Update on the Pathogenesis of Enteropathy-Associated T-Cell Lymphoma
Abstract
1. Introduction
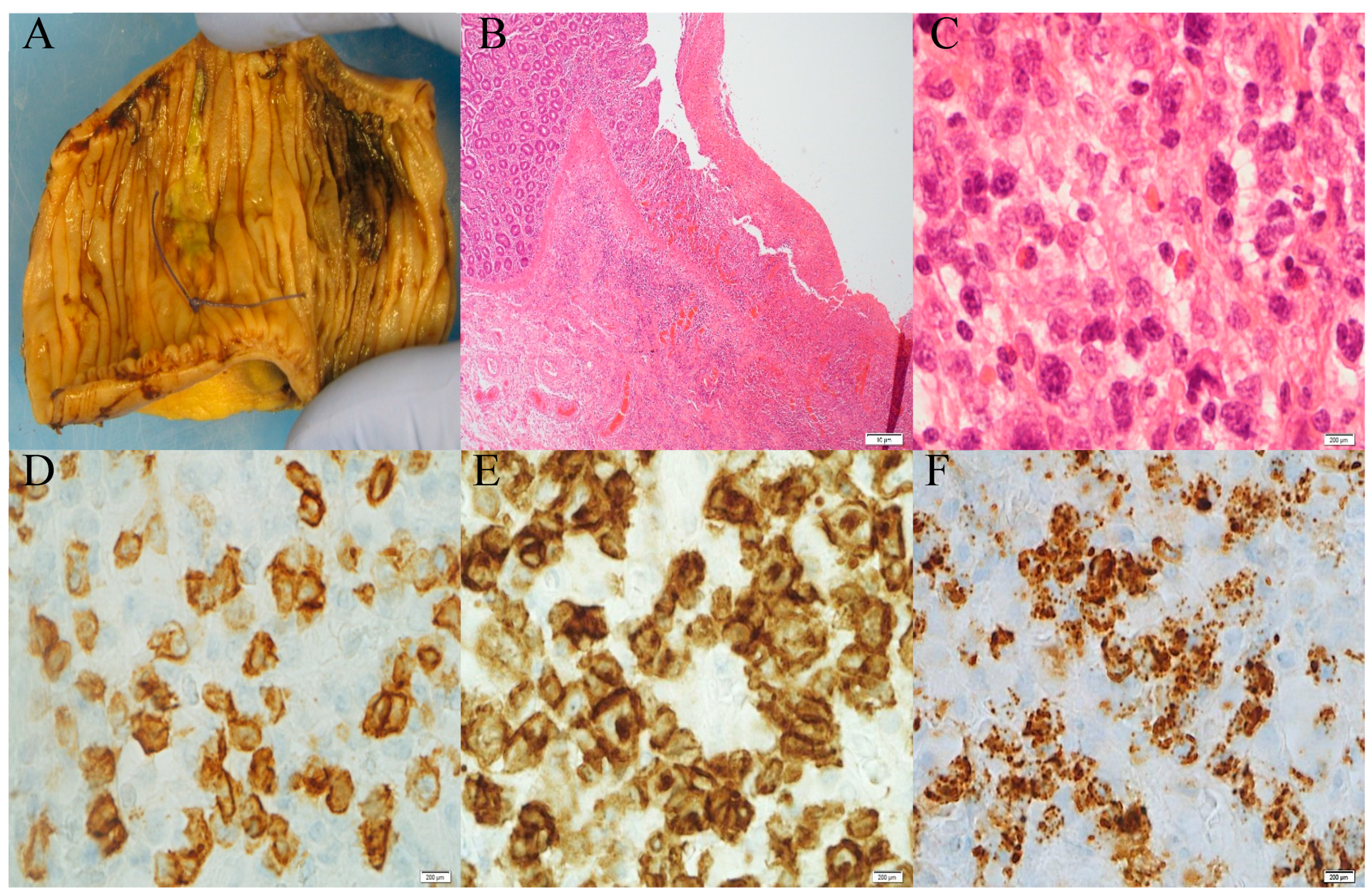
2. Establishment of Clinical Diagnosis
3. Genetics and Environmental Factors
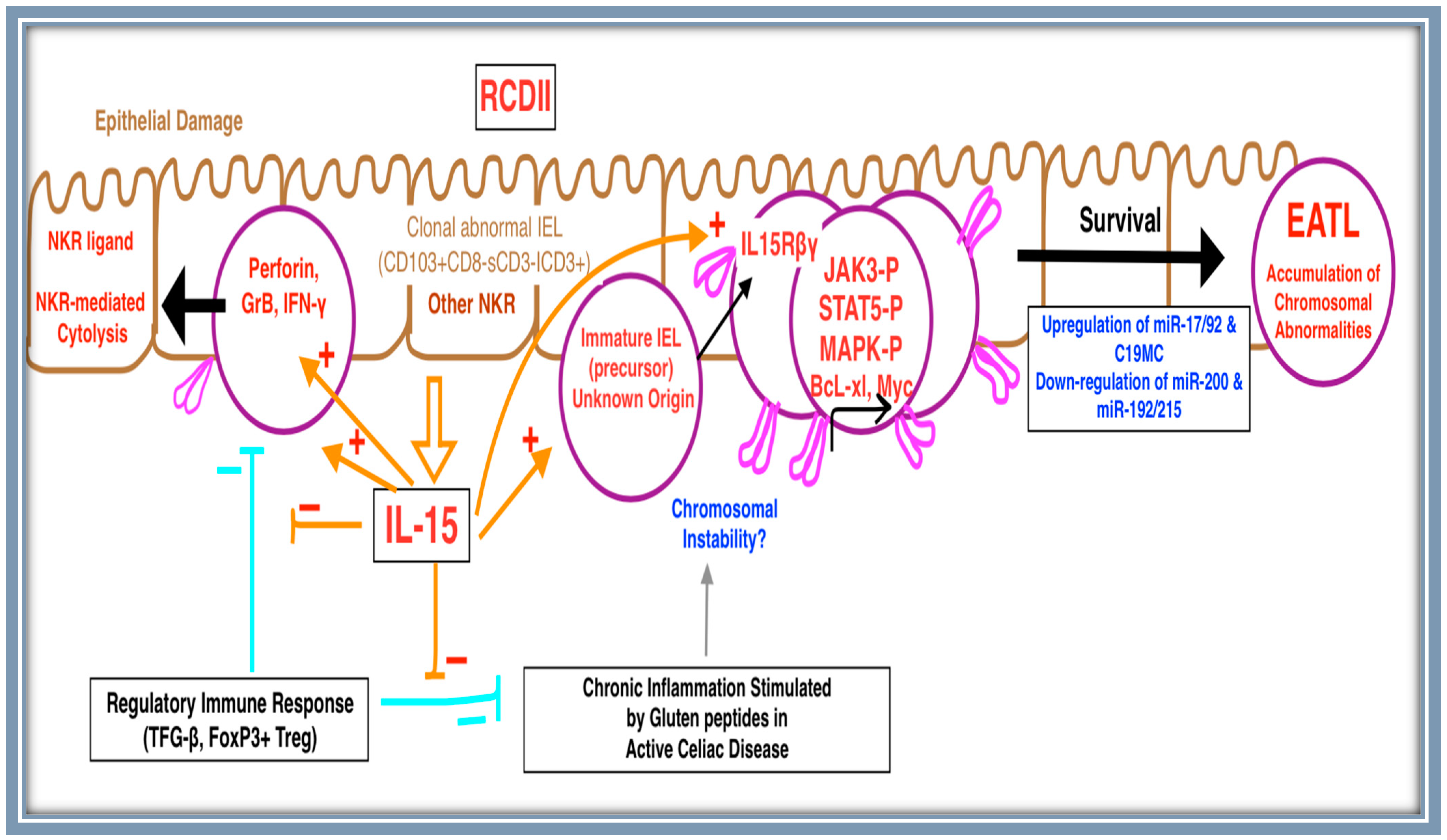
4. Molecular Pathogenesis
4.1. Cell of Origin
4.2. Cytokine Signaling
4.3. Mutational Landscape
4.4. MicroRNA
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferreri, A.J.; Zinzani, P.L.; Govi, S.; Pileri, S.A. Enteropathy-associated T-cell lymphoma. Crit. Rev. Oncol. 2011, 79, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Tomita, S.; Kikuti, Y.Y.; Carreras, J.; Kojima, M.; Ando, K.; Takasaki, H.; Sakai, R.; Takata, K.; Yoshino, T.; Bea, S.; et al. Genomic and immunohistochemical profiles of enteropathy-associated T-cell lymphoma in Japan. Mod. Pathol. 2015, 28, 1286–1296. [Google Scholar] [CrossRef] [PubMed]
- Fei, F.; Reddy, V.; Patel, C.R.; Dhall, D.; Lee, G.; Meng-Jun, X.; Al Diffalha, S. Monomorphic Epitheliotropic Intestinal T-cell Lymphoma: A Study of Four Cases and Review of Literature. Ann. Clin. Lab. Sci. 2020, 50, 806–812. [Google Scholar]
- Ritter, J.; Zimmermann, K.; Jöhrens, K.; Mende, S.; Seegebarth, A.; Siegmund, B.; Hennig, S.; Todorova, K.; Rosenwald, A.; Daum, S.; et al. T-cell repertoires in refractory coeliac disease. Gut 2018, 67, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Mir, B.A.; Majeed, T.; Singh, A.; Rajput, M.S.; Kumar, A.; Chauhan, A. Emerging Biomarkers for Screening and Management of Celiac Disease. BioMed Res. Int. 2022, 2022, 2756242. [Google Scholar] [CrossRef]
- Brown, J.R.G.; Singh, P. Coeliac disease. Paediatr. Int. Child Health 2018, 39, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Delabie, J.; Holte, H.; Vose, J.M.; Ullrich, F.; Jaffe, E.S.; Savage, K.J.; Connors, J.M.; Rimsza, L.; Harris, N.L.; Müller-Hermelink, K.; et al. Enteropathy-associated T-cell lymphoma: Clinical and histological findings from the International Peripheral T-Cell Lymphoma Project. Blood 2011, 118, 148–155. [Google Scholar] [CrossRef]
- Di Sabatino, A.; Biagi, F.; Gobbi, P.G.; Corazza, G.R. How I treat enteropathy-associated T-cell lymphoma. Blood 2012, 119, 2458–2468. [Google Scholar] [CrossRef]
