Asynchronous Federated Learning for Improved Cardiovascular Disease Prediction Using Artificial Intelligence
Abstract
1. Introduction
- Exploring synchronous and asynchronous communication;
- Providing a broad overview of distributed federated learning techniques;
- Proposed an asynchronous FL cardiac prediction (AFLCP);
- Finally, comparing the accuracy and importance of the loss function between synchronous and asynchronous federated learning.
2. Related Work
3. Materials and Method
- Accuracy: By assessing the percentage of cases that were properly classified out of all occurrences, accuracy measures the general correctness of the model’s predictions. In terms of accurately detecting both positive and negative cases, it gives a sign of how effectively the model performs.
- Precision: The ability of the model to properly identify positive instances among all instances predicted to be positive is measured by precision. Indicating the percentage of real positives among all instances anticipated as positive, it focuses on how accurate positive predictions are. When the cost of false positives is significant, precision is advantageous because it ensures a low rate of misclassifying negative instances as positive.
- F1-score: Harmonic mean of recall and precision is the F1-score. It provides a fair assessment of the model’s performance by combining precision and recall into a single statistic. When the dataset is unbalanced or when both false positives and false negatives need to be reduced, it is especially helpful. An increase in the F1-score from 0 to 1 indicates greater overall performance.
3.1. Dataset Descriptions
3.2. Proposed Method
Proposed Algorithm: Asynchronous FL Cardiac Prediction (AFLCP)
- Stap 1.
- E is initialized with the minimum loss threshold.
- Stap 2.
- Xg is initialized with the existing global model.
- Stap 3.
- It involves accessing the learning performance of the model, and at each iteration, the performance is compared with the loss threshold (E) that was established in an earlier step. Because of this, the entire learning process will continue to take place at the distant cloud server for as long as the current loss value is more than E.
- a.
- Aggregating local model parameter updates Xg and loss evaluation to ω.
- Stap 4.
- End of iterations.
- Stap 5.
- Xg value is updated.
- Stap 1.
- The originally trained AI model (Xzi) is applied to the local model of Z nodes.
- Stap 2.
- Initialization of the data size of the threshold to its default value in preparation for the subsequent phases.
- Stap 3.
- Initialization of timestep (γz) and block (Bz) to their default values in preparation for the subsequent phases.
- Stap 4.
- The new private data (Mz) are loaded, and the size of the data is determined by comparing it with the threshold (β) data size.
- Stap 5.
- The classification decision is computed using the R-SVM classifier.
- Stap 6.
- The iteration begins from 1 to L. L is the number of iterations that occur when the whole procedure is being carried out at nodes.
- i.
- If the condition [(ℓ mod Δ) = 0] is satisfied, then it will execute the body of an if statement.
- a.
- When the condition is met, which occurs after iteration, the time ℓ that was calculated is saved in the time-step (γz) list.
- b.
- The local weights (ωz) are extracted from the initial model together with α. Here, α denotes the deep parameter exchange ratio. α indicates the deep parameter ratio that is contributing to the deep exchange rate. The information on the iteration that takes place during the deep exchange with the cloud is stored inside the timestep γ parameter.
- c.
- The extracted weights (ωz), timestep (γz), and block (Bz) are transferred to the cloud server.
- ii.
- If the condition [(ℓ mod Δ) = 0] is not satisfied, then it will execute the body of the else statement.
- a.
- The shallow parameters of the (1 − α) ratio taken from the local model (Xzi) are stored in ωz.
- b.
- The extracted weights (ωz) are passed to the cloud server.
- iii.
- End of if–else statement.
- Stap 7.
- The local model of Z nodes is updated, which makes use of the aggregated model that was retrieved from the server.
- Stap 8.
- All the global model states and information pertaining to timestep ℓ are saved in the Bz so that they may be accessed later.
- Stap 9.
- The loop ends.
- Stap 10.
