; Srigley, J.R. Issues and challenges associated with classifying neoplasms in percutaneous needle biopsies of incidentally found small renal masses. Semin. Diagn. Pathol. 2015, 32, 184–195. [Google Scholar] [CrossRef]
- Kümmerlin, I.; ten Kate, F.; Smedts, F.; Horn, T.; Algaba, F.; Trias, I.; de la Rosette, J.; Laguna, M.P. Core biopsies of renal tumors: A study on diagnostic accuracy, interobserver, and intraobserver variability. Eur. Urol. 2008, 53, 1219–1227. [Google Scholar] [CrossRef]
- Elmore, J.G.; Longton, G.M.; Carney, P.A.; Geller, B.M.; Onega, T.; Tosteson, A.N.A.; Nelson, H.D.; Pepe, M.S.; Allison, K.H.; Schnitt, S.J.; et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 2015, 313, 1122–1132. [Google Scholar] [CrossRef]
- Elmore, J.G.; Barnhill, R.L.; Elder, D.E.; Longton, G.M.; Pepe, M.S.; Reisch, L.M.; Carney, P.A.; Titus, L.J.; Nelson, H.D.; Onega, T.; et al. Pathologists’ diagnosis of invasive melanoma and melanocytic proliferations: Observer accuracy and reproducibility study. BMJ 2017, 357, j2813. [Google Scholar] [CrossRef]
- Shah, M.D.; Parwani, A.V.; Zynger, D.L. Impact of the Pathologist on Prostate Biopsy Diagnosis and Immunohistochemical Stain Usage Within a Single Institution. Am. J. Clin. Pathol. 2017, 148, 494–501. [Google Scholar] [CrossRef]
- Fenstermaker, M.; Tomlins, S.A.; Singh, K.; Wiens, J.; Morgan, T.M. Development and Validation of a Deep-learning Model to Assist With Renal Cell Carcinoma Histopathologic Interpretation. Urology 2020, 144, 152–157. [Google Scholar] [CrossRef] [PubMed]
- van Oostenbrugge, T.J.; Fütterer, J.J.; Mulders, P.F. Diagnostic Imaging for Solid Renal Tumors: A Pictorial Review. Kidney Cancer 2018, 2, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.M.; Lynch, D.T. Renal Oncocytoma; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Leone, A.R.; Kidd, L.C.; Diorio, G.J.; Zargar-Shoshtari, K.; Sharma, P.; Sexton, W.J.; Spiess, P.E. Bilateral benign renal oncocytomas and the role of renal biopsy: Single institution review. BMC Urol. 2017, 17, 6. [Google Scholar] [CrossRef]
- Zhu, M.; Ren, B.; Richards, R.; Suriawinata, M.; Tomita, N.; Hassanpour, S. Development and evaluation of a deep neural network for histologic classification of renal cell carcinoma on biopsy and surgical resection slides. Sci. Rep. 2021, 11, 7080. [Google Scholar] [CrossRef]
- Volpe, A.; Mattar, K.; Finelli, A.; Kachura, J.R.; Evans, A.J.; Geddie, W.R.; Jewett, M.A. Contemporary results of percutaneous biopsy of 100 small renal masses: A single center experience. J. Urol. 2008, 180, 2333–2337. [Google Scholar] [CrossRef]
- Wang, R.; Wolf, J.S.; Wood, D.P.; Higgins, E.J.; Hafez, K.S. Accuracy of Percutaneous Core Biopsy in Management of Small Renal Masses. Urology 2009, 73, 586–590. [Google Scholar] [CrossRef]
- Barwari, K.; de la Rosette, J.J.; Laguna, M.P. The penetration of renal mass biopsy in daily practice: A survey among urologists. J. Endourol. 2012, 26, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B. Emerging immunotherapies for renal cell carcinoma. Ann. Oncol. 2012, 23, viii35–viii40. [Google Scholar] [CrossRef]
- Yanagisawa, T.; Schmidinger, M.; Fajkovic, H.; Karakiewicz, P.I.; Kimura, T.; Shariat, S.F. What is the role of cytoreductive nephrectomy in patients with metastatic renal cell carcinoma? Expert Rev. Anticancer Ther. 2023, 23, 455–459. [Google Scholar] [CrossRef]
- Bertolo, R.; Pecoraro, A.; Carbonara, U.; Amparore, D.; Diana, P.; Muselaers, S.; Marchioni, M.; Mir, M.C.; Antonelli, A.; Badani, K.; et al. Resection Techniques During Robotic Partial Nephrectomy: A Systematic Review. Eur. Urol. Open Sci. 2023, 52, 7–21. [Google Scholar] [CrossRef]
- Tabibu, S.; Vinod, P.K.; Jawahar, C.V. Pan-Renal Cell Carcinoma classification and survival prediction from histopathology images using deep learning. Sci. Rep. 2019, 9, 10509. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, N.; Jiang, L.; Gao, F.; Shao, J.; Wang, T.; Zhang, E.; Yu, H.; Wang, X.; Zheng, J. Clinical use of a machine learning histopathological image signature in diagnosis and survival prediction of clear cell renal cell carcinoma. Int. J. Cancer 2020, 148, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Marostica, E.; Barber, R.; Denize, T.; Kohane, I.S.; Signoretti, S.; Golden, J.A.; Yu, K.-H. Development of a Histopathology Informatics Pipeline for Classification and Prediction of Clinical Outcomes in Subtypes of Renal Cell Carcinoma. Clin. Cancer Res. 2021, 27, 2868–2878. [Google Scholar] [CrossRef] [PubMed]
- Pathology Outlines—WHO Classification. Available online: https://www.pathologyoutlines.com/topic/kidneytumorWHOclass.html (accessed on 24 January 2023).
