Diagnosis of Obstructive Sleep Apnea Using Feature Selection, Classification Methods, and Data Grouping Based Age, Sex, and Race
Abstract
:1. Introduction
- Obstructive sleep apnea (OSA): The most common type of apnea is known as obstructive sleep apnea (OSA), which is identified by two primary characteristics. The first is a continuous reduction in airflow of at least 30% for a duration of 10 seconds, which is accompanied by a minimum oxygen desaturation of 4%. The second is a decrease in airflow of at least 50% for 10 seconds, coupled with a 3% reduction in oxygen saturation [10].
- Mixed sleep apnea (MSA): MSA, also known as complex sleep apnea, represents a combination of obstructive and central sleep apnea disorders, thus presenting a more complex pattern of symptoms and characteristics.
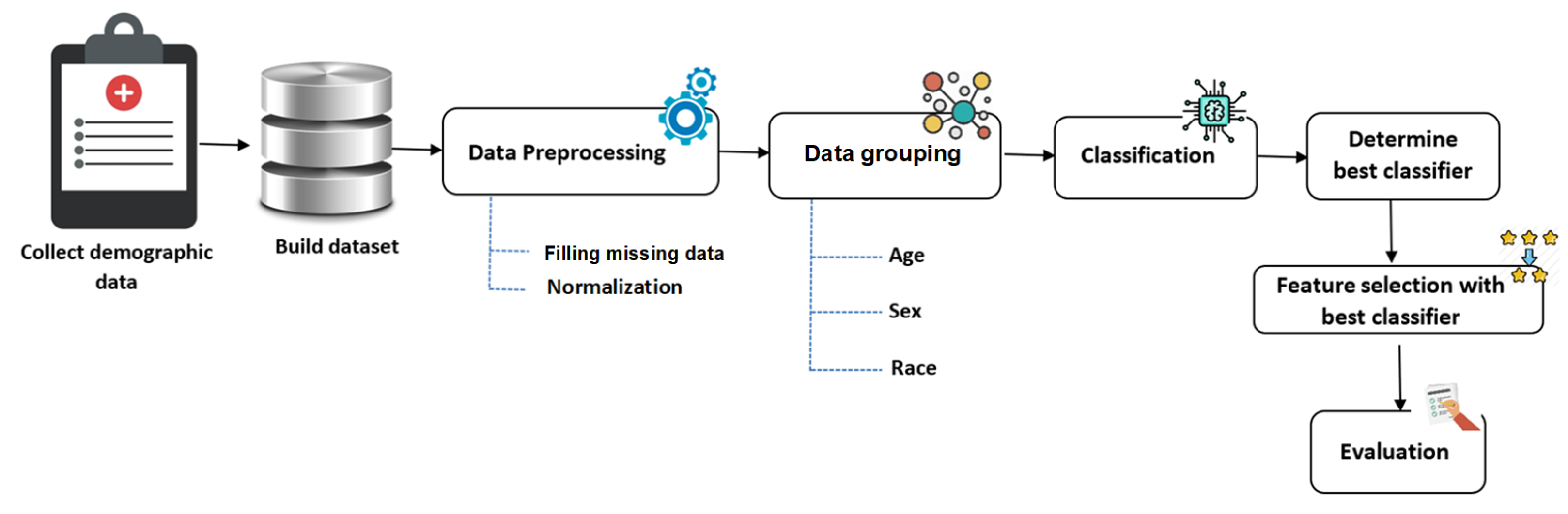
- The OSA data was grouped based on age, sex, and race variables for performance improvement. This type of grouping is novel and has never been presented in this area of research before.
- Various types of the most well-known machine learning algorithms were assessed to determine the best-performing one for the OSA problem. These methods included twelve predefined (fixed) parameter classifiers and two optimized classifiers (using hyperparameter optimization).
- A wrapper feature selection approach using particle swarm optimization (PSO) was employed to determine the most valuable features related to the OSA.
- Experimental results from the actual data (collected from Torr Sleep Center, Texas, USA) confirmed that the proposed method improved the overall performance of the OSA prediction.
2. Proposed Diagnosis Process
3. Sleep Apnea Dataset
4. Data Preprocessing
4.1. Missing Data
- If more than 50% of any rows or columns values are missing, we have to remove the whole row/columns, except where it is feasible to fill in the missing values.
- If only a rational percentage of values are missing, we can adopt simple interpolation methods to fill in those values. Interpolation methods include filling missing values with the mean, median, or mode value of the respective feature.
4.2. Data Normalization
4.3. Role of Grouping in OSA Diagnosis
4.4. Wrapper Feature Selection
4.5. Formulation of Feature Selection Problem
5. Experimental Setup
6. Experimental Results
6.1. Results with All Data
6.2. Data Grouping with Race
6.3. Data Grouping with Gender
6.4. Data Grouping with Age
6.5. Summary Performance with Data Grouping
7. Feature Selection
7.1. Evaluation of BPSO Using Different TFs
7.2. Comparison of BPSO with Well-Known Algorithms
7.3. Relevant Features Selected by BPSO
7.4. Comparison of the BPSO-kNN with CNN, MLP, and kNN*
7.5. Comparison Study
8. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dickens, C. The Posthumous Papers of the Pickwick Club, 1st ed.; Chapman and Hall: London, UK, 1837. [Google Scholar]
- Alruwaili, H.; Ahmed, A.; Fatani, A.; Al-Otaibi, K.; Al-Jahdali, S.; Ali, Y.; Al-Harbi, A.; Baharoon, S.; Khan, M.; Al-Jahdali, H. Symptoms and risk for obstructive sleep apnea among sample of Saudi Arabian adults. Sleep Biol. Rhythm. 2015, 13, 332–341. [Google Scholar] [CrossRef]
- Piriyajitakonkij, M.; Warin, P.; Lakhan, P.; Leelaarporn, P.; Kumchaiseemak, N.; Suwajanakorn, S.; Pianpanit, T.; Niparnan, N.; Mukhopadhyay, S.C.; Wilaiprasitporn, T. SleepPoseNet: Multi-View Learning for Sleep Postural Transition Recognition Using UWB. IEEE J. Biomed. Health Inform. 2020, 25, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Kaditis, A.G.; Alonso Alvarez, M.L.; Boudewyns, A.; Alexopoulos, E.I.; Ersu, R.; Joosten, K.; Larramona, H.; Miano, S.; Narang, I.; Trang, H.; et al. Obstructive sleep disordered breathing in 2- to 18-year-old children: Diagnosis and management. Eur. Respir. J. 2016, 47, 69–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhulst, S.; Kaditis, A. Obstructive sleep apnoea in children. Breathe 2011, 7, 240–247. [Google Scholar] [CrossRef] [Green Version]
- Banluesombatkul, N.; Ouppaphan, P.; Leelaarporn, P.; Lakhan, P.; Chaitusaney, B.; Jaimchariya, N.; Chuangsuwanich, E.; Chen, W.; Phan, H.; Dilokthanakul, N.; et al. MetaSleepLearner: A Pilot Study on Fast Adaptation of Bio-signals-Based Sleep Stage Classifier to New Individual Subject Using Meta-Learning. IEEE J. Biomed. Health Inform. 2020, 25, 1949–1963. [Google Scholar] [CrossRef]
- American Sleep Apnea Association. Available online: https://www.sleepapnea.org/learn/sleep-apnea-information-clinicians/ (accessed on 22 April 2019).
- AASM. American Academy of Sleep Medicine: Economic Burden of Undiagnosed Sleep Apnea in U.S. Is Nearly $150B per Year, 2023. Available online: https://aasm.org/economic-burden-of-undiagnosed-sleep-apnea-in-u-s-is-nearly-150b-per-year/ (accessed on 8 August 2016).