- Once the training has been completed for L iterations, the used data M are deleted from the cache in a permanent manner to improve the safety of the user’s data.
| Algorithm 1: Asynchronous Cardiac Prediction (ACP) |
| Input: Z, Xzi, Δ, L, α Output: Xzo Working at Client Node
Input: ω Output: Xg
|
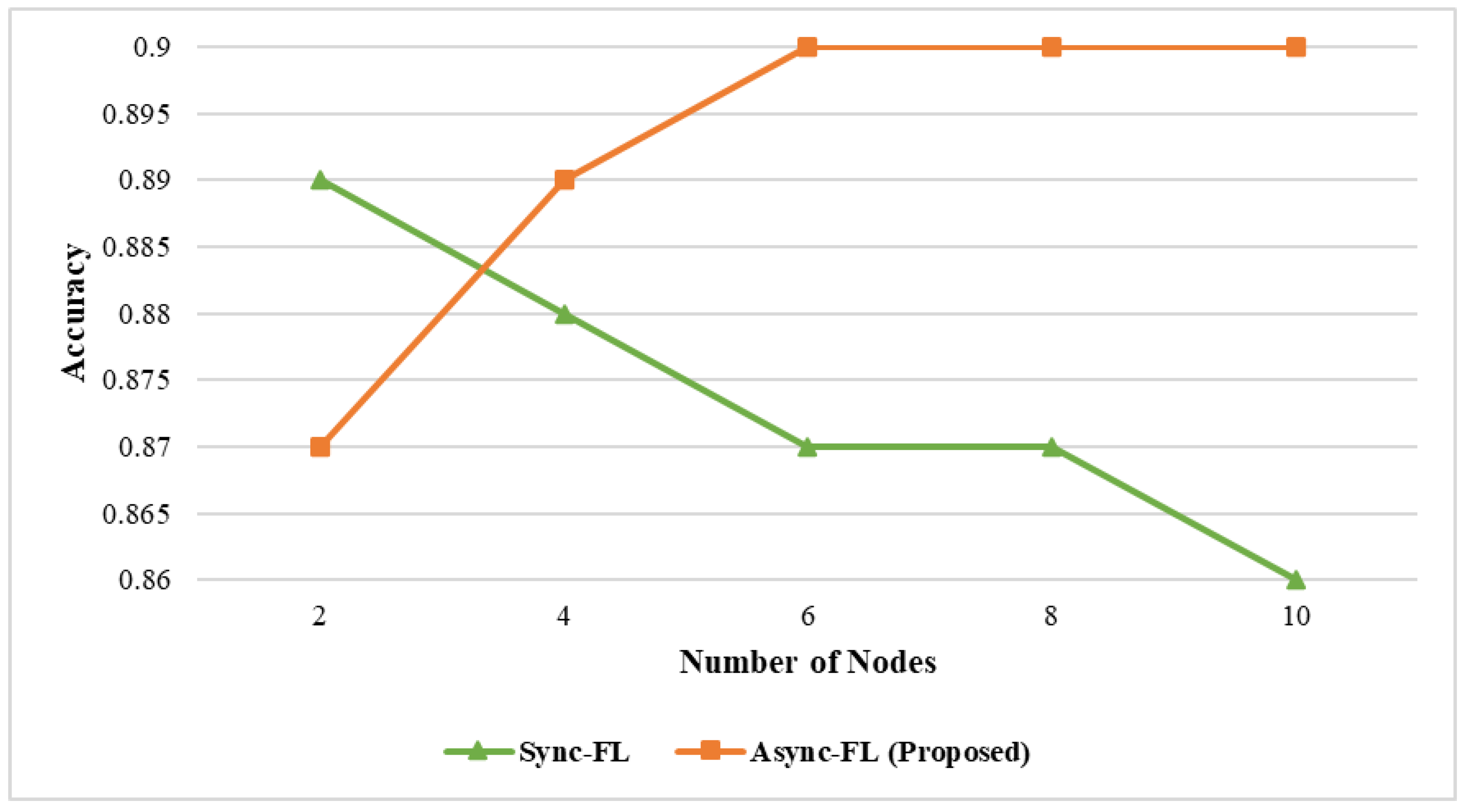
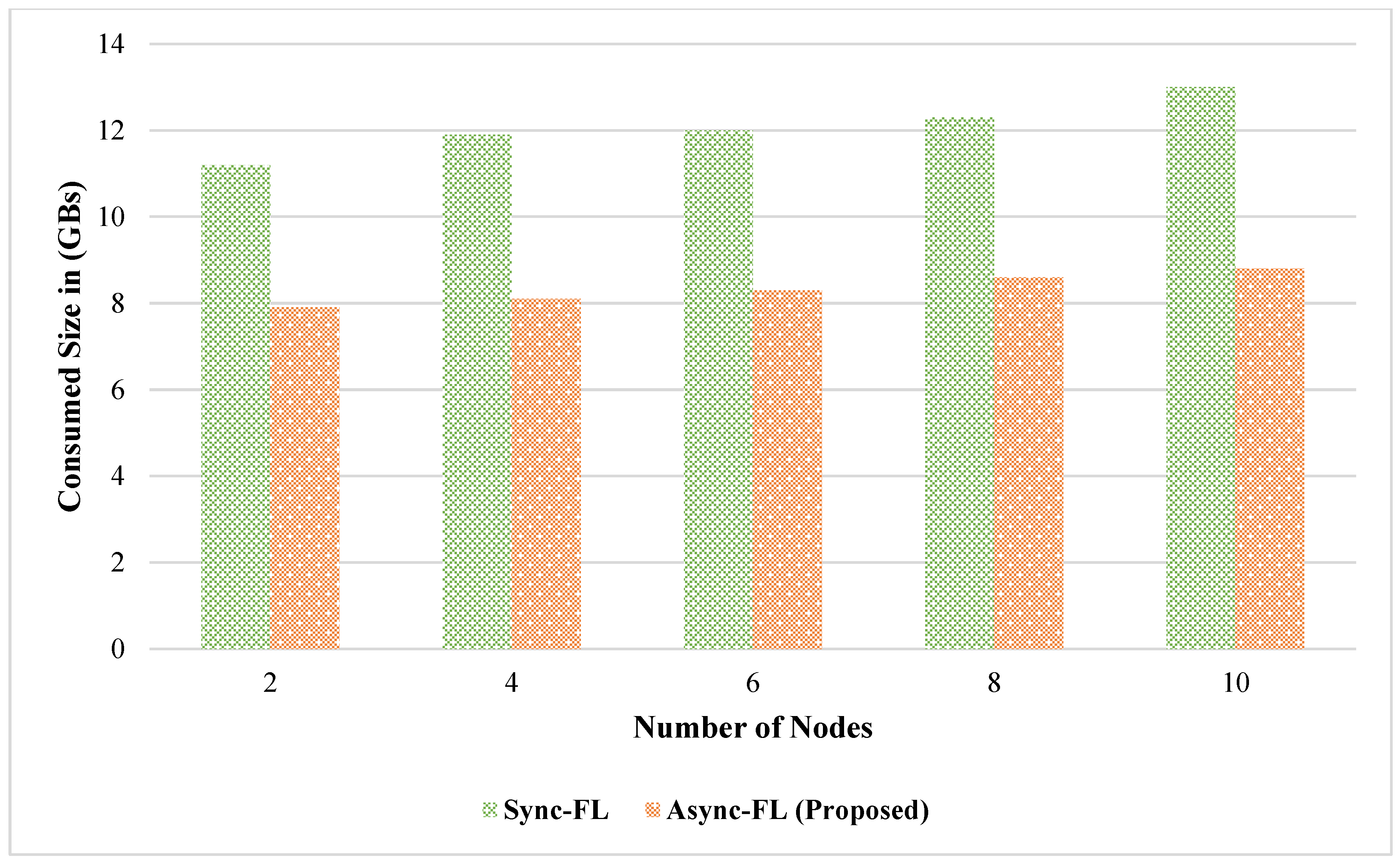
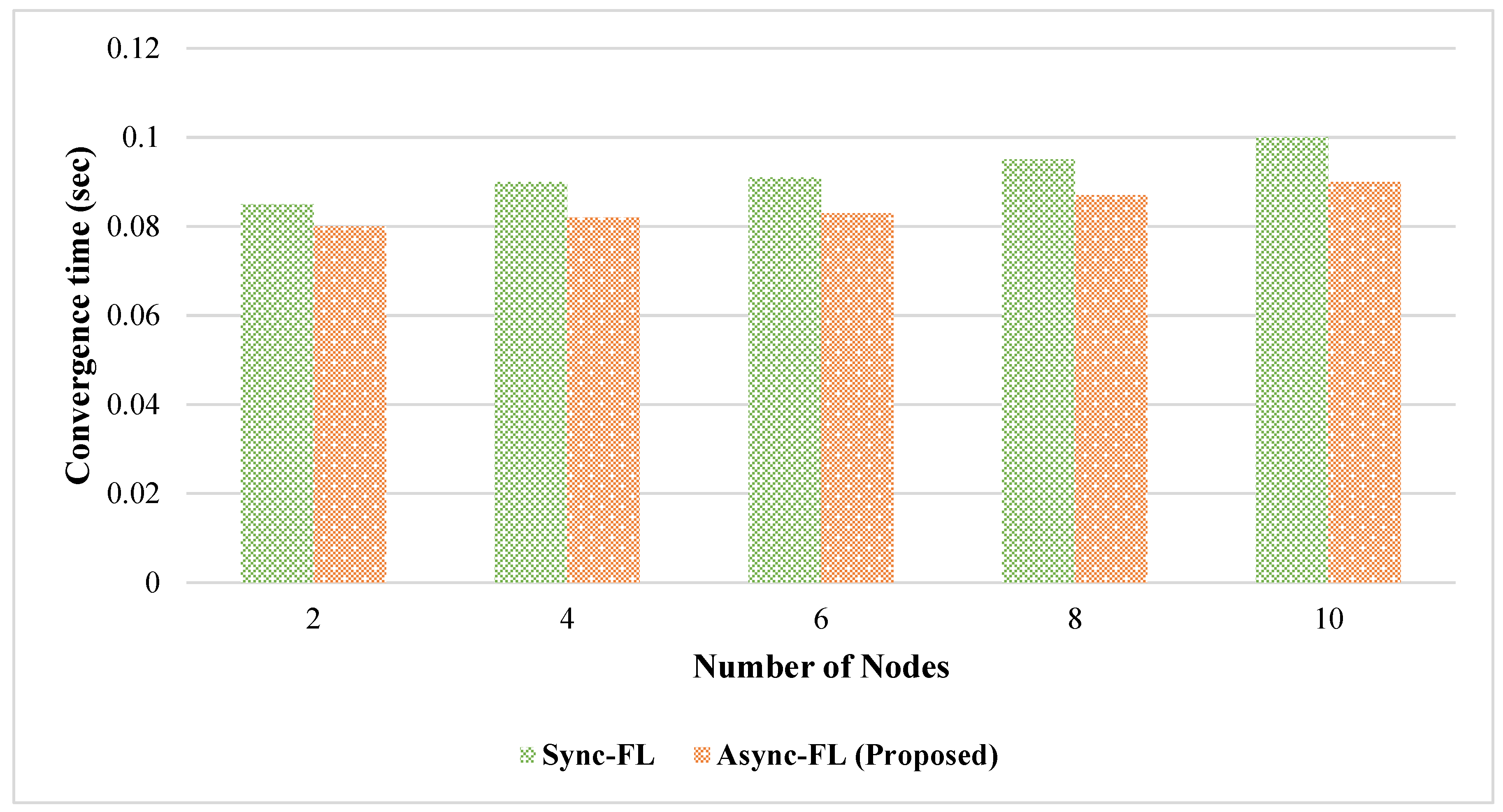
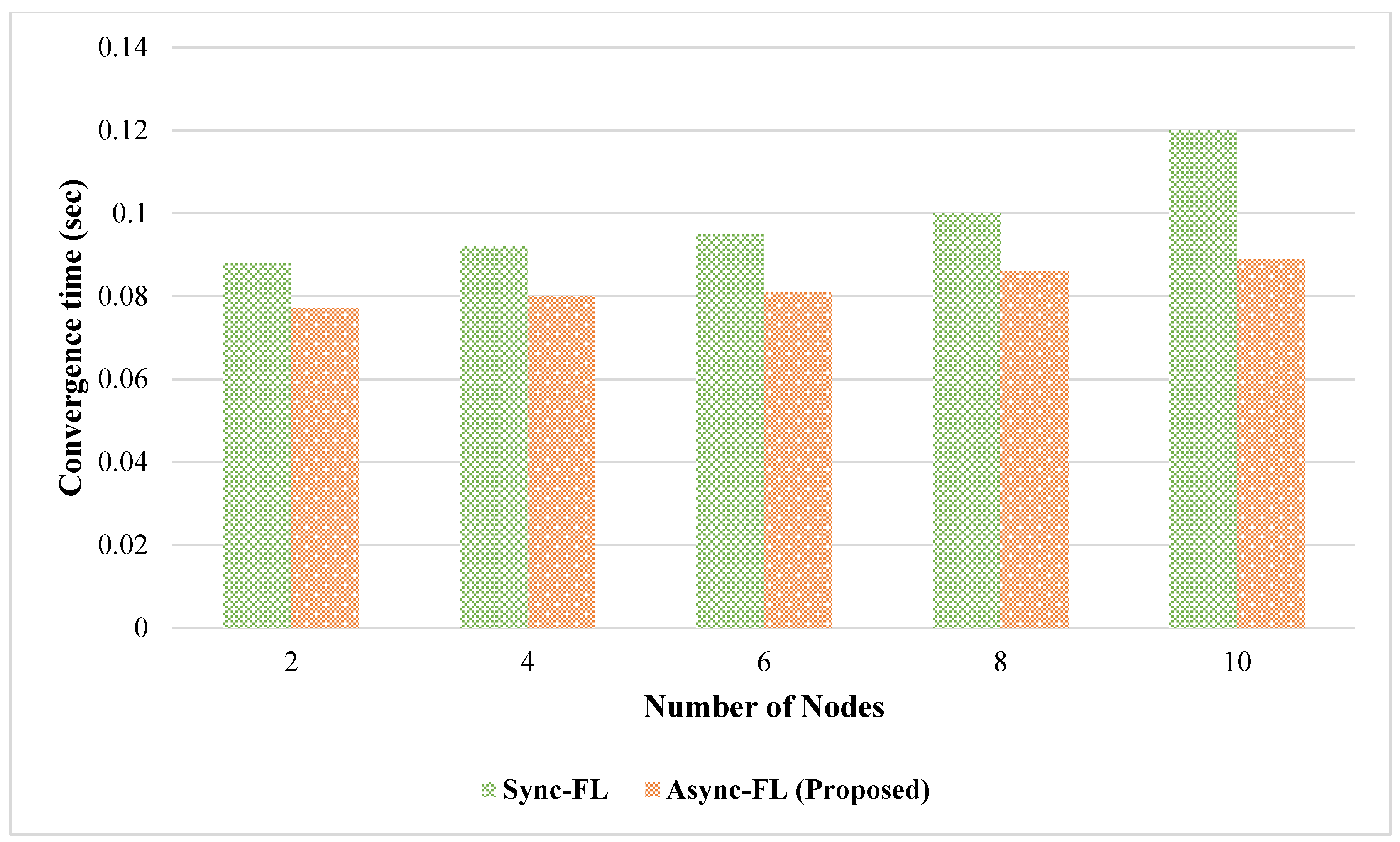
4. Experimental Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moore, W.; Frye, S.A. Review of HIPAA, Part 1: History, Protected Health Information, and Privacy and Security Rules. J. Nucl. Med. Technol. 2019, 47, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gu, Y.; Dvornek, N.; Staib, L.H.; Ventola, P.; Duncan, J.S. Multi-site fMRI analysis using privacy-preserving federated learning and domain adaptation: ABIDE results. Med. Image Anal. 2020, 65, 101765. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, X.; Jin, Y. Communication-Efficient Federated Deep Learning with Layerwise Asynchronous Model Update and Temporally Weighted Aggregation. IEEE Trans. Neural Netw. Learn. Syst. 2020, 31, 4229–4238. [Google Scholar] [CrossRef] [PubMed]
- McMahan, B.; Moore, E.; Ramage, D.; Hampson, S.; Arcas, B.A. Communication-Efficient Learning of Deep Networks from Decentralized Data. In Proceedings of the 20th International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 9–11 May 2017; pp. 1273–1282. [Google Scholar]
- Ramaswamy, S.; Mathews, R.; Rao, K.; Beaufays, F. Federated Learning for Emoji Prediction in a Mobile Keyboard. arXiv 2019, arXiv:1906.04329. [Google Scholar]
- Shyu, C.; Putra, K.T.; Chen, H.; Tsai, Y.; Hossain, K.S.M.T.; Jiang, W.; Shae, Z. A Systematic Review of Federated Learning in the Healthcare Area: From the Perspective of Data Properties and Applications. Appl. Sci. 2021, 11, 11191. [Google Scholar]