- de Leval, L.; Feldman, A.L.; Pileri, S.; Nakamura, S.; Gaulard, P. Extranodal T- and NK-cell lymphomas. Virchows Arch. 2023, 482, 245–264. [Google Scholar] [CrossRef] [PubMed]
- Silano, M.; Agostoni, C.; Sanz, Y.; Guandalini, S. Infant feeding and risk of developing celiac disease: A systematic review. BMJ Open 2016, 6, e009163. [Google Scholar] [CrossRef]
- Kooy-Winkelaar, Y.M.C.; Bouwer, D.; Janssen, G.M.C.; Thompson, A.; Brugman, M.H.; Schmitz, F.; de Ru, A.H.; van Gils, T.; Bouma, G.; van Rood, J.J.; et al. CD4 T-cell cytokines synergize to induce proliferation of malignant and nonmalignant innate intraepithelial lymphocytes. Proc. Natl. Acad. Sci. USA 2017, 114, E980–E989. [Google Scholar] [CrossRef]
- Askling, J.; Linet, M.; Gridley, G.; Halstensen, T.S.; Ekström, K.; Ekbom, A. Cancer incidence in a population-based cohort of individuals hospitalized with celiac disease or dermatitis herpetiformis. Gastroenterology 2002, 123, 1428–1435. [Google Scholar] [CrossRef]
- Al Somali, Z.; Hamadani, M.; Kharfan-Dabaja, M.; Sureda, A.; El Fakih, R.; Aljurf, M. Enteropathy-Associated T cell Lymphoma. Curr. Hematol. Malign-Rep. 2021, 16, 140–147. [Google Scholar] [CrossRef]
- Al–Toma, A.; Goerres, M.S.; Meijer, J.W.; Peña, A.S.; Crusius, J.B.A.; Mulder, C.J. Human Leukocyte Antigen–DQ2 Homozygosity and the Development of Refractory Celiac Disease and Enteropathy-Associated T-Cell Lymphoma. Clin. Gastroenterol. Hepatol. 2006, 4, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Hrdlickova, B.; Mulder, C.J.; Malamut, G.; Meresse, B.; Platteel, M.; Kamatani, Y.; Ricaño-Ponce, I.; van Wanrooij, R.L.; Zorro, M.M.; Bonder, M.J.; et al. A locus at 7p14.3 predisposes to refractory celiac disease progression from celiac disease. Eur. J. Gastroenterol. Hepatol. 2018, 30, 828–837. [Google Scholar] [CrossRef]
- Di Sabatino, A.; Miceli, E.; Dhaliwal, W.; Biancheri, P.; Salerno, R.; Cantoro, L.; Vanoli, A.; De Vincenzi, M.; Blanco, C.D.V.; MacDonald, T.T.; et al. Distribution, Proliferation, and Function of Paneth Cells in Uncomplicated and Complicated Adult Celiac Disease. Am. J. Clin. Pathol. 2008, 130, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Wolters, V.M.; Verbeek, W.H.; Zhernakova, A.; Onland–Moret, C.; Schreurs, M.W.; Monsuur, A.J.; Verduijn, W.; Wijmenga, C.; Mulder, C.J. The MYO9B Gene Is a Strong Risk Factor for Developing Refractory Celiac Disease. Clin. Gastroenterol. Hepatol. 2007, 5, 1399–1405.e2. [Google Scholar] [CrossRef] [PubMed]