- Lang, C.C.; Mancini, D.M. Non-cardiac comorbidities in chronic heart failure. Heart 2007, 93, 665–671. [Google Scholar] [CrossRef] [Green Version]
- Quan, S.; Gillin, J.C.; Littner, M.; Shepard, J. Sleep-related breathing disorders in adults: Recommendations for syndrome definition and measurement techniques in clinical research. editorials. Sleep 1999, 22, 662–689. [Google Scholar] [CrossRef] [Green Version]
- Young, T.; Finn, L.; Kim, H. Nasal obstruction as a risk factor for sleep-disordered breathing. J. Allergy Clin. Immunol. 1997, 99, S757–S762. [Google Scholar] [CrossRef]
- Hung, P.D. Detection of Central Sleep Apnea Based on a Single-Lead ECG. In Proceedings of the 2018 5th International Conference on Bioinformatics Research and Applications, Hong Kong, China, 27–29 December 2018; ACM: New York, NY, USA, 2018; pp. 78–83. [Google Scholar] [CrossRef]
- Daldal, N.; Cömert, Z.; Polat, K. Automatic determination of digital modulation types with different noises using Convolutional Neural Network based on time–frequency information. Appl. Soft Comput. 2020, 86, 105834. [Google Scholar] [CrossRef]
- Bozkurt, F.; Uçar, M.K.; Bozkurt, M.R.; Bilgin, C. Detection of abnormal respiratory events with single channel ECG and hybrid machine learning model in patients with obstructive sleep apnea. IRBM 2020, 41, 241–251. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, M.; Wang, Y.; Liu, S.; Tian, Z.; Jiang, F.; Zhang, H. Obstructive sleep apnea detection using ecg-sensor with convolutional neural networks. Multimed. Tools Appl. 2020, 79, 15813–15827. [Google Scholar] [CrossRef]
- Urtnasan, E.; Park, J.U.; Joo, E.Y.; Lee, K.J. Automated Detection of Obstructive Sleep Apnea Events from a Single-Lead Electrocardiogram Using a Convolutional Neural Network. J. Med. Syst. 2018, 42, 104. [Google Scholar] [CrossRef]
- Faust, O.; Barika, R.; Shenfield, A.; Ciaccio, E.J.; Acharya, U.R. Accurate detection of sleep apnea with long short-term memory network based on RR interval signals. Knowl.-Based Syst. 2021, 212, 106591. [Google Scholar] [CrossRef]
- Schwartz, A.R.; Cohen-Zion, M.; Pham, L.V.; Gal, A.; Sowho, M.; Sgambati, F.P.; Klopfer, T.; Guzman, M.A.; Hawks, E.M.; Etzioni, T.; et al. Brief digital sleep questionnaire powered by machine learning prediction models identifies common sleep disorders. Sleep Med. 2020, 71, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Lakhan, P.; Ditthapron, A.; Banluesombatkul, N.; Wilaiprasitporn, T. Deep Neural Networks with Weighted Averaged Overnight Airflow Features for Sleep Apnea-Hypopnea Severity Classification. In Proceedings of the TENCON 2018—2018 IEEE Region 10 Conference, Jeju, Republic of Korea, 28–31 October 2018; pp. 0441–0445. [Google Scholar] [CrossRef] [Green Version]
- Banluesombatkul, N.; Rakthanmanon, T.; Wilaiprasitporn, T. Single Channel ECG for Obstructive Sleep Apnea Severity Detection Using a Deep Learning Approach. In Proceedings of the TENCON 2018—2018 IEEE Region 10 Conference, Jeju, Republic of Korea, 28–31 October 2018; pp. 2011–2016. [Google Scholar] [CrossRef] [Green Version]
- Herzog, M.; Plößl, S.; Glien, A.; Herzog, B.; Rohrmeier, C.; Kühnel, T.; Plontke, S.; Kellner, P. Evaluation of acoustic characteristics of snoring sounds obtained during drug-induced sleep endoscopy. Sleep Breath. 2015, 19, 1011–1019. [Google Scholar] [CrossRef]
- Alshaer, H.; Hummel, R.; Mendelson, M.; Marshal, T.; Bradley, T.D. Objective Relationship Between Sleep Apnea and Frequency of Snoring Assessed by Machine Learning. J. Clin. Sleep Med. 2019, 15, 463–470. [Google Scholar] [CrossRef]
- Kang, B.; Dang, X.; Wei, R. Snoring and apnea detection based on hybrid neural networks. In Proceedings of the 2017 International Conference on Orange Technologies (ICOT), Singapore, 8–10 December 2017; pp. 57–60. [Google Scholar] [CrossRef]
- Sheta, A.; Turabieh, H.; Braik, M.; Surani, S.R. Diagnosis of Obstructive Sleep Apnea Using Logistic Regression and Artificial Neural Networks Models. In Proceedings of the Future Technologies Conference (FTC) 2019, San Francisco, CA, USA, 24–25 October 2019; Arai, K., Bhatia, R., Kapoor, S., Eds.; Springer International Publishing: Cham, Swizerland, 2020; pp. 766–784. [Google Scholar]
- Aiyer, I.; Shaik, L.; Sheta, A.; Surani, S. Review of Application of Machine Learning as a Screening Tool for Diagnosis of Obstructive Sleep Apnea. Medicina 2022, 58, 1574. [Google Scholar] [CrossRef]
- Surani, S.; Sheta, A.; Turabieh, H.; Subramanian, S. Adaboosting Model for Detecting OSA. Chest 2019, 156, A134–A135. [Google Scholar] [CrossRef]
- Surani, S.; Sheta, A.; Turabieh, H.; Park, J.; Mathur, S.; Katangur, A. Diagnosis of Sleep Apnea Using artificial Neural Network and binary Particle Swarm Optimization for Feature Selection. Chest 2019, 156, A136. [Google Scholar] [CrossRef]
- Haberfeld, C.; Sheta, A.; Hossain, M.S.; Turabieh, H.; Surani, S. SAS Mobile Application for Diagnosis of Obstructive Sleep Apnea Utilizing Machine Learning Models. In Proceedings of the 2020 11th IEEE Annual Ubiquitous Computing, Electronics Mobile Communication Conference (UEMCON), New York, NY, USA, 28–31 October 2020; pp. 0522–0529. [Google Scholar] [CrossRef]
- Rossi, C.; Templier, L.; Miguez, M.; Cruz, J.D.L.; Curto, A.; Albaladejo, A.; Vich, M.L. Comparison of screening methods for obstructive sleep apnea in the context of dental clinics: A systematic review. CRANIO® 2023, 41, 245–263. [Google Scholar] [CrossRef]