- Guruprasad, S.; Mathias, V.L.; Dcunha, W. Heart Disease Prediction Using Machine Learning Techniques. In Proceedings of the 5th International Conference on Electrical, Electronics, Communication, Computer Technologies and Optimization Techniques (ICEECCOT), Mysuru, India, 10–11 December 2021; pp. 762–766. [Google Scholar] [CrossRef]
- Veeramsetty, V.; Kumar, A.T.; Navya, B.; Bhavan, T.; Hrishikesh, Y. Heart disease prediction using machine learning algorithms. AIP Conf. Proc. 2022, 2418, 040013. [Google Scholar] [CrossRef]
- Konecný, J.; McMahan, B.; Ramage, D. Federated optimization: Distributed optimization beyond the datacenter. arXiv 2015, arXiv:1511.03575. [Google Scholar]
- Konecný, J.; McMahan, H.B.; Yu, F.X.; Richtárik, P.; Suresh, A.T.; Bacon, D. Federated learning: Strategies for improving communication efficiency. arXiv 2016, arXiv:1610.05492. [Google Scholar]
- Balaskas, K.; Siozios, K. ECG Analysis and Heartbeat Classification Based on Shallow Neural Networks. In Proceedings of the 8th International Conference on Modern Circuits and Systems Technologies (MOCAST), Thessaloniki, Greece, 13–15 May 2019. [Google Scholar]
- Hannun, A.; Rajpurkar, P.; Haghpanahi, M.; Geoffrey, T.; Bourn, C.; Turakhia, M.; Ng, A. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef]
- Lainscsek, C.; Rowat, P.; Schettino, L.; Lee, D.; Song, D.; Letellier, C.; Poizner, H. Finger tapping movements of Parkinson’s disease patients automatically rated using nonlinear delay differential equations. Chaos: Interdiscip. J. Nonlinear Sci. 2012, 22, 013119. [Google Scholar] [CrossRef]
- Sandeep, R.; Ray, K.C. Sparse representation of ECG signals for automated recognition of cardiac arrhythmias. Expert Syst. Appl. 2018, 105, 49–64. [Google Scholar]