- Perfetti, V.; Baldanti, F.; Lenti, M.V.; Vanoli, A.; Biagi, F.; Gatti, M.; Riboni, R.; Dallera, E.; Paulli, M.; Pedrazzoli, P.; et al. Detection of Active Epstein–Barr Virus Infection in Duodenal Mucosa of Patients With Refractory Celiac Disease. Clin. Gastroenterol. Hepatol. 2016, 14, 1216–1220. [Google Scholar] [CrossRef] [PubMed]
- Foukas, P.G.; Bisig, B.; de Leval, L. Recent advances in upper gastrointestinal lympho-mas: Molecular updates and diagnostic implications. Histopathology 2021, 78, 187–214. [Google Scholar] [CrossRef]
- Ettersperger, J.; Montcuquet, N.; Malamut, G.; Guegan, N.; Lastra, S.L.; Gayraud, S.; Reimann, C.; Vidal, E.; Cagnard, N.; Villarese, P.; et al. Interleukin-15-Dependent T-Cell-like Innate Intraepithelial Lymphocytes Develop in the Intestine and Transform into Lymphomas in Celiac Disease. Immunity 2016, 45, 610–625. [Google Scholar] [CrossRef]
- Tack, G.J.; van Wanrooij, R.L.; Langerak, A.W.; Tjon, J.M.; von Blomberg, B.M.E.; Heideman, D.A.; van Bergen, J.; Koning, F.; Bouma, G.; Mulder, C.J.; et al. Origin and immunophenotype of aberrant IEL in RCDII patients. Mol. Immunol. 2012, 50, 262–270. [Google Scholar] [CrossRef]
- Soderquist, C.R.; Lewis, S.K.; Gru, A.A.; Vlad, G.; Williams, E.S.; Hsiao, S.; Mansukhani, M.M.; Park, D.C.; Bacchi, C.E.; Alobeid, B.; et al. Immunophenotypic Spectrum and Genomic Landscape of Refractory Celiac Disease Type II. Am. J. Surg. Pathol. 2021, 45, 905–916. [Google Scholar] [CrossRef]
- Björck, S.; Lindehammer, S.R.; Fex, M.; Agardh, D. Serum cytokine pattern in young children with screening detected coeliac disease. Clin. Exp. Immunol. 2015, 179, 230–235. [Google Scholar] [CrossRef]
- Malamut, G.; Cording, S.; Cerf-Bensussan, N. Recent advances in celiac disease and refractory celiac disease. F1000Research 2019, 8, 969. [Google Scholar] [CrossRef] [PubMed]
- Malamut, G.; Meresse, B.; Cellier, C.; Cerf-Bensussan, N. Refractory celiac disease: From bench to bedside. Semin. Immunopathol. 2012, 34, 601–613. [Google Scholar] [CrossRef]