- Subramanian, S.; Hesselbacher, S.; Aguilar, R.; Surani, S. The NAMES assessment: A novel combined-modality screening tool for obstructive sleep apnea. Sleep Breath. 2011, 15, 819–826. [Google Scholar] [CrossRef]
- Friedman, M.; Hwang, M.S. Evaluation of the patient with obstructive sleep apnea: Friedman tongue position and staging. Oper. Tech.-Otolaryngol.-Head Neck Surg. 2015, 26, 85–89, Sleep Disordered Breathing: Part 1. [Google Scholar] [CrossRef]
- Kotsiantis, S.; Kanellopoulos, D.; Pintelas, P. Data Preprocessing for Supervised Learning. Int. J. Comput. Sci. 2006, 1, 111–117. [Google Scholar]
- Malley, B.; Ramazzotti, D.; Wu, J.T.y. Data Pre-processing. In Secondary Analysis of Electronic Health Records; Springer International Publishing: Cham, Swizerland, 2016; pp. 115–141. [Google Scholar]
- Fan, C.; Chen, M.; Wang, X.; Wang, J.; Huang, B. A Review on Data Preprocessing Techniques Toward Efficient and Reliable Knowledge Discovery From Building Operational Data. Front. Energy Res. 2021, 9, 77. [Google Scholar] [CrossRef]
- Little, R.J.A.; Rubin, D.B. Statistical Analysis with Missing Data; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1986. [Google Scholar]
- Sterne, J.A.C.; White, I.R.; Carlin, J.B.; Spratt, M.; Royston, P.; Kenward, M.G.; Wood, A.M.; Carpenter, J.R. Multiple imputation for missing data in epidemiological and clinical research: Potential and pitfalls. BMJ 2009, 338, b2393. [Google Scholar] [CrossRef] [PubMed]
- Sola, J.; Sevilla, J. Importance of input data normalization for the application of neural networks to complex industrial problems. IEEE Trans. Nucl. Sci. 1997, 44, 1464–1468. [Google Scholar] [CrossRef]
- Sathya Durga, V.; Jeyaprakash, T. An Effective Data Normalization Strategy for Academic Datasets using Log Values. In Proceedings of the 2019 International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 17–19 July 2019; pp. 610–612. [Google Scholar] [CrossRef]
- Mohsenin, V.; Yaggi, H.K.; Shah, N.; Dziura, J. The effect of gender on the prevalence of hypertension in obstructive sleep apnea. Sleep Med. 2009, 10, 759–762. [Google Scholar] [CrossRef]
- Freitas, L.S.; Drager, L.F. Gender and cardiovascular impact of obstructive sleep apnea: Work in progress! J. Thorac. Dis. 2017, 9, 3579–3582. [Google Scholar] [CrossRef] [Green Version]
- Ralls, F.M.; Grigg-Damberger, M. Roles of gender, age, race/ethnicity, and residential socioeconomics in obstructive sleep apnea syndromes. Curr. Opin. Pulm. Med. 2012, 18, 568–573. [Google Scholar] [CrossRef]
- Slaats, M.; Vos, W.; Van Holsbeke, C.; De Backer, J.; Loterman, D.; De Backer, W.; Boudewyns, A.; Verhulst, S. The role of ethnicity in the upper airway in a Belgian paediatric population with obstructive sleep apnoea. Eur. Respir. J. 2017, 50. [Google Scholar] [CrossRef]
- Cano-Pumarega, I.; Barbé, F.; Esteban, A.; Martínez-Alonso, M.; Egea, C.; Durán-Cantolla, J.; Montserrat, J.M.; Muria, B.; Sánchez de la Torre, M.; Abad Fernández, A. Sleep Apnea and Hypertension: Are There Sex Differences? The Vitoria Sleep Cohort. Chest 2017, 152, 742–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohsenin, V. Gender Differences in the Expression of Sleep-Disordered Breathing: Role of Upper Airway Dimensions. Chest 2001, 120, 1442–1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Motoda, H. Feature Selection for Knowledge Discovery and Data Mining; Springer Science & Business Media: Berlin, Germany, 2012; Volume 454. [Google Scholar]
- Chantar, H.K.; Corne, D.W. Feature subset selection for Arabic document categorization using BPSO-KNN. In Proceedings of the 2011 Third World Congress on Nature and Biologically Inspired Computing, Salamanca, Spain, 19–21 October 2011; pp. 546–551. [Google Scholar]
- Chantar, H.; Thaher, T.; Turabieh, H.; Mafarja, M.; Sheta, A. BHHO-TVS: A Binary Harris Hawks Optimizer with Time-Varying Scheme for Solving Data Classification Problems. Appl. Sci. 2021, 11, 6516. [Google Scholar] [CrossRef]
- Tumar, I.; Hassouneh, Y.; Turabieh, H.; Thaher, T. Enhanced Binary Moth Flame Optimization as a Feature Selection Algorithm to Predict Software Fault Prediction. IEEE Access 2020, 8, 8041–8055. [Google Scholar] [CrossRef]
- Wang, A.; An, N.; Chen, G.; Li, L.; Alterovitz, G. Accelerating wrapper-based feature selection with K-nearest-neighbor. Knowl.-Based Syst. 2015, 83, 81–91. [Google Scholar] [CrossRef]
- Saeys, Y.; Iñaki, I.; Pedro, L.n. A review of feature selection techniques in bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef] [Green Version]
- Dash, M.; Liu, H. Feature selection for classification. Intell. Data Anal. 1997, 1, 131–156. [Google Scholar] [CrossRef]
- Siedlecki, W.; Sklansky, J. On automatic feature selection. Int. J. Pattern Recognit. Artif. Intell. 1988, 2, 197–220. [Google Scholar] [CrossRef]
- Langley, P. Selection of relevant features in machine learning. In Proceedings of the AAAI Fall Symposium on Relevance, New Orleans, LA, USA, 4–6 November 1994. [Google Scholar]
- Lai, C.; Reinders, M.J.; Wessels, L. Random subspace method for multivariate feature selection. Pattern Recognit. Lett. 2006, 27, 1067–1076. [Google Scholar] [CrossRef]
- Talbi, E. Metaheuristics From Design to Implementation; Wiley Online Library: Hoboken, NJ, USA, 2009. [Google Scholar]
- Lu, J.J.; Zhang, M. Heuristic Search. In Encyclopedia of Systems Biology; Springer: New York, NY, USA, 2013; pp. 885–886. [Google Scholar] [CrossRef]
- Kennedy, J.; Eberhart, R.C. A discrete binary version of the particle swarm algorithm. In Proceedings of the 1997 IEEE International Conference on Systems, Man, and Cybernetics, Computational Cybernetics and Simulation, Orlando, FL, USA, 12–15 October 1997; Volume 5, pp. 4104–4108. [Google Scholar]