- Dohare, A.K.; Kumar, V.; Kumar, R. Detection of myocardial infarction in 12 lead ECG using support vector machine. Appl. Soft Comput. 2018, 64, 138–147. [Google Scholar] [CrossRef]
- Azariadi, D.; Tsoutsouras, V.; Xydis, S.; Soudris, D. ECG signal analysis and arrhythmia detection on IoT wearable medical devices. In Proceedings of the 5th International Conference on Modern Circuits and Systems Technologies (MOCAST), Thessaloniki, Greece, 12–14 May 2016. [Google Scholar]
- Zhang, M.; Wang, Y.; Luo, T. Federated Learning for Arrhythmia Detection of Non-IID ECG. In Proceedings of the IEEE 6th International Conference on Computer and Communications (ICCC), Chengdu, China, 11–14 December 2020. [Google Scholar]
- Yaqoob, M.M.; Nazir, M.; Yousafzai, A.; Khan, M.A.; Shaikh, A.A.; Algarni, A.D.; Elmannai, H. Modified Artificial Bee Colony Based Feature Optimized Federated Learning for Heart Disease Diagnosis in Healthcare. Appl. Sci. 2022, 12, 12080. [Google Scholar] [CrossRef]
- Sakib, S.; Fouda, M.M.; Fadlullah, Z.M.; Abualsaud, K.; Yaacoub, E.; Guizani, M. Asynchronous Federated Learning-based ECG Analysis for Arrhythmia Detection. In Proceedings of the IEEE International Mediterranean Conference on Communications and Networking (MeditCom), Chengdu, China, 11–14 December 2021; pp. 277–282. [Google Scholar] [CrossRef]
- Lv, Z.; Yu, Z.; Xie, S.; Alamri, A. Deep Learning-based Smart Predictive Evaluation for Interactive Multimedia-enabled Smart Healthcare. ACM Trans. Multimed. Comput. Commun. Appl. 2022, 18, 43. [Google Scholar] [CrossRef]
- Lu, S.; Yang, B.; Xiao, Y.; Liu, S.; Liu, M.; Yin, L.; Zheng, W. Iterative reconstruction of low-dose CT based on differential sparse. Biomed. Signal Process. Control. 2023, 79, 104204. [Google Scholar] [CrossRef]
