Human Parainfluenza Virus (HPIV) Detection in Hospitalized Children with Acute Respiratory Tract Infection in the Western Cape, South Africa during 2014–2022 Reveals a Shift in Dominance of HPIV 3 and 4 Infections
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Testing of Clinical Samples
2.3. Typing of HPIV Positive Samples Collected during 2020–2022
2.4. Subtyping of HPIV 4 Positive Samples Collected during 2014–2017
2.5. Phylogenetic Analysis of HPIV 4 Positive Sequences
2.6. Comparing Respiratory Virus Data Generated from Anyplex RV16 and Allplex Essential Assays
3. Results
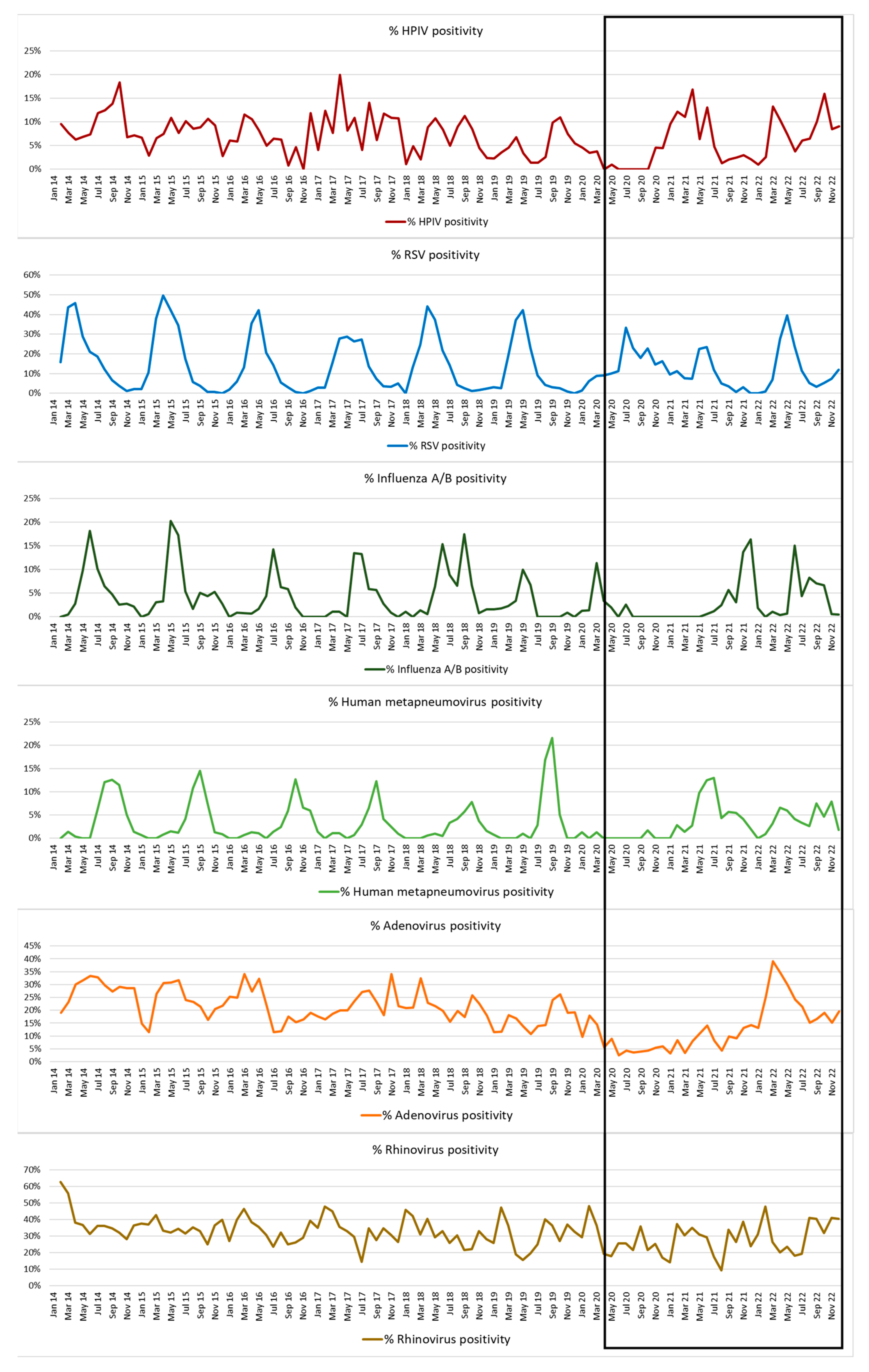
3.1. Respiratory Virus Detections during 2014–2022
3.2. HPIV-Positive Samples
3.3. HPIV Co-Infections
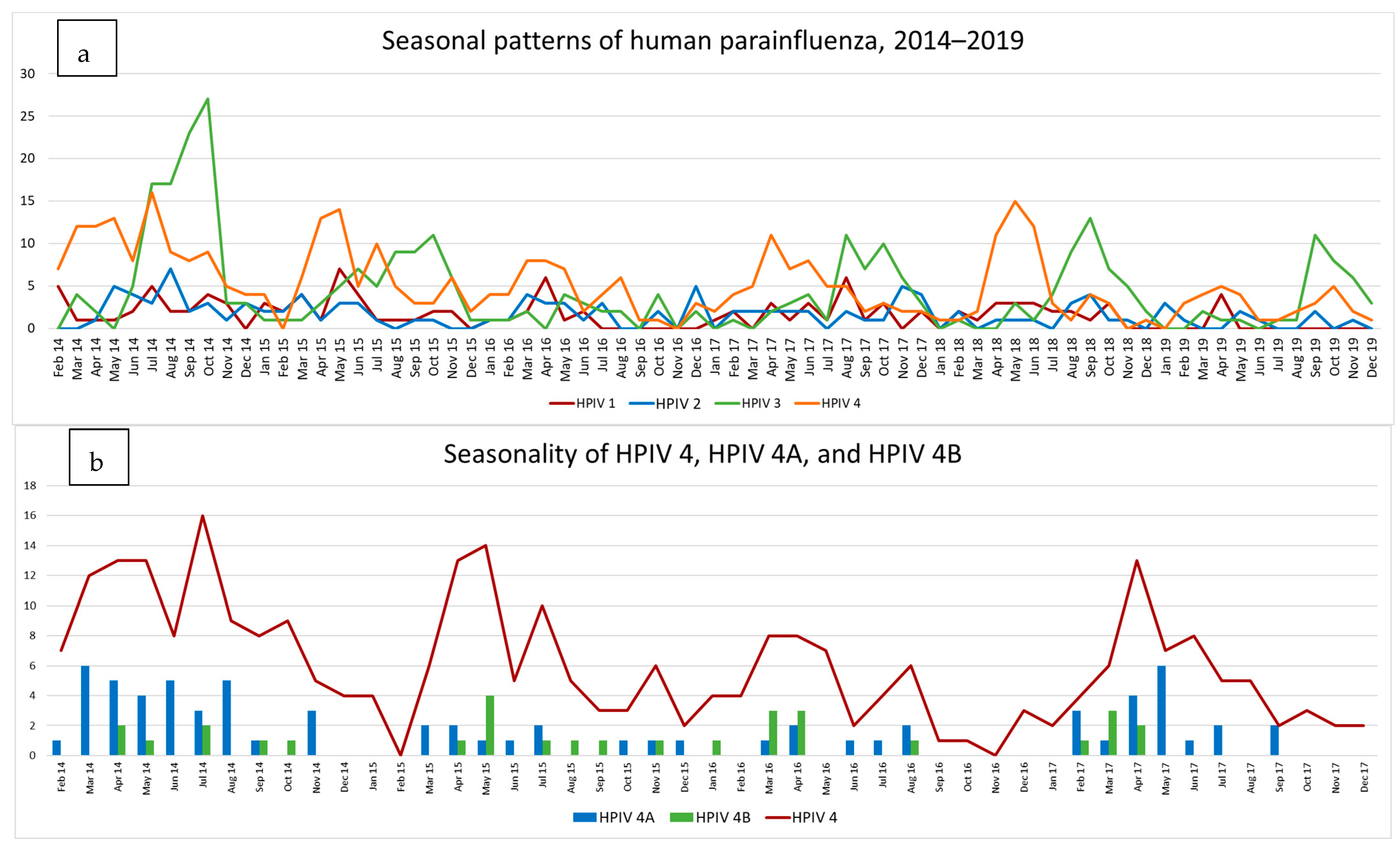
3.4. HPIV Activity during 2014–2019
3.4.1. Type Specific Prevalence
3.4.2. HPIV 4A and B