- Cimadamore, A.; Cheng, L.; Scarpelli, M.; Massari, F.; Mollica, V.; Santoni, M.; Lopez-Beltran, A.; Montironi, R.; Moch, H. Towards a new WHO classification of renal cell tumor: What the clinician needs to know—A narrative review. Transl. Androl. Urol. 2021, 10, 1506–1520. [Google Scholar] [CrossRef]
- Weng, S.; DiNatale, R.G.; Silagy, A.; Mano, R.; Attalla, K.; Kashani, M.; Weiss, K.; Benfante, N.E.; Winer, A.G.; Coleman, J.A.; et al. The Clinicopathologic and Molecular Landscape of Clear Cell Papillary Renal Cell Carcinoma: Implications in Diagnosis and Management. Eur. Urol. 2020, 79, 468–477. [Google Scholar] [CrossRef]
- Williamson, S.R.; Eble, J.N.; Cheng, L.; Grignon, D.J. Clear cell papillary renal cell carcinoma: Differential diagnosis and extended immunohistochemical profile. Mod. Pathol. 2013, 26, 697–708. [Google Scholar] [CrossRef]
- Abdeltawab, H.A.; Khalifa, F.A.; Ghazal, M.A.; Cheng, L.; El-Baz, A.S.; Gondim, D.D. A deep learning framework for automated classification of histopathological kidney whole-slide images. J. Pathol. Inform. 2022, 13, 100093. [Google Scholar] [CrossRef]
- Faust, K.; Roohi, A.; Leon, A.J.; Leroux, E.; Dent, A.; Evans, A.J.; Pugh, T.J.; Kalimuthu, S.N.; Djuric, U.; Diamandis, P. Unsupervised Resolution of Histomorphologic Heterogeneity in Renal Cell Carcinoma Using a Brain Tumor–Educated Neural Network. JCO Clin. Cancer Inform. 2020, 4, 811–821. [Google Scholar] [CrossRef]
- Renal Cell Carcinoma EAU Guidelines on 2022. Available online: https://uroweb.org/guidelines/renal-cell-carcinoma (accessed on 1 February 2023).
- Gelb, A.B. Communication Union Internationale Contre le Cancer (UICC) and the American Joint Renal Cell Carcinoma Committee on Cancer (AJCC) Current Prognostic Factors BACKGROUND. Renal Cell Carcinomas Include Several Distinct Entities with a Range. Available online: 10.1002/(sici)1097-0142(19970901)80:5<994::aid-cncr27>3.0.co;2-q (accessed on 1 February 2023).
- Beksac, A.T.; Paulucci, D.J.; Blum, K.A.; Yadav, S.S.; Sfakianos, J.P.; Badani, K.K. Heterogeneity in renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2017, 35, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Dall’Oglio, M.F.; Ribeiro-Filho, L.A.; Antunes, A.A.; Crippa, A.; Nesrallah, L.; Gonçalves, P.D.; Leite, K.R.M.; Srougi, M. Microvascular Tumor Invasion, Tumor Size and Fuhrman Grade: A Pathological Triad for Prognostic Evaluation of Renal Cell Carcinoma. J. Urol. 2007, 178, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Tsui, K.-H.; Shvarts, O.; Smith, R.B.; Figlin, R.A.; Dekernion, J.B.; Belldegrun, A. Prognostic indicators for renal cell carcinoma: A multivariate analysis of 643 patients using the revised 1997 tnm staging criteria. J. Urol. 2000, 163, 1090–1095. [Google Scholar] [CrossRef] [PubMed]
- Ficarra, V.; Righetti, R.; Pilloni, S.; D’amico, A.; Maffei, N.; Novella, G.; Zanolla, L.; Malossini, G.; Mobilio, G. Prognostic Factors in Patients with Renal Cell Carcinoma: Retrospective Analysis of 675 Cases. Eur. Urol. 2002, 41, 190–198. [Google Scholar] [CrossRef]
- Scopus Preview—Scopus—Document Details—Prognostic Significance of Morphologic Parameters in Renal Cell Carcinoma. Available online: https://www.scopus.com/record/display.uri?eid=2-s2.0-2642552183&origin=inward&txGid=18f4bff1afabc920febe75bb222fbbab (accessed on 18 January 2023).
- Prognostic Value of Nuclear Grade of Renal Cell Carcinoma. Available online: https://acsjournals.onlinelibrary.wiley.com/doi/epdf/10.1002/1097-0142(19951215)76:12%3C2543::AID-CNCR2820761221%3E3.0.CO;2-S?src=getftr (accessed on 18 January 2023).