- Qin, X.; Ban, Y.; Wu, P.; Yang, B.; Liu, S.; Yin, L.; Liu, M.; Zheng, W. Improved Image Fusion Method Based on Sparse Decomposition. Electronics 2022, 11, 2321. [Google Scholar] [CrossRef]
- Liu, H.; Liu, M.; Li, D.; Zheng, W.; Yin, L.; Wang, R. Recent Advances in Pulse-Coupled Neural Networks with Applications in Image Processing. Electronics 2022, 11, 3264. [Google Scholar] [CrossRef]
- Arif, S.Z.; Yaqoob, M.M.; Rehman, A.; Jamil, F. Word sense disambiguation for Urdu text by machine learning. Int. J. Comput. Sci. Inf. Secur. 2016, 14, 738–757. [Google Scholar]
- Yaqoob, M.M.; Alsulami, M.; Khan, M.A.; Alsadie, D.; Saudagar, A.K.J.; AlKhathami, M. Federated Machine Learning for Skin Lesion Diagnosis: An Asynchronous and Weighted Approach. Diagnostics 2023, 13, 1964. [Google Scholar] [CrossRef]
- Yaqoob, M.M.; Khurshid, W.; Liu, L.; Arif, S.Z.; Khan, I.A.; Khalid, O.; Nawaz, R. Adaptive Multi-Cost Routing Protocol to Enhance Lifetime for Wireless Body Area Network. Comput. Mater. Contin. 2022, 72, 1089–1103. [Google Scholar] [CrossRef]
- Qadri, S.F.; Lin, H.; Shen, L.; Ahmad, M.; Qadri, S.; Khan, S.; Khan, M.; Zareen, S.S.; Akbar, M.A.; Bin Heyat, B.; et al. CT-Based Automatic Spine Segmentation Using Patch-Based Deep Learning. Int. J. Intell. Syst. 2023, 2023, 2345835. [Google Scholar] [CrossRef]
- Handelman, G.S.; Kok, H.K.; Chandra, R.V.; Razavi, A.H.; Huang, S.; Brooks, M.; Lee, M.J.; Asadi, H. Peering Into the Black Box of Artificial Intelligence: Evaluation Metrics of Machine Learning Methods. Am. J. Roentgenol. 2019, 212, 38–43. [Google Scholar] [CrossRef] [PubMed]
- IqbalMalik, K.; Yaqoob, M.M. An Analytical Survey on Routing Protocols for Wireless Sensor Network (WSN). Int. J. Comput. Appl. 2014, 975, 8887. [Google Scholar] [CrossRef]
- Kochi, A.N.; Tagliari, A.P.; Forleo, G.B.; Fassini, G.M.; Tondo, C. Cardiac and arrhythmic complications in patients with COVID-19. J. Cardiovasc. Electrophysiol. 2020, 31, 1003–1008. [Google Scholar] [CrossRef]