- Dorigo, M.; Birattari, M.; Stutzle, T. Ant colony optimization. IEEE Comput. Intell. Mag. 2006, 1, 28–39. [Google Scholar] [CrossRef]
- Yang, X.S. Firefly Algorithms for Multimodal Optimization. In Stochastic Algorithms: Foundations and Applications; Watanabe, O., Zeugmann, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 169–178. [Google Scholar]
- Mirjalili, S. The ant lion optimizer. Adv. Eng. Softw. 2015, 83, 80–98. [Google Scholar] [CrossRef]
- Mirjalili, S.; Lewis, A. The whale optimization algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey wolf optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef] [Green Version]
- Mafarja, M.; Mirjalili, S. Hybrid Whale Optimization Algorithm with Simulated Annealing for Feature Selection. Neurocomputing 2017, 260, 302–312. [Google Scholar] [CrossRef]
- Chantar, H.; Mafarja, M.; Alsawalqah, H.; Heidari, A.A.; Aljarah, I.; Faris, H. Feature selection using binary grey wolf optimizer with elite-based crossover for Arabic text classification. Neural Comput. Appl. 2020, 32, 12201–12220. [Google Scholar] [CrossRef]
- Zhang, L.; Mistry, K.; Lim, C.; Neoh, S. Feature selection using firefly optimization for classification and regression models. Decis. Support Syst. 2018, 106, 64–85. [Google Scholar] [CrossRef] [Green Version]
- Deriche, M. Feature Selection using Ant Colony Optimization. In Proceedings of the 2009 6th International Multi-Conference on Systems, Signals and Devices, Djerba, Tunisia, 23–26 March 2009; pp. 1–4. [Google Scholar] [CrossRef]
- Zawbaa, H.M.; Emary, E.; Parv, B. Feature selection based on antlion optimization algorithm. In Proceedings of the 2015 Third World Conference on Complex Systems (WCCS), Marrakech, Morocco, 23–25 November 2015; pp. 1–7. [Google Scholar] [CrossRef]
- Rostami, M.; Berahmand, K.; Nasiri, E.; Forouzandeh, S. Review of swarm intelligence-based feature selection methods. Eng. Appl. Artif. Intell. 2021, 100, 104210. [Google Scholar] [CrossRef]
- Thaher, T.; Mafarja, M.; Turabieh, H.; Castillo, P.A.; Faris, H.; Aljarah, I. Teaching Learning-Based Optimization With Evolutionary Binarization Schemes for Tackling Feature Selection Problems. IEEE Access 2021, 9, 41082–41103. [Google Scholar] [CrossRef]
- Eberhart, R.; Kennedy, J. Particle swarm optimization. In Proceedings of the IEEE International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar]
- Mirjalili, S.; Lewis, A. S-shaped versus V-shaped transfer functions for binary particle swarm optimization. Swarm Evol. Comput. 2013, 9, 1–14. [Google Scholar] [CrossRef]
- Mafarja, M.; Aljarah, I.; Faris, H.; Hammouri, A.I.; Ala’M, A.Z.; Mirjalili, S. Binary grasshopper optimisation algorithm approaches for feature selection problems. Expert Syst. Appl. 2019, 117, 267–286. [Google Scholar] [CrossRef]
- Thaher, T.; Heidari, A.A.; Mafarja, M.; Dong, J.S.; Mirjalili, S. Binary Harris Hawks optimizer for high-dimensional, low sample size feature selection. In Evolutionary Machine Learning Techniques; Springer: Singapore, 2020; pp. 251–272. [Google Scholar]
- Wolpert, D.H. The Lack of A Priori Distinctions Between Learning Algorithms. Neural Comput. 1996, 8, 1341–1390. [Google Scholar] [CrossRef]
- Faris, H.; Heidari, A.A.; Ala’M, A.Z.; Mafarja, M.; Aljarah, I.; Eshtay, M.; Mirjalili, S. Time-varying hierarchical chains of salps with random weight networks for feature selection. Expert Syst. Appl. 2020, 140, 112898. [Google Scholar] [CrossRef]
- Rashedi, E.; Nezamabadi-Pour, H.; Saryazdi, S. BGSA: Binary gravitational search algorithm. Nat. Comput. 2010, 9, 727–745. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, D. Binary whale optimization algorithm and its application to unit commitment problem. Neural Comput. Appl. 2020, 32, 2095–2123. [Google Scholar] [CrossRef]
- Emary, E.; Zawbaa, H.M.; Hassanien, A.E. Binary grey wolf optimization approaches for feature selection. Neurocomputing 2016, 172, 371–381. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Yang, X.S. Binary bat algorithm. Neural Comput. Appl. 2014, 25, 663–681. [Google Scholar] [CrossRef]








| Attributes | Data Type | |
|---|---|---|
| f1 | Race | Categorical |
| f2 | Age | Numeric |
| f3 | Sex | Categorical |
| f4 | BMI | Categorical |
| f5 | Epworth | Numeric |
| f6 | Wast | Numeric |
| f7 | Hip | Numeric |
| f8 | RDI | Numeric |
| f9 | Neck | Numeric |
| f10 | M.Friedman | Numeric |
| f11 | Co-morbid | Categorical |
| f12 | Snoring | Categorical |
| f13 | Daytime sleepiness | Categorical |
| f14 | DM | Categorical |
| f15 | HTN | Categorical |
| f16 | CAD | Categorical |
| f17 | CVA | Categorical |
| f18 | TST | Numeric |
| f19 | Sleep Effic | Numeric |
| f20 | REM AHI | Numeric |
| f21 | NREM AHI | Numeric |
| f22 | Supine AHI | Numeric |
| f23 | Apnea Index | Numeric |
| f24 | Hypopnea Index | Numeric |
| f25 | Berlin Q | Categorical |
| f26 | Arousal index | Numeric |
| f27 | Awakening Index | Numeric |
| f28 | PLM Index | Numeric |
| f29 | Mins. SaO | Numeric |
| f30 | Mins. SaO Desats | Numeric |
| f31 | Lowest SaO | Numeric |
| class | Witnessed apnea | Categorical |
| Range | Description |
|---|---|
| 0–5 | Lower normal daytime sleepiness |
| 6–10 | Higher normal daytime sleepiness |
| 11–12 | Mild level of sleepiness experienced during the daytime |
| 13–15 | Moderate level of sleepiness experienced during the daytime |
| 16–24 | Significant level of sleepiness experienced during the daytime |
| Datasets | No. Features | No. Samples | Negative | Positive | |