- Bektas, S.; Bahadir, B.; Kandemir, N.O.; Barut, F.; Gul, A.E.; Ozdamar, S.O. Intraobserver and Interobserver Variability of Fuhrman and Modified Fuhrman Grading Systems for Conventional Renal Cell Carcinoma. Kaohsiung J. Med. Sci. 2009, 25, 596–600. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1016/S1607-551X(09)70562-5 (accessed on 18 January 2023). [CrossRef]
- Yeh, F.-C.; Parwani, A.V.; Pantanowitz, L.; Ho, C. Automated grading of renal cell carcinoma using whole slide imaging. J. Pathol. Inform. 2014, 5, 23. [Google Scholar] [CrossRef]
- Paner, G.P.; Stadler, W.M.; Hansel, D.E.; Montironi, R.; Lin, D.W.; Amin, M.B. Updates in the Eighth Edition of the Tumor-Node-Metastasis Staging Classification for Urologic Cancers. Eur. Urol. 2018, 73, 560–569. [Google Scholar] [CrossRef]
- Holdbrook, D.A.; Singh, M.; Choudhury, Y.; Kalaw, E.M.; Koh, V.; Tan, H.S.; Kanesvaran, R.; Tan, P.H.; Peng, J.Y.S.; Tan, M.-H.; et al. Automated Renal Cancer Grading Using Nuclear Pleomorphic Patterns. JCO Clin. Cancer Inform. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Qayyum, T.; McArdle, P.; Orange, C.; Seywright, M.; Horgan, P.; Oades, G.; Aitchison, M.; Edwards, J. Reclassification of the Fuhrman grading system in renal cell carcinoma-does it make a difference? SpringerPlus 2013, 2, 378. [Google Scholar] [CrossRef]
- Tian, K.; Rubadue, C.A.; Lin, D.; Veta, M.; Pyle, M.E.; Irshad, H.; Heng, Y.J. Automated clear cell renal carcinoma grade classification with prognostic significance. PLoS ONE 2019, 14, e0222641. [Google Scholar] [CrossRef]
- Song, J.; Xiao, L.; Lian, Z. Contour-Seed Pairs Learning-Based Framework for Simultaneously Detecting and Segmenting Various Overlapping Cells/Nuclei in Microscopy Images. IEEE Trans. Image Process. 2018, 27, 5759–5774. [Google Scholar] [CrossRef]
- Arjumand, W.; Sultana, S. Role of VHL gene mutation in human renal cell carcinoma. Tumor Biol. 2012, 33, 9–16. Available online: https://link.springer.com/article/10.1007/s13277-011-0257-3 (accessed on 18 January 2023). [CrossRef] [PubMed]
- Nogueira, M.; Kim, H.L. Molecular markers for predicting prognosis of renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2008, 26, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Roussel, E.; Beuselinck, B.; Albersen, M. Tailoring treatment in metastatic renal cell carcinoma. Nat. Rev. Urol. 2022, 19, 455–456. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, T.; Lee, C.-H.; Hsieh, J.J. A systematic review of predictive and prognostic biomarkers for VEGF-targeted therapy in renal cell carcinoma. Cancer Treat. Rev. 2014, 40, 533–547. [Google Scholar] [CrossRef]
- Rodriguez-Vida, A.; Strijbos, M.; Hutson, T.X. Predictive and prognostic biomarkers of targeted agents and modern immunotherapy in renal cell carcinoma. ESMO Open 2016, 1, e000013. [Google Scholar] [CrossRef]
- Motzer, R.J.; Robbins, P.B.; Powles, T.; Albiges, L.; Haanen, J.B.; Larkin, J.; Mu, X.J.; Ching, K.A.; Uemura, M.; Pal, S.K.; et al. Avelumab plus axitinib versus sunitinib in advanced renal cell carcinoma: Biomarker analysis of the phase 3 JAVELIN Renal 101 trial. Nat. Med. 2020, 26, 1733–1741. [Google Scholar] [CrossRef]
- Schimmel, H.; Zegers, I.; Emons, H. Standardization of protein biomarker measurements: Is it feasible? Scand. J. Clin. Lab. Investig. 2010, 70, 27–33. [Google Scholar] [CrossRef]
- Mayeux, R. Biomarkers: Potential uses and limitations. NeuroRx 2004, 1, 182–188. [Google Scholar] [CrossRef]
- Singh, N.P.; Bapi, R.S.; Vinod, P. Machine learning models to predict the progression from early to late stages of papillary renal cell carcinoma. Comput. Biol. Med. 2018, 100, 92–99. [Google Scholar] [CrossRef]
- Bhalla, S.; Chaudhary, K.; Kumar, R.; Sehgal, M.; Kaur, H.; Sharma, S.; Raghava, G.P.S. Gene expression-based biomarkers for discriminating early and late stage of clear cell renal cancer. Sci. Rep. 2017, 7, srep44997. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, F.G.; Silveira, H.C.S.; Júnior, J.N.A.; da Silveira, R.A.; Zucca, L.E.; Cárcano, F.M.; Sanches, A.O.N.; Neder, L.; Scapulatempo-Neto, C.; Serrano, S.V.; et al. Somatic Copy Number Alterations and Associated Genes in Clear-Cell Renal-Cell Carcinoma in Brazilian Patients. Int. J. Mol. Sci. 2021, 22, 2265. [Google Scholar] [CrossRef] [PubMed]