- Sakib, S.; Fouda, M.M.; Fadlullah, Z.M.; Nasser, N.; Alasmary, W. A Proof-of-Concept of Ultra-Edge Smart IoT Sensor: A Continuous and Lightweight Arrhythmia Monitoring Approach. IEEE Access 2021, 9, 26093–26106. [Google Scholar] [CrossRef]
- Yaqoob, M.M.; Nazir, M.; Khan, M.A.; Qureshi, S.; Al-Rasheed, A. Hybrid Classifier-Based Federated Learning in Health Service Providers for Cardiovascular Disease Prediction. Appl. Sci. 2023, 13, 1911. [Google Scholar] [CrossRef]
- Valarmathi, R.; Sheela, T. Heart disease prediction using hyper parameter optimization (HPO) tuning. Biomed. Signal Process. Control. 2021, 70, 103033. [Google Scholar] [CrossRef]
- Ansarullah, S.I.; Saif, S.M.; Andrabi, S.A.B.; Kumhar, S.H.; Kirmani, M.M.; Kumar, P. An Intelligent and Reliable Hyperparameter Optimization Machine Learning Model for Early Heart Disease Assessment Using Imperative Risk Attributes. J. Healthc. Eng. 2022, 2022, 9882288. [Google Scholar] [CrossRef]
- Li, J.P.; Haq, A.U.; Din, S.U.; Khan, J.; Khan, A.; Saboor, A. Heart Disease Identification Method Using Machine Learning Classification in E-Healthcare. IEEE Access 2020, 8, 107562–107582. [Google Scholar] [CrossRef]
- Soni, M.; Nayak, N.R.; Selvakumar, V.; Pande, S.D. Recurrent neural network model for identifying epilepsy based neurological auditory disorder. In Artificial Intelligence for Neurological Disorders; Academic Press: Cambridge, MA, USA, 2023; pp. 91–105. [Google Scholar] [CrossRef]
- Madhavan, M.V.; Khamparia, A.; Pande, S.D. An augmented customized deep learning approach for brain tumour identification. Imaging Sci. J. 2023, 71, 331–342. [Google Scholar] [CrossRef]




| S. No | Attributes | Value |
|---|---|---|
| 1 | Age in years | >79 = 2, 61–79 = 1, 51–60 = 0, 35–50 = −1, <35 = −2 |