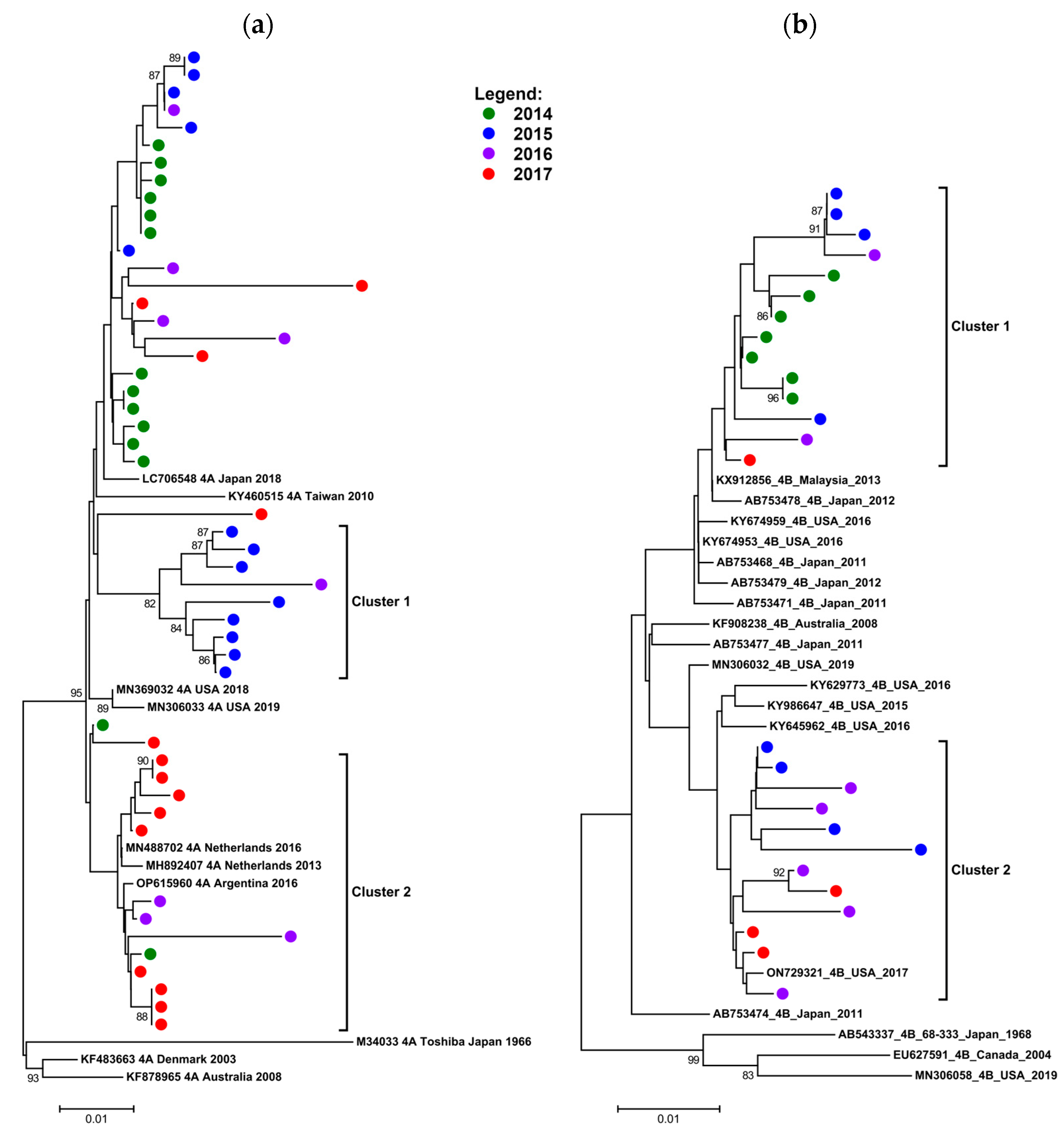
3.4.3. Phylogenetic Data on HPIV 4A and B (2014–2017)
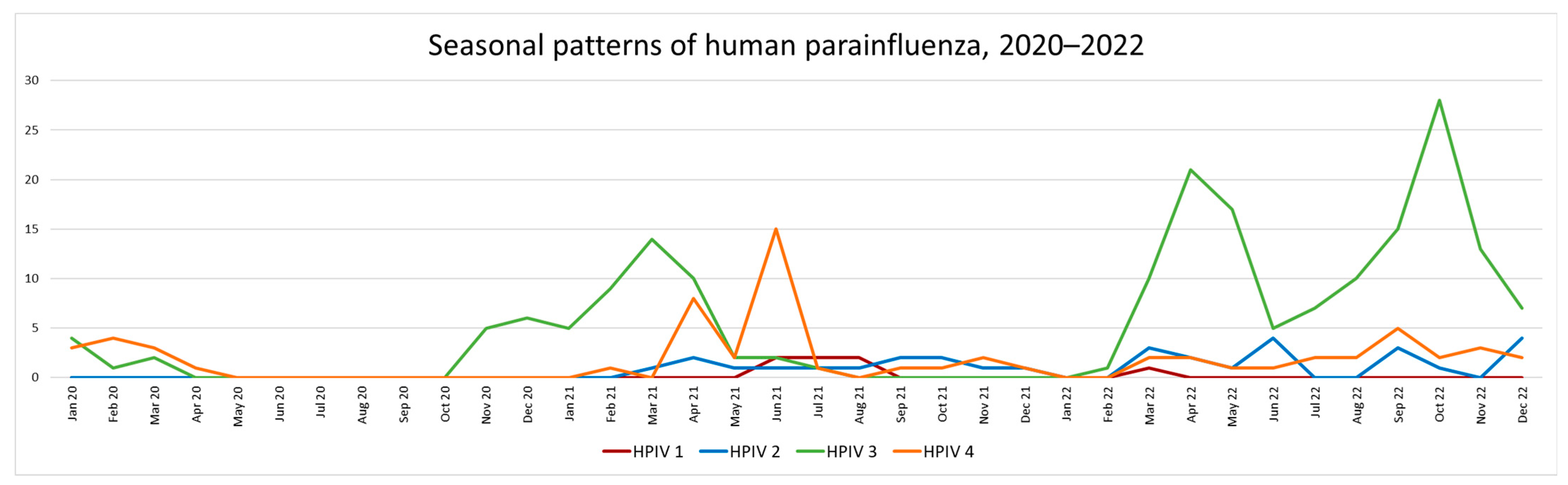
3.5. HPIV Activity during 2020–2022 (Pandemic Period)
Type Specific Prevalence 2020–2022
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, L.; Oza, S.; Hogan, D.; Perin, J.; Rudan, I.; Lawn, J.E.; Cousens, S.; Mathers, C.; Black, R.E. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: An updated systematic analysis. Lancet 2015, 385, 430–440. [Google Scholar] [CrossRef]
- The Pneumonia Etiology Research for Child Health (PERCH) Study Group. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: The PERCH multi-country case-control study. Lancet 2019, 394, 757–779. [Google Scholar] [CrossRef] [Green Version]
- Zar, H.J.; Barnett, W.; Stadler, A.; Gardner-Lubbe, S.; Myer, L.; Nicol, M.P. Aetiology of childhood pneumonia in a well vaccinated South