- D’Avella, C.; Abbosh, P.; Pal, S.K.; Geynisman, D.M. Mutations in renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2018, 38, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Farrukh, M.; Ali, M.A.; Naveed, M.; Habib, R.; Khan, H.; Kashif, T.; Zubair, H.; Saeed, M.; Butt, S.K.; Niaz, R.; et al. Efficacy and Safety of Checkpoint Inhibitors in Clear Cell Renal Cell Carcinoma: A Systematic Review of Clinical Trials. Hematol. Oncol. Stem. Cell Ther. 2023, 16, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Go, H.; Kang, M.J.; Kim, P.-J.; Lee, J.-L.; Park, J.Y.; Park, J.-M.; Ro, J.Y.; Cho, Y.M. Development of Response Classifier for Vascular Endothelial Growth Factor Receptor (VEGFR)-Tyrosine Kinase Inhibitor (TKI) in Metastatic Renal Cell Carcinoma. Pathol. Oncol. Res. 2017, 25, 51–58. [Google Scholar] [CrossRef]
- Padmanabhan, R.K.; Somasundar, V.H.; Griffith, S.D.; Zhu, J.; Samoyedny, D.; Tan, K.S.; Hu, J.; Liao, X.; Carin, L.; Yoon, S.S.; et al. An Active Learning Approach for Rapid Characterization of Endothelial Cells in Human Tumors. PLoS ONE 2014, 9, e90495. [Google Scholar] [CrossRef]
- Ing, N.; Huang, F.; Conley, A.; You, S.; Ma, Z.; Klimov, S.; Ohe, C.; Yuan, X.; Amin, M.B.; Figlin, R.; et al. A novel machine learning approach reveals latent vascular phenotypes predictive of renal cancer outcome. Sci. Rep. 2017, 7, 13190. [Google Scholar] [CrossRef]
- Herman, J.G.; Latif, F.; Weng, Y.; Lerman, M.I.; Zbar, B.; Liu, S.; Samid, D.; Duan, D.S.; Gnarra, J.R.; Linehan, W.M. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl. Acad. Sci. USA 1994, 91, 9700–9704. [Google Scholar] [CrossRef]
- Yamana, K.; Ohashi, R.; Tomita, Y. Contemporary Drug Therapy for Renal Cell Carcinoma—Evidence Accumulation and Histological Implications in Treatment Strategy. Biomedicines 2022, 10, 2840. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, J.; Kong, W.; Huang, J.; Dong, B.; Huang, Y.; Xue, W.; Zhang, J. LSD1 inhibition suppresses the growth of clear cell renal cell carcinoma via upregulating P21 signaling. Acta Pharm. Sin. B 2018, 9, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, H.; Chen, Z.; Jiang, H.; Liao, L.; Fan, S.; Xing, J.; Xie, Y.; Chen, S.; Ding, H.; et al. Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma. Oncogenesis 2018, 7, 83. [Google Scholar] [CrossRef]
- Joosten, S.C.; Smits, K.M.; Aarts, M.J.; Melotte, V.; Koch, A.; Tjan-Heijnen, V.C.; Van Engeland, M. Epigenetics in renal cell cancer: Mechanisms and clinical applications. Nat. Rev. Urol. 2018, 15, 430–451. [Google Scholar] [CrossRef]
- Zheng, H.; Momeni, A.; Cedoz, P.-L.; Vogel, H.; Gevaert, O. Whole slide images reflect DNA methylation patterns of human tumors. npj Genom. Med. 2020, 5, 11. [Google Scholar] [CrossRef]
- Singh, N.P.; Vinod, P.K. Integrative analysis of DNA methylation and gene expression in papillary renal cell carcinoma. Mol. Genet. Genom. 2020, 295, 807–824. [Google Scholar] [CrossRef] [PubMed]
- Guida, A.; Le Teuff, G.; Alves, C.; Colomba, E.; Di Nunno, V.; Derosa, L.; Flippot, R.; Escudier, B.; Albiges, L. Identification of international metastatic renal cell carcinoma database consortium (IMDC) intermediate-risk subgroups in patients with metastatic clear-cell renal cell carcinoma. Oncotarget 2020, 11, 4582–4592. [Google Scholar] [CrossRef] [PubMed]
- Zigeuner, R.; Hutterer, G.; Chromecki, T.; Imamovic, A.; Kampel-Kettner, K.; Rehak, P.; Langner, C.; Pummer, K. External validation of the Mayo Clinic stage, size, grade, and necrosis (SSIGN) score for clear-cell renal cell carcinoma in a single European centre applying routine pathology. Eur. Urol. 2010, 57, 102–111. [Google Scholar] [CrossRef]
- Prediction of Progression after Radical Nephrectomy for Patients with Clear Cell Renal Cell Carcinoma: A Stratification Tool for Prospective Clinical Trials—PubMed n.d. Available online: https://pubmed.ncbi.nlm.nih.gov/12655523/ (accessed on 5 March 2023).
- Zisman, A.; Pantuck, A.J.; Dorey, F.; Said, J.W.; Shvarts, O.; Quintana, D.; Gitlitz, B.J.; Dekernion, J.B.; Figlin, R.A.; Belldegrun, A.S. Improved prognostication of renal cell carcinoma using an integrated staging system. J. Clin. Oncol. 2001, 19, 1649–1657. [Google Scholar] [CrossRef]
- Lubbock, A.L.R.; Stewart, G.D.; O’mahony, F.C.; Laird, A.; Mullen, P.; O’donnell, M.; Powles, T.; Harrison, D.J.; Overton, I.M. Overcoming intratumoural heterogeneity for reproducible molecular risk stratification: A case study in advanced kidney cancer. BMC Med. 2017, 15, 118. [Google Scholar] [CrossRef]
- Heng, D.Y.; Xie, W.; Regan, M.M.; Harshman, L.C.; Bjarnason, G.A.; Vaishampayan, U.N.; Mackenzie, M.; Wood, L.; Donskov, F.; Tan, M.-H.; et al. External validation and comparison with other models of the International Metastatic Renal-Cell Carcinoma Database Consortium prognostic model: A population-based study. Lancet Oncol. 2013, 14, 141–148. [Google Scholar] [CrossRef]