| 2 | Sex | Female = 0, Male = 1 |
| 3 | Chest Pain Type (4 different values) | 0–0.3 = -1, 0.9–1.2 = 0, 1.8–2.1 = 1, 2.7–3.0 = 2 |
| 4 | Resting BP | Above 139 mmHg = High = 1 120–139 mmHg = Normal = 0 Below 120 mmHg = Low = −1 |
| 5 | Serum Cholesterol | >240 mg/dL = High = 1 200–239 mg/dL = Normal = 0 <200 mg/dL = Low = −1 |
| 6 | Fasting blood sugar>120mg/dl (Boolean) | True = 1 False = 0 |
| 7 | Resting electrocardiographic result (3 values) | Hypertrophy = 2 ST T = 1 Normal = 0 |
| 8 | Diabetes | Yes = 1 No = 0 |
| 9 | Exercise-induced angina | Yes = 1 No = 0 |
| 10 | ST depression induced by exercise | Up = 2 Flat = 1 Down = 0 |
| 11 | Slope of peak exercise ST segment | <0.5 mm = Normal = 0 >0.5 mm = High = 1 |
| 12 | Smoke | Yes = 1 No = 0 |
| 13 | Status of heart (3 possible values) | Reversible defect = 7, Normal = 3, fixed defect = 6 |
| 14 | Heart disease (target) | Yes = 1 No = 0 |
| 15 | Number of major vessels colored by fluoroscopy | Vessel 0 = 0 vessel 1 = 1 vessel 2 = 2 vessel 3 = 3 |
| 16 | Maximum heart rate | <69 bpm = Low = −1 70–90 bpm = Normal = 0 >91 bpm = High = 1 |
| Symbol | Description |
|---|---|
| Xzi | Initial model for node Z |
| Xzo | Output local model from node Z |
| Δ | Iteration time |
| L | Total iterations |
| α | Deep parameter exchange ratio |
| B | Block |
| β | Minimum acceptable size of ECG |
| γ | Timestep |
| ℓ | Iteration |
| Z | Number of nodes |
| ωz | Local weights extracted from Xzi |
| Mz | New data obtained |
| ω | Global weight |
| Xg | Existing global model |
| E | Minimum loss threshold |
| Method | Datasets | Metrices | Number of Nodes | ||||
|---|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | |||
| Sync-FL | DS1 | Accuracy | 0.888 | 0.889 | 0.889 | 0.878 | 0.877 |
| Precision | 0.878 | 0.879 | 0.874 | 0.868 | 0.871 | ||
| F1-Score | 0.880 | 0.882 | 0.881 | 0.872 | 0.872 | ||