African birth cohort: A nested case-control study of the Drakenstein Child Health Study. Lancet Respir. Med. 2016, 4, 463–472. [Google Scholar] [CrossRef] [Green Version]
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.; Lee, B.; Rota, P.; et al. Ictv Report Consortium, ICTV Virus Taxonomy Profile: Paramyxoviridae. J. Gen. Virol. 2019, 100, 1593–1594. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Deloria-Knoll, M.; Madhi, S.A.; Cohen, C.; Arguelles, V.L.; Basnet, S.; Bassat, Q.; Brooks, W.A.; Echavarria, M.; et al. Global burden of acute lower respiratory infection associated with human parainfluenza virus in children younger than 5 years for 2018: A systematic review and meta-analysis. Lancet Glob. Health 2021, 9, e1077–e1087. [Google Scholar] [CrossRef] [PubMed]
- Barratt, K.; Anderson, T.P.; Fahey, J.A.; Jennings, L.C.; Werno, A.M.; Murdoch, D.R. Comparison of the fast track diagnostics respiratory 21 and Seegene Allplex multiplex polymerase chain reaction assays for the detection of respiratory viruses. Br. J. Biomed. Sci. 2017, 74, 85–89. Available online: http://www.embase.com/search/results?subaction=viewrecord&from=export&id=L614467297%0A (accessed on 23 June 2023). [CrossRef]
- Virant, M.J.; Uršič, T.; Kogoj, R.; Korva, M.; Petrovec, M.; Avšič-Županc, T. Evaluation of Two Broadly Used Commercial Methods for Detection of Respiratory Viruses with a Recently Added New Target for Detection of SARS-CoV-2. Viruses 2022, 14, 1530. [Google Scholar] [CrossRef]
- Parsons, J. A Molecular Epidemiological Study of Human Parainfluenza Virus 4 in the Western Cape, South Africa. Master’s Thesis, University of Cape Town, Cape Town, South Africa, 2021. Available online: http://hdl.handle.net/11427/36147 (accessed on 23 June 2023).