- Erdem, S.; Capitanio, U.; Campi, R.; Mir, M.C.; Roussel, E.; Pavan, N.; Kara, O.; Klatte, T.; Kriegmair, M.C.; Degirmenci, E.; et al. External validation of the VENUSS prognostic model to predict recurrence after surgery in non-metastatic papillary renal cell carcinoma: A multi-institutional analysis. Urol. Oncol. Semin. Orig. Investig. 2022, 40, 198.e9–198.e17. [Google Scholar] [CrossRef]
- Di Nunno, V.; Mollica, V.; Schiavina, R.; Nobili, E.; Fiorentino, M.; Brunocilla, E.; Ardizzoni, A.; Massari, F. Improving IMDC Prognostic Prediction Through Evaluation of Initial Site of Metastasis in Patients With Metastatic Renal Cell Carcinoma. Clin. Genitourin. Cancer 2020, 18, e83–e90. [Google Scholar] [CrossRef]
- Huang, S.-C.; Pareek, A.; Seyyedi, S.; Banerjee, I.; Lungren, M.P. Fusion of medical imaging and electronic health records using deep learning: A systematic review and implementation guidelines. npj Digit. Med. 2020, 3, 136. [Google Scholar] [CrossRef] [PubMed]
- Wessels, F.; Schmitt, M.; Krieghoff-Henning, E.; Kather, J.N.; Nientiedt, M.; Kriegmair, M.C.; Worst, T.S.; Neuberger, M.; Steeg, M.; Popovic, Z.V.; et al. Deep learning can predict survival directly from histology in clear cell renal cell carcinoma. PLoS ONE 2022, 17, e0272656. [Google Scholar] [CrossRef]
- Chen, S.; Jiang, L.; Gao, F.; Zhang, E.; Wang, T.; Zhang, N.; Wang, X.; Zheng, J. Machine learning-based pathomics signature could act as a novel prognostic marker for patients with clear cell renal cell carcinoma. Br. J. Cancer 2021, 126, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhang, J.; Han, Y.; Wang, X.; Ye, X.; Meng, Y.; Parwani, A.; Han, Z.; Feng, Q.; Huang, K. Integrative Analysis of Histopathological Images and Genomic Data Predicts Clear Cell Renal Cell Carcinoma Prognosis. Cancer Res. 2017, 77, e91–e100. [Google Scholar] [CrossRef]
- Ning, Z.; Pan, W.; Chen, Y.; Xiao, Q.; Zhang, X.; Luo, J.; Wang, J.; Zhang, Y. Integrative analysis of cross-modal features for the prognosis prediction of clear cell renal cell carcinoma. Bioinformatics 2020, 36, 2888–2895. [Google Scholar] [CrossRef]
- Schulz, S.; Woerl, A.-C.; Jungmann, F.; Glasner, C.; Stenzel, P.; Strobl, S.; Fernandez, A.; Wagner, D.-C.; Haferkamp, A.; Mildenberger, P.; et al. Multimodal Deep Learning for Prognosis Prediction in Renal Cancer. Front. Oncol. 2021, 11, 788740. [Google Scholar] [CrossRef] [PubMed]
- Khene, Z.; Kutikov, A.; Campi, R.; the EAU-YAU Renal Cancer Working Group. Machine learning in renal cell carcinoma research: The promise and pitfalls of ‘renal-izing’ the potential of artificial intelligence. BJU Int. 2023. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Carbonara, U.; Campi, R. Re: Criteria for the Translation of Radiomics into Clinically Useful Tests. Eur. Urol. 2023, 84, 142–143. [Google Scholar] [CrossRef]
- Shortliffe, E.H.; Sepúlveda, M.J. Clinical Decision Support in the Era of Artificial Intelligence. JAMA 2018, 320, 2199–2200. [Google Scholar] [CrossRef] [PubMed]
- Durán, J.M.; Jongsma, K.R. Who is afraid of black box algorithms? On the epistemological and ethical basis of trust in medical AI. J. Med. Ethic 2021, 47, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Teo, Y.Y.; Danilevsky, A.; Shomron, N. Overcoming Interpretability in Deep Learning Cancer Classification. Methods Mol. Biol. 2021, 2243, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Moore, T.; Wong, W.-K.; Stumpf, S.; Oberst, I.; McIntosh, K.; Burnett, M. End-user feature labeling: Supervised and semi-supervised approaches based on locally-weighted logistic regression. Artif. Intell. 2013, 204, 56–74. [Google Scholar] [CrossRef]
- Krzywinski, M.; Altman, N. Points of significance: Power and sample size. Nat. Methods 2013, 10, 1139–1140. [Google Scholar] [CrossRef]
- Button, K.S.; Ioannidis, J.P.A.; Mokrysz, C.; Nosek, B.A.; Flint, J.; Robinson, E.S.J.; Munafò, M.R. Power failure: Why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013, 14, 365–376. [Google Scholar] [CrossRef]
- Wang, L. Heterogeneous Data and Big Data Analytics. Autom. Control Inf. Sci. 2017, 3, 8–15. [Google Scholar] [CrossRef]
- Borodinov, N.; Neumayer, S.; Kalinin, S.V.; Ovchinnikova, O.S.; Vasudevan, R.K.; Jesse, S. Deep neural networks for understanding noisy data applied to physical property extraction in scanning probe microscopy. npj Comput. Mater. 2019, 5, 25. [Google Scholar] [CrossRef]
- Bin Goh, W.W.; Wong, L. Dealing with Confounders in Omics Analysis. Trends Biotechnol. 2018, 36, 488–498. [Google Scholar] [CrossRef]
- Tougui, I.; Jilbab, A.; El Mhamdi, J. Impact of the Choice of Cross-Validation Techniques on the Results of Machine Learning-Based Diagnostic Applications. Health Inform. Res. 2021, 27, 189–199. [Google Scholar] [CrossRef]