| DS2 | Accuracy | 0.893 | 0.889 | 0.886 | 0.895 | 0.863 | |
| Precision | 0.885 | 0.868 | 0.859 | 0.881 | 0.838 | ||
| F1-Score | 0.867 | 0.862 | 0.867 | 0.865 | 0.849 | ||
| Async-FL | DS1 | Accuracy | 0.869 | 0.879 | 0.891 | 0.879 | 0.878 |
| Precision | 0.873 | 0.878 | 0.883 | 0.875 | 0.877 | ||
| F1-Score | 0.869 | 0.876 | 0.887 | 0.875 | 0.875 | ||
| DS2 | Accuracy | 0.871 | 0.886 | 0.895 | 0.895 | 0.899 | |
| Precision | 0.841 | 0.856 | 0.889 | 0.881 | 0.891 | ||
| F1-Score | 0.852 | 0.868 | 0.867 | 0.869 | 0.874 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.A.; Alsulami, M.; Yaqoob, M.M.; Alsadie, D.; Saudagar, A.K.J.; AlKhathami, M.; Farooq Khattak, U. Asynchronous Federated Learning for Improved Cardiovascular Disease Prediction Using Artificial Intelligence. Diagnostics 2023, 13, 2340. https://doi.org/10.3390/diagnostics13142340
Khan MA, Alsulami M, Yaqoob MM, Alsadie D, Saudagar AKJ, AlKhathami M, Farooq Khattak U. Asynchronous Federated Learning for Improved Cardiovascular Disease Prediction Using Artificial Intelligence. Diagnostics. 2023; 13(14):2340. https://doi.org/10.3390/diagnostics13142340
Chicago/Turabian StyleKhan, Muhammad Amir, Musleh Alsulami, Muhammad Mateen Yaqoob, Deafallah Alsadie, Abdul Khader Jilani Saudagar, Mohammed AlKhathami, and Umar Farooq Khattak. 2023. "Asynchronous Federated Learning for Improved Cardiovascular Disease Prediction Using Artificial Intelligence" Diagnostics 13, no. 14: 2340. https://doi.org/10.3390/diagnostics13142340
APA StyleKhan, M. A., Alsulami, M., Yaqoob, M. M., Alsadie, D., Saudagar, A. K. J., AlKhathami, M., & Farooq Khattak, U. (2023). Asynchronous Federated Learning for Improved Cardiovascular Disease Prediction Using Artificial Intelligence. Diagnostics, 13(14), 2340. https://doi.org/10.3390/diagnostics13142340