- Di Sabatino, A.; Ciccocioppo, R.; Cupelli, F.; Cinque, B.; Millimaggi, D.; Clarkson, M.M.; Paulli, M.; Cifone, M.G.; Corazza, G.R. Epithelium derived interleukin 15 regulates intraepithelial lymphocyte Th1 cytokine production, cytotoxicity, and survival in coeliac disease. Gut 2006, 55, 469–477. [Google Scholar] [CrossRef]
- Chander, U.; Leeman-Neill, R.J.; Bhagat, G. Pathogenesis of Enteropathy-Associated T Cell Lymphoma. Curr. Hematol. Malign-Rep. 2018, 13, 308–317. [Google Scholar] [CrossRef]
- Malamut, G.; El Machhour, R.; Montcuquet, N.; Martin-Lannerée, S.; Dusanter-Fourt, I.; Verkarre, V.; Mention, J.-J.; Rahmi, G.; Kiyono, H.; Butz, E.A.; et al. IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease–associated inflammation and lymphomagenesis. J. Clin. Investig. 2010, 120, 2131–2143. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, F.; Kooy-Winkelaar, Y.; Wiekmeijer, A.-S.; Brugman, M.H.; Mearin, M.L.; Mulder, C.; Lopes, S.C.d.S.; Mummery, C.L.; Staal, F.J.; van Bergen, J.; et al. The composition and differentiation potential of the duodenal intraepithelial innate lymphocyte compartment is altered in coeliac disease. Gut 2016, 65, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, F.; Tjon, J.M.-L.; van Bergen, J.; Koning, F. Dendritic cells promote expansion and survival of aberrant TCR-negative intraepithelial lymphocyte lines from refractory celiac disease type II patients. Mol. Immunol. 2014, 58, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, A.B.; Ondrejka, S.L.; McKinney, M.; Rempel, R.E.; Goodlad, J.R.; Teh, C.H.; Leppa, S.; Mannisto, S.; Kovanen, P.E.; Tse, E.; et al. Enteropathy-associated T cell lymphoma subtypes are characterized by loss of function of SETD2. J. Exp. Med. 2017, 214, 1371–1386. [Google Scholar] [CrossRef]
- Cording, S.; Lhermitte, L.; Malamut, G.; Berrabah, S.; Trinquand, A.; Guegan, N.; Villarese, P.; Kaltenbach, S.; Meresse, B.; Khater, S.; et al. Oncogenetic landscape of lymphomagenesis in coeliac disease. Gut 2021, 71, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Vogel, T.P.; Milner, J.D.; Cooper, M.A. The Ying and Yang of STAT3 in Human Disease. J. Clin. Immunol. 