- Veta, M.; Heng, Y.J.; Stathonikos, N.; Bejnordi, B.E.; Beca, F.; Wollmann, T.; Rohr, K.; Shah, M.A.; Wang, D.; Rousson, M.; et al. Predicting breast tumor proliferation from whole-slide images: The TUPAC16 challenge. Med. Image Anal. 2019, 54, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Sirinukunwattana, K.; Pluim, J.P.; Chen, H.; Qi, X.; Heng, P.-A.; Guo, Y.B.; Wang, L.Y.; Matuszewski, B.J.; Bruni, E.; Sanchez, U.; et al. Gland segmentation in colon histology images: The glas challenge contest. Med. Image Anal. 2017, 35, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y. Color standardization and optimization in Whole Slide Imaging. Diagn. Pathol. 2011, 6, S15. [Google Scholar] [CrossRef]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.-M.; Ciompi, F.; van der Laak, J. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.Y.; Phua, K.; Wong, L.; Bin Goh, W.W. Extensions of the External Validation for Checking Learned Model Interpretability and Generalizability. Patterns 2020, 1, 100129. [Google Scholar] [CrossRef] [PubMed]
- Mühlbauer, J.; Egen, L.; Kowalewski, K.-F.; Grilli, M.; Walach, M.T.; Westhoff, N.; Nuhn, P.; Laqua, F.C.; Baessler, B.; Kriegmair, M.C. Radiomics in Renal Cell Carcinoma—A Systematic Review and Meta-Analysis. Cancers 2021, 13, 1348. [Google Scholar] [CrossRef]
- Lu, C.; Shiradkar, R.; Liu, Z. Integrating pathomics with radiomics and genomics for cancer prognosis: A brief review. Chin. J. Cancer Res. 2021, 33, 563–573. [Google Scholar] [CrossRef]
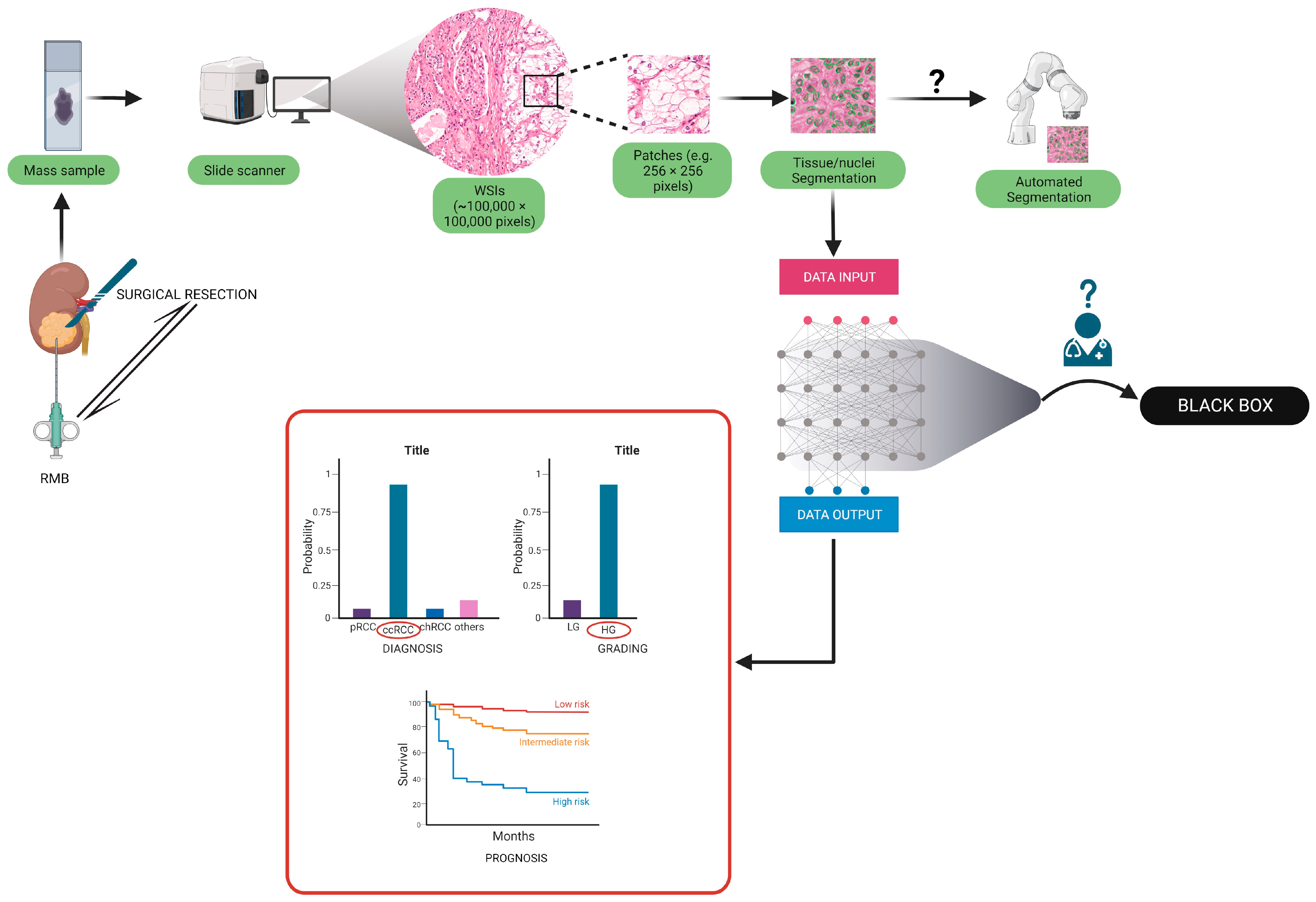
| Group | Aim | Number of Patients | Training Process/Methodologies | Accuracy on the Test Set | External Validation (N of Patients) | Accuracy on the External Validation Cohort | Algorithm |
|---|---|---|---|---|---|---|---|
| Yeh et al. [84] | RCC grading | 39 ccRCC | Pixels from the nuclei were manually selected to further train a SVM classifier to recognize nuclei. A person with no special training in pathology engaged in training the classifier with an interactive interface. | AUC: 0.97 | N.A | N.A. | WSI analysis with an automatic stain recognition algorithm. An SVM classifier was trained to recognize nuclei. Sizes of the recognized nuclei were estimated, and the spatial distribution of nuclear size was calculated using Kernel regression. |
| Holdbrook et al. [86] | (1) RCC grading; (2) survival prediction. | 59 ccRCC | A cascade detector of prominent nucleoli (constructed by stacking 20 classifiers sequentially) was trained with WSI images to extract image patches for subsequent analysis. This pipeline used two nucleoli detectors to extract prominent nucleoli image patches. | (1) F-score: 0.78–0.83 grade prediction; (2) High degree of correlation (R = 0.59) with a multigene score. | N.A. | N.A. | An automated image classification pipeline was used to detect and analyze prominent nucleoli in WSIs and classify them as either low or high grade. The pipeline employed ML and image pixel intensity-based feature extraction methods for nuclear analysis. Multiple classification systems were used for patch classification (SVM, logistic regression and AdaBoost). |
| Tian et al. [88] | (1) RCC grading, (2) survival prediction | 395 ccRCC | Seven ML classification methods were used to categorize grades based on nuclei histomics features were evaluated. Among these methods, LASSO regression demonstrated the highest performance with a built-in feature selection capability. LASSO regression and its optimal hyper parameter selected the final list of histomics features most associated with grade. | (1) 84.6% sensitivity and 81.3% specificity grade prediction; (2) predicted grade associated with overall survival (HR: 2.05; 95% CI 1.21–3.47). | N.A. | N.A. | Nuclear segmentation occurred, and 72 features were extracted. Features associated with grade were identified via a LASSO model using data from cases with concordancet between TCGA and Pathologist 1. Discordant cases were additionally reviewed by Pathologist 2. Prognostic efficacy of the predicted grades was evaluated using a Cox proportional hazard model in an extended test set created by combining the test set and discordant cases. |
| Group | Aim | Number of Patients | Training Process/Methodologies | Accuracy on the Test Set | External Validation (N of Patients) | Accuracy on the External Validation Cohort | Algorithm |
|---|---|---|---|---|---|---|---|