- Mizuta, K.; Tsukagoshi, H.; Ikeda, T.; Aoki, Y.; Abiko, C.; Itagaki, T.; Nagano, M.; Noda, M.; Kimura, H. Molecular evolution of the haemagglutinin- neuraminidase gene in human parainfluenza virus type 3 isolates from children with acute respiratory illness in Yamagata prefecture, Japan. J. Med. Microbiol. 2014, 63, 570–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, N.; Ji, Y.; Xie, Z.; Wang, H.; Wang, H.; An, J.; Zhang, X.; Zhang, Y.; Zhu, Z.; Cui, A.; et al. Human Parainfluenza Virus-Associated Respiratory Tract Infection among Children and Genetic Analysis of HPIV-3 Strains in Beijing, China. PLoS ONE 2012, 7, e43893. [Google Scholar] [CrossRef] [Green Version]
- Košutić-Gulija, T.; Slovic, A.; Ljubin-Sternak, S.; Mlinarić-Galinović, G.; Fočić, D. Genetic analysis of human parainfluenza virus type 3 obtained in Croatia, 2011–2015. J. Med. Microbiol. 2017, 66, 502–510. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NTo Title. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rafeek, R.A.M.; Divarathna, M.V.M.; Noordeen, F. A review on disease burden and epidemiology of childhood parainfluenza virus infections in Asian countries. Rev. Med. Virol. 2021, 31, e2164. [Google Scholar] [CrossRef]
- Frost, H.M.; Robinson, C.C.; Dominguez, S.R. Epidemiology and clinical presentation of parainfluenza type 4 in children: A 3-year comparative study to parainfluenza types 1–3. J. Infect. Dis. 2014, 209, 695–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.E.; Park, J.Y.; Lee, M.K.; Lim, I.S. Characteristics of human parainfluenza virus type 4 infection in hospitalized children in Korea. Pediatr. Int. 2020, 62, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; Suh, W.; Han, S.B. Seasonal epidemiological and clinical characteristics of pediatric patients with human parainfluenza virus infection by serotype: A retrospective study. Virol. J. 2022, 19, 141. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Biere, B.; Grenz, M.; Wolff, T.; Schweiger, B.; Dürrwald, R.; Reiche, J. Virological surveillance and molecular characterization of human parainfluenzavirus infection in children with acute respiratory illness: Germany, 2015–2019. Microorganisms 2021, 9, 1508. [Google Scholar] [CrossRef]
- Burke, C.W.; Bridges, O.; Brown, S.; Rahija, R.; Russell, C.J. Mode of Parainfluenza Virus Transmission Determines the Dynamics of Primary Infection and Protection from Reinfection. PLoS Pathog. 2013, 9, e1003786. [Google Scholar] [CrossRef] [Green Version]
- Tempia, S.; Moyes, J.; Cohen, A.L.; Walaza, S.; McMorrow, M.L.; Treurnicht, F.K.; Hellferscee, O.; Wolter, N.; von Gottberg, A.; Dawood, H.; et al. The national burden of influenza-like illness and severe respiratory illness overall and associated with nine respiratory viruses in South Africa, 2013–2015. Influenza Other Respi. Viruses 2022, 16, 438–451. [Google Scholar] [CrossRef]
- Degroote, N.P.; Haynes, A.K.; Taylor, C.; Killerby, M.E.; Dahl, R.M.; Mustaquim, D.; Gerber, S.I.; Watson, J.T. Human parainfluenza virus circulation, United States, 2011–2019. J. Clin. Virol. 2020, 124, 104261. [Google Scholar] [CrossRef]