2015, 35, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Roberti, A.; Dobay, M.P.; Bisig, B.; Vallois, D.; Boéchat, C.; Lanitis, E.; Bouchindhomme, B.; Parrens, M.C.; Bossard, C.; Quintanilla-Martinez, L.; et al. Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations. Nat. Commun. 2016, 7, 12602. [Google Scholar] [CrossRef] [PubMed]
- Manso, R.; Rodriguez, M.; Chamizo, C.; Pérez, N.; Alonso-Alonso, R.; Minguez, P.A.; Borregon, J.; Baez-Duran, E.; Pozo, E.M.C.; Piris, M.A.; et al. Intestinal T-Cell Lymphomas: Molecular Integrative Analysis Recognizes Different Therapeutic Targets for Each Subtype. Hematol. Oncol. 2021, 39, 396–425. [Google Scholar] [CrossRef]
- Gooderham, M.J.; Forman, S.B.; Bissonnette, R.; Beebe, J.S.; Zhang, W.; Banfield, C.; Zhu, L.; Papacharalambous, J.; Vincent, M.S.; Peeva, E. Efficacy and Safety of Oral Janus Kinase 1 Inhibitor Abrocitinib for Patients With Atopic Dermatitis: A Phase 2 Randomized Clin-ical Trial. JAMA Dermatol. 2020, 156, 104. [Google Scholar] [CrossRef]
- Bao, X.; Ren, T.; Huang, Y.; Ren, C.; Yang, K.; Zhang, H.; Guo, W. Bortezomib induces apoptosis and suppresses cell growth and metastasis by inactivation of Stat3 signaling in chondrosarcoma. Int. J. Oncol. 2017, 50, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Nicolae, A.; Xi, L.; Pham, T.H.; Pham, T.-A.; Navarro, W.; Meeker, H.G.; Pittaluga, S.; Jaffe, E.S.; Raffeld, M. Mutations in the JAK/STAT and RAS signaling pathways are common in intestinal T-cell lymphomas. Leukemia 2016, 30, 2245–2247. [Google Scholar] [CrossRef]
- Clarke, L.; Adduri, R.S.; Smyth, P.; Quinn, F.; Jeffers, M.; Dunne, B.; O’Leary, J.; McKiernan, S.; Vandenberghe, E.; Pyne, S.; et al. Potentially important miRNAs in enteropathy-associated T-cell lymphoma pathogenesis: A pilot study. Leuk. Res. Rep. 2018, 10, 52–54. [Google Scholar] [CrossRef]
- Vaira, V.; Gaudioso, G.; Laginestra, M.A.; Terrasi, A.; Agostinelli, C.; Bosari, S.; Di Sabatino, A.; Vanoli, A.; Paulli, M.; Ferrero, S.; et al. Deregulation of miRNAs-cMYC circuits is a key event in refractory celiac disease type-2 lymphomagenesis. Clin. Sci. 2020, 134, 1151–1166. [Google Scholar] [CrossRef]