| Marostica et al. [68] | (1) RCC diagnosis; (2) RCC subtyping; (3) CNAs identification; (4) RCC survival prediction; (5) Tumor mutation burden prediction. | (1) and (2): 537 ccRCC, 288 pRCC, and 103 chRCC; (3) 528 ccRCC, 288 pRCC, and 66 chRCC; (4) 269 stage I ccRCC; (5) 302 ccRCC. | (1) Weak supervision approach used for malignant region identification; (2) Same transfer learning approach trained for 15 epochs; (3) Independent models for ccRCC, pRCC, and chRCC were developed; (4) 10-fold cross-validation was employed. Upsampling of uncensored data points was performed in each fold’s training set to enhance the model training process. | (1)AUC: 0.990 ccRCC, 1.00 pRCC, 0.9998 chRCC; (2) AUC: 0.953 (3) ccRCC KRAS CNA: AUC = 0.724, pRCC somatic mutations: AUC: 0.419–0.684; (4) Short vs. long-term survivors log-rank test P = 0.02, n = 269; (5) Spearman’s correlation coefficient: 0.419 | (1) and (2) 841 ccRCC, 41 pRCC, and 31 chRCC. | (1) 0.964–0.985 ccRCC; (2) 0.782–0.993 | (1) Three DCNN architectures (VGG-16, Inception-v3, and ResNet-50) were compared for each task. (2) Same transfer learning approach as above was used. The hyperparameters of DCNNs were optimized via Talos. (3) Two transfer learning approaches were used: gene-specific binary classification and multi-task classification for all genes for CNAs. DCNNs were used for associations between genetic mutations and WSI images. (4) DCNN models used image patches as inputs, predicting binary values for each patient. Grad-CAM was generated to identify the regions of greatest importance for survival prediction. |
| Go et al. [104] | RCC VEGFR-TKI response classifier; survival prediction. | 101 m-ccRCC | ML approaches were applied to establish a predictive classifying model for VEGFR-TKI response. A 10-fold-cross-validated SVM method and decision tree analysis were used for modeling | Apparent accuracy of the model: 87.5%; C-index = 0.7001 for PFS; C-index of 0.6552 for OS | N.A. | N.A. | Features that showed the statistical differences between the good and bad-response groups were selected, and the most appropriate cut-off for each feature was calculated. Secondary feature selection was performed using SVM to develop the most efficient model, i.e., the model showing the highest accuracy with the least number of features |
| Ing et al. [106] | (1) RCC vascular phenotypes; (2) survival prediction; (3) identification of prognostic gene signature; (4) prediction models. | (1), (2), and (3): 64 ccRCC; (4) 301 ccRCC. | A stochastic backwards feature selection method with 1500 iterations was applied to identify the subset of VF with the highest predictive power. Two GLMNET models were trained: one model was trained on VF-risk groups, and the other model was trained using a 24-month disease-free status as the ground truth for a validation cohort. | (1) AUC = 0.79; (2) log-rank p = 0.019, HR = 2.4; (3) Wilcoxon rank-sum test p < 0.0511; (4) C-Index: Stage = 0.7, Stage + 14VF = 0.74, Stage + 14GT = 0.74. | N.A. | N.A. | Quantitative analysis of tumor vasculature and developement of a gene signature. The algorithms trained in this framework classified with SVM and random forest classifiers, i.e., endothelial cells, and generated a VAM within a WSI. By quantifying the VAMs, nine VFs were identified, which showed a predictive value for DFS in a discovery cohort. Correlation analysis showed that a 14-gene expression signature related to the 9VF was discovered. The two GLMNET were developed based on these 14 genes, separating independent cohorts into groups with good or poor DFS, which were assessed via Kaplan–Meier plots. |
| Zheng et al. [112] | RCC methylation profile | 326 RCC (also tested on glioma) | In total, 30 sets of training/testing data were generated. Binary classifiers were fitted on the training set, and the best parameters were selected using 5-fold cross-validation. Logistic regression with LASSO regularization, random forest, SVM, Adaboost, Naive Bayes, and a two-layer FCNN were used with optimized parameters. | Average AUC and F1 score higher than 0.6 | N.A. | N.A. | To demonstrate that DNA methylation can be predicted based on morphometric features, different classical ML models were tested. Binary classifiers for each task were evaluated using accuracy, precision, recall, F1-score, ROC curve, AUC score, and precision–recall curves. Scores from 30 training/testing data sets were averaged per task. For logistic regression, feature importance analysis was conducted to rank the influence of morphometric features on the prediction task. |
| Group | Aim | Number of Patients | Training Process/Methodologies | Accuracy on the Test Set | External Validation (N of Patients) | Accuracy on the External Validation Cohort | Algorithm |
|---|---|---|---|---|---|---|---|