|---|---|---|---|---|---|
| Original Dataset | 31 | 274 | 149 | 125 | |
| Race | Caucasian | 30 | 151 | 92 | 59 |
| Hispanic | 30 | 123 | 57 | 66 | |
| Gender | Females | 30 | 118 | 85 | 33 |
| Males | 30 | 156 | 64 | 92 | |
| Age | Age ≤ 50 | 31 | 109 | 55 | 54 |
| Age > 50 | 31 | 165 | 94 | 71 | |
| Name | Transfer Function Formula |
|---|---|
| S1 | |
| S2 | |
| S3 | |
| S4 |
| Preset Classifier | Description | Parameter | Value |
|---|---|---|---|
| FDT | Fine Decision Tree | Maximum number of splits | 100 |
| Split criterion | Gini’s diversity index | ||
| CDT | Coarse Decision Tree | Maximum number of splits | 100 |
| Split criterion | Gini’s diversity index | ||
| LDA | Linear Discrimenant Analaysis | Discriminant type | linear |
| LR | Logistic Regression | - | - |
| GNB | Gaussian Naïve Bayes | Distribution name | Gaussian |
| KNB | Kernel Naïve Bayes | Distribution name | Kernel |
| Kernel type | Gaussian | ||
| LSVM | Linear Support Vector Machine | Kernel function | Linear |
| Kernel scale | Automatic | ||
| Box contraint level | 1 | ||
| standardize data | TRUE | ||
| MGSVM | Medium Gaussian SVM | Kernel function | Gaussian |
| Kernel scale | 5.6 | ||
| Box contraint level | 1 | ||
| Standardize data | TRUE | ||
| CGSVM | Coarse Gaussian SVM | Kernel function | Gaussian |
| Kernel scale | 22 | ||
| Box contraint level | 1 | ||
| Standardize data | TRUE | ||
| CKNN | Cosine K-Nearest Neighbor | Number of neighbors | 10 |
| Distance metric | cosine | ||
| Distance weight | equal | ||
| Standardize data | TRUE | ||
| WKNN | Weighted kNN | Number of neighbors | 10 |
| Distance metric | Euclidean | ||
| Distance weight | Squared Inverse | ||
| Standardize data | TRUE | ||
| Ensemble | Subspace Discriminant | Ensemble method | Subspace |
| Learner type | Discriminant | ||
| Number of learners | 30 | ||
| Subspace dimension | 16 |
| Dataset | Number of Neighbors | Distance Metric | Distance Weight | Standardize Data |
|---|---|---|---|---|
| All | 16 | Spearman | Inverse | TRUE |
| Caucasian | 6 | Correlation | Squared Inverse | TRUE |
| Hispanic | 32 | Cityblock | Squared Inverse | TRUE |
| Females | 4 | Hamming | Squared Inverse | FALSE |
| Males | 22 | Cityblock | Equal | FALSE |
| Age ≤ 50 | 14 | Cosine | Squared Inverse | TRUE |
| Age > 50 | 31 | 16 | Squared Inverse | TRUE |
| Dataset | Kernel Function | Kernel Scale | Box Contraint Level | Standardized Data |
|---|---|---|---|---|
| All | Polynomial (degree = 2) | 1 | 0.002351927 | TRUE |
| Caucasian | Linear | 1 | 0.18078 | TRUE |
| Hispanic | Linear | 1 | 0.01115743 | TRUE |
| Females | Gaussian | 2.990548535 | 122.3491994 | FALSE |
| Males | Linear | 1 | 0.001000015 | FALSE |
| Age ≤ 50 | Gaussian | 415.5625146 | 341.7329909 | FALSE |
| Age > 50 | Gaussian | 26.42211158 | 7.503025335 | TRUE |
| Classifier | Accuracy | TPR | TNR | AUC | Precision | F-Score | G-Mean | Mean Rank |
|---|---|---|---|---|---|---|---|---|
| DT | 0.5876 | 0.6309 | 0.5360 | 0.5834 | 0.6184 | 0.6246 | 0.5815 | 8.97 |
| LDA | 0.6861 | 0.7584 | 0.6000 | 0.6792 | 0.6933 | 0.7244 | 0.6746 | 5.79 |
| LR | 0.6861 | 0.7383 | 0.6240 | 0.6811 | 0.7006 | 0.7190 | 0.6787 | 5.57 |
| NB | 0.6642 | 0.7785 | 0.5280 | 0.6533 | 0.6629 | 0.7160 | 0.6411 | 7.71 |
| SVM | 0.6898 | 0.8188 | 0.5360 | 0.6774 | 0.6778 | 0.7416 | 0.6625 | 5.71 |
| kNN | 0.6934 | 0.7517 | 0.6240 | 0.6878 | 0.7044 | 0.7273 | 0.6849 | 4.21 |
| Ensemble | 0.7044 | 0.8188 | 0.5680 | 0.6934 | 0.6932 | 0.7508 | 0.6820 | 3.93 |
| SVM* | 0.7226 | 0.7919 | 0.6400 | 0.7160 | 0.7239 | 0.7564 | 0.7119 | 2.14 |
| kNN* | 0.7409 | 0.8322 | 0.6320 | 0.7321 | 0.7294 | 0.7774 | 0.7252 | 1.14 |
| Classifier | Accuracy | TPR | TNR | AUC | Precision | F-Score | G-Mean | Mean Rank |
|---|---|---|---|---|---|---|---|---|
| FDT | 0.7285 | 0.7935 | 0.6271 | 0.7103 | 0.7684 | 0.7807 | 0.7054 | 3.29 |
| LDA | 0.6908 | 0.7957 | 0.5254 | 0.6606 | 0.7255 | 0.7590 | 0.6466 | 6.71 |
| LR | 0.6887 | 0.7609 | 0.5763 | 0.6686 | 0.7368 | 0.7487 | 0.6622 | 6.21 |
| GNB | 0.6887 | 0.8261 | 0.4746 | 0.6503 | 0.7103 | 0.7638 | 0.6261 | 7.93 |
| MGSVM | 0.7219 | 0.8913 | 0.4576 | 0.6745 | 0.7193 | 0.7961 | 0.6387 | 5.79 |
| CKNN | 0.7483 | 0.9457 | 0.4407 | 0.6932 | 0.7250 | 0.8208 | 0.6455 | 4.36 |
| Ensemble | 0.7219 | 0.8696 | 0.4915 | 0.6805 | 0.7273 | 0.7921 | 0.6538 | 5.07 |
| SVM* | 0.7351 | 0.8478 | 0.5593 | 0.7036 | 0.7500 | 0.7959 | 0.6886 | 3.36 |
| kNN* | 0.7483 | 0.8804 | 0.5424 | 0.7114 | 0.7500 | 0.8100 | 0.6910 | 2.29 |
| Data | Accuracy | TPR | TNR | AUC | Precision | F-Score | G-Mean | Mean Rank |
|---|---|---|---|---|---|---|---|---|
| CDT | 0.6260 | 0.5263 | 0.7121 | 0.6192 | 0.6122 | 0.5660 | 0.6122 | 8.29 |
| LDA | 0.6423 | 0.5614 | 0.7121 | 0.6368 | 0.6275 | 0.5926 | 0.6323 | 6.50 |
| LR | 0.6260 | 0.5614 | 0.6818 | 0.6216 | 0.6038 | 0.5818 | 0.6187 | 8.00 |
| GNB | 0.6504 | 0.6140 | 0.6818 | 0.6479 | 0.6250 | 0.6195 | 0.6470 | 5.64 |
| MGSVM | 0.6829 | 0.5614 | 0.7879 | 0.6746 | 0.6957 | 0.6214 | 0.6651 | 2.86 |
| WKNN | 0.6585 | 0.5439 | 0.7576 | 0.6507 | 0.6596 | 0.5962 | 0.6419 | 5.07 |
| Ensemble | 0.6585 | 0.5965 | 0.7121 | 0.6543 | 0.6415 | 0.6182 | 0.6517 | 4.57 |
| SVM* | 0.6748 | 0.6316 | 0.7121 | 0.6719 | 0.6545 | 0.6429 | 0.6706 | 3.00 |
| kNN* | 0.7724 | 0.6316 | 0.8939 | 0.7628 | 0.8372 | 0.7200 | 0.7514 | 1.07 |
| Classifier | Accuracy | TPR | TNR | AUC | Precision | F-Score | G-Mean | Mean Rank |
|---|---|---|---|---|---|---|---|---|
| CDT | 0.7119 | 0.8824 | 0.2727 | 0.5775 | 0.7576 | 0.8152 | 0.4906 | 4.86 |
| LDA | 0.6695 | 0.8118 | 0.3030 | 0.5574 | 0.7500 | 0.7797 | 0.4960 | 5.57 |