- Xiao, N.-G.; Duan, Z.-J.; Xie, Z.-P.; Zhong, L.-L.; Zeng, S.Z.; Huang, H.; Gao, H.-C. Human parainfluenza virus types 1–4 in hospitalised children with acute lower respiratory infections in China. J. Med. Virol. 2016, 88, 2085–2091. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Li, K.S.M.; Chau, K.Y.; So, L.Y.; Lee, R.A.; Lau, Y.L.; Chan, K.H.; Lim, W.W.L.; Woo, P.C.Y.; Yuen, K.Y. Clinical and molecular epidemiology of human parainfluenza virus 4 infections in Hong Kong: Subtype 4B as common as subtype 4A. J. Clin. Microbiol. 2009, 47, 1549–1552. [Google Scholar] [CrossRef] [Green Version]
- Chellapuri, A.; Smitheman, M.; Chappell, J.G.; Clark, G.; Howson-Wells, H.C.; Berry, L.; Ball, J.K.; Irving, W.L.; Tarr, A.W.; McClure, C.P. Human parainfluenza 2 & 4: Clinical and genetic epidemiology in the UK, 2013–2017, reveals distinct disease features and co-circulating genomic subtypes. Influenza Other Respi. Viruses 2022, 16, 1122–1132. [Google Scholar] [CrossRef]
- Liu, P.; Xu, M.; Cao, L.; Su, L.; Lu, L.; Dong, N.; Jia, R.; Zhu, X.; Xu, J. Impact of COVID-19 pandemic on the prevalence of respiratory viruses in children with lower respiratory tract infections in China. Virol. J. 2021, 18, 159. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Autore, G.; Ramundo, G.; Esposito, S. Epidemiology of Respiratory Infections during the COVID-19 Pandemic. Viruses 2023, 15, 1160. [Google Scholar] [CrossRef]
- Lee, H.; Kim, S.H.; Cho, S.J.; Lee, Y.U.; Lee, K.; Lee, Y.P.; Seo, J.; Chung, Y.S. Genetic Analysis of HPIV3 That Emerged during the SARS-CoV-2 Pandemic in Gwangju, South Korea. Viruses 2022, 14, 1446. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parsons, J.; Korsman, S.; Smuts, H.; Hsiao, N.-Y.; Valley-Omar, Z.; Gelderbloem, T.; Hardie, D. Human Parainfluenza Virus (HPIV) Detection in Hospitalized Children with Acute Respiratory Tract Infection in the Western Cape, South Africa during 2014–2022 Reveals a Shift in Dominance of HPIV 3 and 4 Infections. Diagnostics 2023, 13, 2576. https://doi.org/10.3390/diagnostics13152576
Parsons J, Korsman S, Smuts H, Hsiao N-Y, Valley-Omar Z, Gelderbloem T, Hardie D. Human Parainfluenza Virus (HPIV) Detection in Hospitalized Children with Acute Respiratory Tract Infection in the Western Cape, South Africa during 2014–2022 Reveals a Shift in Dominance of HPIV 3 and 4 Infections. Diagnostics. 2023; 13(15):2576. https://doi.org/10.3390/diagnostics13152576
Chicago/Turabian StyleParsons, Jane, Stephen Korsman, Heidi Smuts, Nei-Yuan Hsiao, Ziyaad Valley-Omar, Tathym Gelderbloem, and Diana Hardie. 2023. "Human Parainfluenza Virus (HPIV) Detection in Hospitalized Children with Acute Respiratory Tract Infection in the Western Cape, South Africa during 2014–2022 Reveals a Shift in Dominance of HPIV 3 and 4 Infections" Diagnostics 13, no. 15: 2576. https://doi.org/10.3390/diagnostics13152576
APA StyleParsons, J., Korsman, S., Smuts, H., Hsiao, N.-Y., Valley-Omar, Z., Gelderbloem, T., & Hardie, D. (2023). Human Parainfluenza Virus (HPIV) Detection in Hospitalized Children with Acute Respiratory Tract Infection in the Western Cape, South Africa during 2014–2022 Reveals a Shift in Dominance of HPIV 3 and 4 Infections. Diagnostics, 13(15), 2576. https://doi.org/10.3390/diagnostics13152576