| Ning et al. [126] | RCC prognosis prediction | 209 ccRCC | The training procedures employed 10-fold cross-validation. Survival distributions of low- and high-risk groups were estimated using the Kaplan–Meier estimator and compared via the log-rank test. The performance of prognostic prediction was assessed using the C-index. | Mean C-index = 0.832 (0.761–0.903) | N.A | N.A. | Two CNNs with identical structures were employed to extract deep features from CT and histopathological images. Histological patches were carefully reviewed by two pathologists to confirm coverage of tumor cells. Global pooling and fully connected layers were utilized at the end of the network to integrate information from all feature maps and make predictions. The BFPS algorithm was employed for feature selection. |
| Cheng et al. [125] | RCC prognosis prediction | 410 ccRCC | A two-level cross-validation strategy was used to validate our method. In the first level, a single patient was chosen as the test set, with the rest used as training sets. The second level was a 10-fold cross-validation performed in the training set to select the best regularization parameter. A regularized Cox proportional hazards model was built on the training set using the selected parameter and based on the model; risk indices of all patients were also calculated. | Log-rank test p values < 0.05 | N.A. | N.A. | The unsupervised segmentation method for cell nuclei and features extraction was used. lmQCM was used to perform gene coexpression network analysis. The LASSO-Cox model for prognosis prediction calculated the risk index for each patient based on their cellular morphologic features and eigengenes |
| Schulz et al. [127] | RCC prognosis prediction | 248 ccRCC | Unimodal training was conducted. This method was followed by multimodal training, which used the pre-trained weights from unimodal training. Training lasted for 200–400 epochs, and the best model was selected based on the convergence of training and validation curves. The standard Cox loss function was employed for survival analysis, while the cross-entropy loss function was used for binary classification tasks. | A mean C-index of 0.7791 and a mean accuracy of 83.43%. (prognosis prediction) | 18 ccRCC | Mean C-index reached 0.799 ± 0.060 with a maximum of 0.8662. The accuracy averaged at 79.17% ± 9.8% with a maximum of 94.44%. | CNN consisting of one individual 18-layer residual network (ResNet) per image modality (histopathology slides, CT scans, MR scans) and a dense layer for genomic data. The network outputs were then combined using an attention layer, which assigned weights to each output based on its relevance to the task at hand. The combined outputs were passed through a fully connected network. Depending on the specific case, either C-index calculation or binary classification for 5YSS was performed. The 5YSS category included patients who either survived for longer than 60 months or passed away within five years of diagnosis. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Distante, A.; Marandino, L.; Bertolo, R.; Ingels, A.; Pavan, N.; Pecoraro, A.; Marchioni, M.; Carbonara, U.; Erdem, S.; Amparore, D.; et al. Artificial Intelligence in Renal Cell Carcinoma Histopathology: Current Applications and Future Perspectives. Diagnostics 2023, 13, 2294. https://doi.org/10.3390/diagnostics13132294
Distante A, Marandino L, Bertolo R, Ingels A, Pavan N, Pecoraro A, Marchioni M, Carbonara U, Erdem S, Amparore D, et al. Artificial Intelligence in Renal Cell Carcinoma Histopathology: Current Applications and Future Perspectives. Diagnostics. 2023; 13(13):2294. https://doi.org/10.3390/diagnostics13132294
Chicago/Turabian StyleDistante, Alfredo, Laura Marandino, Riccardo Bertolo, Alexandre Ingels, Nicola Pavan, Angela Pecoraro, Michele Marchioni, Umberto Carbonara, Selcuk Erdem, Daniele Amparore, and et al. 2023. "Artificial Intelligence in Renal Cell Carcinoma Histopathology: Current Applications and Future Perspectives" Diagnostics 13, no. 13: 2294. https://doi.org/10.3390/diagnostics13132294
APA StyleDistante, A., Marandino, L., Bertolo, R., Ingels, A., Pavan, N., Pecoraro, A., Marchioni, M., Carbonara, U., Erdem, S., Amparore, D., Campi, R., Roussel, E., Caliò, A., Wu, Z., Palumbo, C., Borregales, L. D., Mulders, P., & Muselaers, C. H. J., on behalf of the EAU Young Academic Urologists (YAU) Renal Cancer Working Group. (2023). Artificial Intelligence in Renal Cell Carcinoma Histopathology: Current Applications and Future Perspectives. Diagnostics, 13(13), 2294. https://doi.org/10.3390/diagnostics13132294