| LR | 0.6102 | 0.7529 | 0.2424 | 0.4977 | 0.7191 | 0.7356 | 0.4272 | 8.00 |
| KNB | 0.6356 | 0.7176 | 0.4242 | 0.5709 | 0.7625 | 0.7394 | 0.5518 | 5.00 |
| MGSVM | 0.7203 | 1.0000 | 0.0000 | 0.5000 | 0.7203 | 0.8374 | 0.0000 | 5.79 |
| WKNN | 0.7458 | 0.9765 | 0.1515 | 0.5640 | 0.7477 | 0.8469 | 0.3846 | 4.36 |
| Ensemble | 0.7458 | 0.8824 | 0.3939 | 0.6381 | 0.7895 | 0.8333 | 0.5896 | 2.43 |
| SVM* | 0.7203 | 1.0000 | 0.0000 | 0.5000 | 0.7203 | 0.8374 | 0.0000 | 5.79 |
| kNN* | 0.7373 | 0.9176 | 0.2727 | 0.5952 | 0.7647 | 0.8342 | 0.5003 | 3.21 |
| Classifier | Accuracy | TPR | TNR | AUC | Precision | F-Score | G-Mean | Mean Rank |
|---|---|---|---|---|---|---|---|---|
| CDT | 0.5256 | 0.2969 | 0.6848 | 0.4908 | 0.3958 | 0.3393 | 0.4509 | 8.64 |
| LDA | 0.6538 | 0.5938 | 0.6957 | 0.6447 | 0.5758 | 0.5846 | 0.6427 | 4.07 |
| LR | 0.6474 | 0.5938 | 0.6848 | 0.6393 | 0.5672 | 0.5802 | 0.6376 | 5.14 |
| KNB | 0.6234 | 0.6406 | 0.6111 | 0.6259 | 0.5395 | 0.5857 | 0.6257 | 5.86 |
| LSVM | 0.6346 | 0.5313 | 0.7065 | 0.6189 | 0.5574 | 0.5440 | 0.6126 | 6.79 |
| CKNN | 0.6282 | 0.6094 | 0.6413 | 0.6253 | 0.5417 | 0.5735 | 0.6251 | 6.50 |
| Ensemble | 0.6603 | 0.5469 | 0.7391 | 0.6430 | 0.5932 | 0.5691 | 0.6358 | 4.29 |
| SVM* | 0.6667 | 0.6094 | 0.7065 | 0.6579 | 0.5909 | 0.6000 | 0.6562 | 2.57 |
| kNN* | 0.6987 | 0.6250 | 0.7500 | 0.6875 | 0.6349 | 0.6299 | 0.6847 | 1.14 |
| Classifier | Accuracy | TPR | TNR | AUC | Precision | F-Score | G-Mean | Mean Rank |
|---|---|---|---|---|---|---|---|---|
| FDT | 0.7064 | 0.7091 | 0.7037 | 0.7064 | 0.7091 | 0.7091 | 0.7064 | 4.00 |
| LDA | 0.6514 | 0.6727 | 0.6296 | 0.6512 | 0.6491 | 0.6607 | 0.6508 | 7.29 |
| LR | 0.6697 | 0.7091 | 0.6296 | 0.6694 | 0.6610 | 0.6842 | 0.6682 | 6.29 |
| KNB | 0.6239 | 0.5636 | 0.6852 | 0.6244 | 0.6458 | 0.6019 | 0.6214 | 8.00 |
| LSVM | 0.7064 | 0.8000 | 0.6111 | 0.7056 | 0.6769 | 0.7333 | 0.6992 | 4.93 |
| CKNN | 0.6514 | 0.8182 | 0.4815 | 0.6498 | 0.6164 | 0.7031 | 0.6276 | 7.07 |
| Ensemble | 0.7156 | 0.7818 | 0.6481 | 0.7150 | 0.6935 | 0.7350 | 0.7119 | 3.57 |
| SVM* | 0.7523 | 0.7818 | 0.7222 | 0.7520 | 0.7414 | 0.7611 | 0.7514 | 1.64 |
| kNN* | 0.7431 | 0.8364 | 0.6481 | 0.7423 | 0.7077 | 0.7667 | 0.7363 | 2.21 |
| Classifier | Accuracy | TPR | TNR | AUC | Precision | F-Score | G-Mean | Mean Rank |
|---|---|---|---|---|---|---|---|---|
| CDT | 0.6061 | 0.6702 | 0.5211 | 0.5957 | 0.6495 | 0.6597 | 0.5910 | 8.57 |
| LDA | 0.6667 | 0.7234 | 0.5915 | 0.6575 | 0.7010 | 0.7120 | 0.6542 | 5.36 |
| LR | 0.6606 | 0.7234 | 0.5775 | 0.6504 | 0.6939 | 0.7083 | 0.6463 | 6.43 |
| KNB | 0.6788 | 0.7979 | 0.5211 | 0.6595 | 0.6881 | 0.7389 | 0.6448 | 5.93 |
| CGSVM | 0.6727 | 0.9149 | 0.3521 | 0.6335 | 0.6515 | 0.7611 | 0.5676 | 6.29 |
| CKNN | 0.6909 | 0.7766 | 0.5775 | 0.6770 | 0.7087 | 0.7411 | 0.6697 | 3.43 |
| Ensemble | 0.6909 | 0.7979 | 0.5493 | 0.6736 | 0.7009 | 0.7463 | 0.6620 | 4.29 |
| SVM* | 0.7091 | 0.8511 | 0.5211 | 0.6861 | 0.7018 | 0.7692 | 0.6660 | 3.14 |
| KNN* | 0.7333 | 0.8617 | 0.5634 | 0.7125 | 0.7232 | 0.7864 | 0.6968 | 1.57 |
| Classifier | All | Caucasian | Hispanic | Females | Males | Age ≤ 50 | Age > 50 | Average Rank |
|---|---|---|---|---|---|---|---|---|
| DT | 8.97 | 3.29 | 8.29 | 4.86 | 8.64 | 4.00 | 8.57 | 6.66 |
| LDA | 5.79 | 6.71 | 6.50 | 5.57 | 4.07 | 7.29 | 5.36 | 5.90 |
| LR | 5.57 | 6.21 | 8.00 | 8.00 | 5.14 | 6.29 | 6.43 | 6.52 |
| NB | 7.71 | 7.93 | 5.64 | 5.00 | 5.86 | 8.00 | 5.93 | 6.58 |
| SVM | 5.71 | 5.79 | 2.86 | 5.79 | 6.79 | 4.93 | 6.29 | 5.45 |
| KNN | 4.21 | 4.36 | 5.07 | 4.36 | 6.50 | 7.07 | 3.43 | 5.00 |
| Ensemble | 3.93 | 5.07 | 4.57 | 2.43 | 4.29 | 3.57 | 4.29 | 4.02 |
| SVM* | 2.14 | 3.36 | 3.00 | 5.79 | 2.57 | 1.64 | 3.14 | 3.09 |
| KNN* | 1.14 | 2.29 | 1.07 | 3.21 | 1.14 | 2.21 | 1.57 | 1.80 |
| Classifier | All | Caucasian | Hispanic | Females | Males | Age ≤ 50 | Age > 50 |
|---|---|---|---|---|---|---|---|
| DT | 0.5876 | 0.7285 | 0.6260 | 0.7119 | 0.5256 | 0.7064 | 0.6061 |
| LDA | 0.6861 | 0.6908 | 0.6423 | 0.6695 | 0.6538 | 0.6514 | 0.6667 |
| LR | 0.6861 | 0.6887 | 0.6260 | 0.6102 | 0.6474 | 0.6697 | 0.6606 |
| NB | 0.6642 | 0.6887 | 0.6504 | 0.6356 | 0.6234 | 0.6239 | 0.6788 |
| SVM | 0.6898 | 0.7219 | 0.6829 | 0.7203 | 0.6346 | 0.7064 | 0.6727 |
| kNN | 0.6934 | 0.7483 | 0.6585 | 0.7458 | 0.6282 | 0.6514 | 0.6909 |
| Ensemble | 0.7044 | 0.7219 | 0.6585 | 0.7458 | 0.6603 | 0.7156 | 0.6909 |
| SVM* | 0.7226 | 0.7351 | 0.6748 | 0.7203 | 0.6667 | 0.7523 | 0.7091 |
| kNN* | 0.7409 | 0.7483 | 0.7724 | 0.7373 | 0.6987 | 0.7431 | 0.7333 |
| mean Rank | 3.44 | 1.33 | 5.00 | 3.44 | 6.44 | 3.78 | 4.56 |
| Classifier | All | Caucasion | Hispanic | Females | Males | Age ≤ 50 | Age > 50 | Average Rank |
|---|---|---|---|---|---|---|---|---|
| DT | 1.7018 | 0.1440 | 0.1093 | 0.1185 | 0.1123 | 0.0911 | 0.1280 | 1.29 |
| LDA | 1.6479 | 0.1742 | 0.1670 | 0.1720 | 0.1729 | 0.1400 | 0.1654 | 2.14 |
| LR | 0.3802 | 0.3067 | 0.2393 | 0.2837 | 0.2654 | 0.2357 | 0.2433 | 4.00 |
| NB | 3.6635 | 2.1350 | 3.0485 | 2.7358 | 3.8645 | 4.1244 | 3.2748 | 6.86 |
| SVM | 2.9095 | 0.2939 | 0.2926 | 0.2804 | 0.2520 | 0.2577 | 0.2710 | 4.57 |
| KNN | 1.7064 | 0.1768 | 0.1940 | 0.1648 | 0.2247 | 0.1761 | 0.1751 | 3.00 |
| Ensemble | 4.2913 | 1.9599 | 2.1277 | 1.8031 | 1.9468 | 2.0706 | 2.0714 | 6.14 |
| Dataset | Measure | BPSO1 | BPSO2 | BPSO3 | BPSO4 |
|---|---|---|---|---|---|
| All | AVG | 0.7511 | 0.7518 | 0.7464 | 0.7449 |
| STD | 0.0075 | 0.0045 | 0.0039 | 0.0040 | |
| Caucasian | AVG | 0.7967 | 0.7954 | 0.7841 | 0.7808 |
| STD | 0.0136 | 0.0114 | 0.0084 | 0.0091 | |
| Females | AVG | 0.8034 | 0.7941 | 0.7949 | 0.7907 |
| STD | 0.0078 | 0.0070 | 0.0067 | 0.0098 | |
| Age > 50 | AVG | 0.7836 | 0.7782 | 0.7752 | 0.7655 |
| STD | 0.0111 | 0.0111 | 0.0092 | 0.0064 | |
| Hispanic | AVG | 0.7870 | 0.7862 | 0.7797 | 0.7740 |
| STD | 0.0064 | 0.0067 | 0.0071 | 0.0107 | |
| Age ≤ 50 | AVG | 0.8321 | 0.8211 | 0.8092 | 0.7945 |
| STD | 0.0168 | 0.0099 | 0.0149 | 0.0131 | |
| Males | AVG | 0.7513 | 0.7205 | 0.7186 | 0.7180 |
| STD | 0.0159 | 0.0081 | 0.0047 | 0.0043 | |
| Mean Rank | F-test | 1.14 | 2.00 | 2.86 | 4.00 |
| Dataset | Measure | BPSO1 | BPSO2 | BPSO3 | BPSO4 |
|---|---|---|---|---|---|
| All | AVG | 17.6 | 16.4 | 17.7 | 16.9 |
| STD | 2.5473 | 3.0984 | 2.8304 | 2.6854 | |
| Caucasian | AVG | 13.0 | 13.7 | 14.9 | 14.5 |
| STD | 3.2660 | 2.9458 | 3.4785 | 2.7988 | |
| Females | AVG | 14.1 | 14.4 | 14.8 | 13.2 |
| STD | 1.8529 | 2.2211 | 2.0440 | 3.1903 | |
| Age > 50 | AVG | 14.8 | 15.0 | 15.4 | 16.0 |
| STD | 2.6583 | 2.4944 | 1.8974 | 3.4641 | |
| Hispanic | AVG | 14.6 | 14.7 | 15.4 | 16.2 |
| STD | 0.6992 | 2.0575 | 2.5473 | 2.8206 | |
| Age ≤ 50 | AVG | 14.8 | 15.6 | 15.4 | 15.5 |
| STD | 1.3166 | 1.7764 | 2.3190 | 2.5927 | |
| Males | AVG | 12.5 | 12.8 | 14.8 | 13.6 |
| STD | 2.8771 | 3.1903 | 2.4404 | 1.3499 | |
| Mean Rank | F-test | 1.43 | 2.29 | 3.43 | 2.86 |
| Dataset | Measure | BPSO1 | BPSO2 | BPSO3 | BPSO4 |
|---|---|---|---|---|---|
| All | AVG | 464.0 | 481.7 | 466.2 | 467.8 |
| STD | 4.8617 | 3.6674 | 3.3655 | 3.2812 | |
| caucasion | AVG | 368.1 | 374.5 | 371.8 | 374.0 |
| STD | 4.3402 | 4.2569 | 3.6641 | 3.5733 | |
| females | AVG | 245.8 | 247.9 | 247.9 | 248.4 |
| STD | 2.1698 | 2.1417 | 1.7136 | 2.1340 | |
| age > 50 | AVG | 266.9 | 270.2 | 268.7 | 270.0 |
| STD | 2.5395 | 2.2341 | 2.4543 | 1.5417 | |
| hispanic | AVG | 260.5 | 262.9 | 262.1 | 263.9 |
| STD | 2.9500 | 2.1613 | 1.8231 | 2.4382 | |
| age ≤ 50 | AVG | 261.4 | 262.1 | 264.8 | 263.7 |
| STD | 2.3115 | 1.7837 | 2.1478 | 2.0247 | |
| males | AVG | 382.7 | 251.8 | 250.3 | 250.9 |
| STD | 46.5774 | 1.8658 | 1.8342 | 1.7393 | |
| mean rank | F-test | 1.43 | 3.21 | 2.21 | 3.14 |
| Dataset | Measure | BPSO1 | BHHO | BGSA | BWOA | BGWO | BBA | BALO | BMFO |
|---|---|---|---|---|---|---|---|---|---|
| All | AVG | 0.7511 | 0.7515 | 0.7245 | 0.7507 | 0.7372 | 0.6624 | 0.7474 | 0.7504 |
| STD | 0.0075 | 0.0095 | 0.0086 | 0.0042 | 0.0064 | 0.0472 | 0.0038 | 0.0039 | |
| Caucasian | AVG | 0.7967 | 0.7821 | 0.7430 | 0.7815 | 0.7623 | 0.7060 | 0.7781 | 0.7821 |
| STD | 0.0136 | 0.0049 | 0.0061 | 0.0054 | 0.0049 | 0.0356 | 0.0056 | 0.0058 | |
| Females | AVG | 0.8034 | 0.8068 | 0.7517 | 0.8093 | 0.7864 | 0.6949 | 0.8017 | 0.8059 |
| STD | 0.0078 | 0.0067 | 0.0106 | 0.0072 | 0.0088 | 0.0344 | 0.0091 | 0.0063 | |
| Age > 50 | AVG | 0.7836 | 0.7703 | 0.7236 | 0.7691 | 0.7473 | 0.6945 | 0.7649 | 0.7721 |
| STD | 0.0111 | 0.0078 | 0.0122 | 0.0097 | 0.0081 | 0.0192 | 0.0110 | 0.0077 | |
| Hispanic | AVG | 0.7870 | 0.7805 | 0.7382 | 0.7789 | 0.7683 | 0.6805 | 0.7813 | 0.7772 |
| STD | 0.0064 | 0.0094 | 0.0120 | 0.0064 | 0.0103 | 0.0617 | 0.0071 | 0.0042 | |
| Age ≤ 50 | AVG | 0.8321 | 0.7991 | 0.7376 | 0.7991 | 0.7661 | 0.6661 | 0.7835 | 0.7853 |
| STD | 0.0168 | 0.0110 | 0.0189 | 0.0091 | 0.0108 | 0.0604 | 0.0151 | 0.0064 | |
| Males | AVG | 0.7513 | 0.7224 | 0.6949 | 0.7154 | 0.7090 | 0.6430 | 0.7160 | 0.7160 |
| STD | 0.0159 | 0.0053 | 0.0106 | 0.0033 | 0.0033 | 0.0327 | 0.0031 | 0.0031 | |
| Mean Rank | F-test | 1.57 | 2.29 | 7.00 | 3.36 | 6.00 | 8.00 | 4.36 | 3.43 |
| Dataset | BPSO (the Best Performaing Method) vs. | ||||||
|---|---|---|---|---|---|---|---|
| BHHO | BGSA | BWOA | BGWO | BBA | BALO | BMFO | |
| All | |||||||
| Caucasian | |||||||
| Females | |||||||
| Age > 50 | |||||||
| Hispanic | |||||||
| Age ≤ 50 | |||||||
| Males | |||||||
| Dataset | Measure | BPSO1 | BHHO | BGSA | BWOA | BGWO | BBA | BALO | BMFO |
|---|---|---|---|---|---|---|---|---|---|
| All | AVG | 17.6 | 22.8 | 18.1 | 22.2 | 27.4 | 13 | 25.1 | 24.2 |
| STD | 2.55 | 2.10 | 2.23 | 2.66 | 1.17 | 2.11 | 1.37 | 1.81 | |
| Caucasian | AVG | 13.0 | 19.9 | 14.6 | 21.5 | 26.2 | 12.1 | 24.6 | 21.7 |
| STD | 3.27 | 3.28 | 2.63 | 2.32 | 0.79 | 2.33 | 1.35 | 1.89 | |
| Females | AVG | 14.1 | 23 | 15 | 22.8 | 27 | 13.4 | 24 | 24.8 |
| STD | 1.85 | 1.56 | 1.89 | 2.39 | 1.56 | 2.17 | 1.33 | 1.69 | |
| Age > 50 | AVG | 14.8 | 18.6 | 16.9 | 21.3 | 27.3 | 15.1 | 25.6 | 22.5 |
| STD | 2.66 | 5.87 | 2.88 | 3.47 | 1.64 | 2.42 | 1.65 | 2.12 | |
| Hispanic | AVG | 14.6 | 19.6 | 17.3 | 20.7 | 25.2 | 13 | 23.3 | 21.1 |
| STD | 0.70 | 3.78 | 3.53 | 3.59 | 1.23 | 2.45 | 1.89 | 2.38 | |
| Age ≤ 50 | AVG | 14.8 | 18.3 | 16.4 | 19 | 25.1 | 12.6 | 23.4 | 19.5 |
| STD | 1.32 | 2.11 | 2.80 | 1.83 | 1.20 | 2.46 | 1.51 | 1.18 | |
| Males | AVG | 12.5 | 19.1 | 14.9 | 18.6 | 25.5 | 11.3 | 22.5 | 21.4 |
| STD | 2.88 | 5.24 | 1.60 | 3.75 | 1.84 | 4.57 | 1.96 | 2.27 | |
| Mean Rank | F-test | 1.86 | 4.43 | 3.00 | 4.57 | 8.00 | 1.14 | 6.86 | 6.14 |
| Dataset | BPSO (the Best Performaing Method) vs. | ||||||
|---|---|---|---|---|---|---|---|
| BHHO | BGSA | BWOA | BGWO | BBA | BALO | BMFO | |
| Caucasian | |||||||
| Females | |||||||
| Age > 50 | |||||||
| Hispanic | |||||||
| Age ≤ 50 | |||||||
| Males | |||||||
| Dataset | Measure | BPSO1 | BHHO | BGSA | BWOA | BGWO | BBA | BALO | BMFO |
|---|---|---|---|---|---|---|---|---|---|
| all | AVG | 464.05 | 798.02 | 465.45 | 476.24 | 475.84 | 468.76 | 474.47 | 468.78 |
| STD | 4.862 | 9.414 | 4.992 | 4.813 | 5.720 | 5.622 | 7.119 | 6.179 | |
| caucasion | AVG | 368.14 | 613.92 | 376.20 | 378.73 | 377.41 | 376.77 | 375.30 | 374.28 |
| STD | 4.340 | 5.295 | 3.015 | 3.928 | 4.045 | 3.191 | 5.114 | 4.507 | |
| females | AVG | 245.75 | 401.38 | 248.90 | 248.18 | 249.38 | 250.27 | 247.90 | 247.08 |
| STD | 2.170 | 4.390 | 2.088 | 2.839 | 2.664 | 1.802 | 2.385 | 2.445 | |
| age > 50 | AVG | 266.88 | 441.47 | 267.54 | 272.05 | 272.21 | 269.97 | 269.77 | 269.41 |
| STD | 2.540 | 4.207 | 2.024 | 2.318 | 2.357 | 2.169 | 3.116 | 2.618 | |
| hispanic | AVG | 260.48 | 431.94 | 264.26 | 261.83 | 264.53 | 265.01 | 262.17 | 261.13 |
| STD | 2.950 | 4.529 | 1.983 | 2.527 | 2.019 | 3.110 | 2.884 | 1.943 | |
| age ≤ 50 | AVG | 261.38 | 429.46 | 266.24 | 263.29 | 266.10 | 265.21 | 262.29 | 262.48 |
| STD | 2.311 | 3.038 | 2.525 | 1.785 | 2.330 | 2.237 | 3.266 | 1.700 | |
| males | AVG | 382.68 | 409.66 | 250.95 | 249.72 | 251.26 | 255.21 | 249.02 | 249.54 |
| STD | 46.577 | 4.520 | 1.540 | 2.240 | 1.781 | 3.173 | 2.732 | 2.101 | |
| mean rank | F-test | 1.86 | 8.00 | 4.14 | 4.86 | 6.00 | 5.43 | 3.14 | 2.57 |
| Dataset | KNN* | BPSO-KNN | Improvement Rate | |||
|---|---|---|---|---|---|---|
| Accuracy | No. Features | Accuracy | No. Features | Features Reduction | Accuracy | |
| All | 0.7409 | 31 | 0.7628 | 18 | 41.94% | 2.19% |
| Caucasion | 0.7483 | 30 | 0.8080 | 13 | 56.67% | 5.96% |
| Hispanic | 0.7724 | 30 | 0.7968 | 14 | 53.33% | 2.44% |
| Females | 0.7373 | 30 | 0.8136 | 13 | 56.67% | 7.63% |
| Males | 0.6987 | 30 | 0.7885 | 11 | 63.33% | 8.97% |
| Age ≤ 50 | 0.7431 | 31 | 0.8624 | 15 | 51.61% | 11.93% |
| Age > 50 | 0.7333 | 31 | 0.8061 | 17 | 45.16% | 7.27% |
| Dataset | Accuracy | #Features | f1 | f2 | f3 | f4 | f5 | f6 | f7 | f8 | f9 | f10 | f11 | f12 | f13 | f14 | f15 | f16 | f17 | f18 | f19 | f20 | f21 | f22 | f23 | f24 | f25 | f26 | f27 | f28 | f29 | f30 | f31 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| all | 0.7628 | 18 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| caucasian | 0.8080 | 13 | - | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 |
| Hispanic | 0.7968 | 14 | - | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| females | 0.8136 | 13 | 1 | 0 | - | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| males | 0.7885 | 11 | 0 | 0 | - | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 |
| age<=50 | 0.8624 | 15 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 |
| age>50 | 0.8061 | 17 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| Dataset | CNN | MLP | kNN* | BPSO-kNN | ||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Time | Accuracy | Time | Accuracy | Time | Accuracy | Time | |
| All | 0.6105 | 291.656 | 0.5438 | 0.789 | 0.7409 | 1.706 | 0.7628 | 464.050 |
| Caucasion | 0.7283 | 204.591 | 0.6159 | 3.282 | 0.7483 | 0.177 | 0.8080 | 368.142 |
| Hispanic | 0.6513 | 180.510 | 0.5285 | 2.341 | 0.7724 | 0.194 | 0.7968 | 245.754 |
| Females | 0.7023 | 208.605 | 0.6102 | 3.398 | 0.7373 | 0.165 | 0.8136 | 266.882 |
| Males | 0.6263 | 174.349 | 0.5769 | 2.969 | 0.6987 | 0.225 | 0.7885 | 260.481 |
| Age ≤ 50 | 0.6427 | 177.557 | 0.5780 | 2.917 | 0.7431 | 0.176 | 0.8624 | 261.377 |
| Age > 50 | 0.6629 | 219.354 | 0.5455 | 3.483 | 0.7333 | 0.175 | 0.8061 | 382.676 |
| Results of NAMES [30] | Proposed (BPSO-KNN) | Haberfeld et al. [28] | Surani et al. [27] | ||||
|---|---|---|---|---|---|---|---|
| Combination | AUC | Dataset | Average AUC | SVM | LR | LR | ANN |
| NC + MF + CM + ESS + S + BMI | 0.6577 | all | 0.7438 | ||||
| NC + MF + CM + ESS + S + M | 0.6572 | caucasion | 0.7690 | ||||
| NC + MF + CM + ESS + S + BMI + M (NAMES2) | 0.6690 | hispanic | 0.7811 | ||||
| NC + MF + M + ESS + S | 0.6583 | females | 0.6707 | 0.6220 | 0.6080 | 0.7030 | 0.5830 |
| BMI + MF + CM + ESS + S + M | 0.6436 | males | 0.7318 | 0.6070 | 0.6070 | 0.7130 | 0.6360 |
| (NC + MF) × 2 + CM + ESS + S | 0.6661 | age ≤ 50 | 0.8320 | ||||
| (NC + BMI) × 2 + M + ESS + S | 0.6433 | ag > 50 | 0.7684 | ||||
| (NC + MF) × 2 + M + ESS + S | 0.6484 | ||||||
| (NC + BMI) × 2 + CM + ESS + S | 0.6426 | ||||||
| (NC + MF + BMI)×2 + CM + ESS + S + M | 0.6478 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheta, A.; Thaher, T.; Surani, S.R.; Turabieh, H.; Braik, M.; Too, J.; Abu-El-Rub, N.; Mafarjah, M.; Chantar, H.; Subramanian, S. Diagnosis of Obstructive Sleep Apnea Using Feature Selection, Classification Methods, and Data Grouping Based Age, Sex, and Race. Diagnostics 2023, 13, 2417. https://doi.org/10.3390/diagnostics13142417
Sheta A, Thaher T, Surani SR, Turabieh H, Braik M, Too J, Abu-El-Rub N, Mafarjah M, Chantar H, Subramanian S. Diagnosis of Obstructive Sleep Apnea Using Feature Selection, Classification Methods, and Data Grouping Based Age, Sex, and Race. Diagnostics. 2023; 13(14):2417. https://doi.org/10.3390/diagnostics13142417
Chicago/Turabian StyleSheta, Alaa, Thaer Thaher, Salim R. Surani, Hamza Turabieh, Malik Braik, Jingwei Too, Noor Abu-El-Rub, Majdi Mafarjah, Hamouda Chantar, and Shyam Subramanian. 2023. "Diagnosis of Obstructive Sleep Apnea Using Feature Selection, Classification Methods, and Data Grouping Based Age, Sex, and Race" Diagnostics 13, no. 14: 2417. https://doi.org/10.3390/diagnostics13142417
APA StyleSheta, A., Thaher, T., Surani, S. R., Turabieh, H., Braik, M., Too, J., Abu-El-Rub, N., Mafarjah, M., Chantar, H., & Subramanian, S. (2023). Diagnosis of Obstructive Sleep Apnea Using Feature Selection, Classification Methods, and Data Grouping Based Age, Sex, and Race. Diagnostics, 13(14), 2417. https://doi.org/10.3390/diagnostics13142417