- Bhansali, R.S.; Barta, S.K. SOHO State of the Art Updates and Next Questions|Challenging Cases in Rare T-Cell Lymphomas. Clin. Lymphoma Myeloma Leuk. 2023. [Google Scholar] [CrossRef] [PubMed]
- Veloza, L.; Cavalieri, D.; Missiaglia, E.; Ledoux-Pilon, A.; Bisig, B.; Pereira, B.; Bonnet, C.; Poullot, E.; Quintanilla-Martinez, L.; Dubois, R.; et al. Monomorphic epitheliotropic intestinal T-cell lymphoma comprises morphologic and genomic heterogeneity impacting outcome. Haematologica 2023, 108, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, M.; Kong, W.-J.; Cui, M.; Gao, F. Association between intestinal neoplasms and celiac disease: A review. World J. Gastrointest. Oncol. 2021, 13, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
| Pathway | Somatic Mutations | Primary Mechanisms of Mutation | RCDII-EATL Mutation Frequency | De Novo EATL Mutation Frequency | Clinical Significance |
|---|---|---|---|---|---|
| JAK/STAT | JAK1 [32] STAT3 [32] SOCS1 [32] SOCS3 [32] | Gain-of-function mutations | 48% 38% 7% 8% | 32% (double mutation with STAT3) 32% (double mutation with JAK1) | Therapeutic and potential diagnostic value [32] |
| NF-κB | TNFAIP3 [32] TNIP3 [32] | Nonsense or frameshift mutations | 13% 9% | 28% | Potential prognostic value [32] |
| Gene Regulation | KMT2D [32] TET2 [32] POT1 [32] | Loss of function (frameshift, nonsense, or missense mutations) | 22% 30% | 37% 32% 26% | Potential prognostic value [32] |
| SETD2 [31] | Frameshift or nonsense | 10–20% | Diagnostic Value [31] | ||
| Gene Expression | DDX3X [32] | Missense mutation | 20% | 32% | Potential prognostic value [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah, S.A.A.; Goa, P.; Vandenberghe, E.; Flavin, R. Update on the Pathogenesis of Enteropathy-Associated T-Cell Lymphoma. Diagnostics 2023, 13, 2629. https://doi.org/10.3390/diagnostics13162629
Abdullah SAA, Goa P, Vandenberghe E, Flavin R. Update on the Pathogenesis of Enteropathy-Associated T-Cell Lymphoma. Diagnostics. 2023; 13(16):2629. https://doi.org/10.3390/diagnostics13162629
Chicago/Turabian StyleAbdullah, Shahed Azzam Ahmed, Patricia Goa, Elisabeth Vandenberghe, and Richard Flavin. 2023. "Update on the Pathogenesis of Enteropathy-Associated T-Cell Lymphoma" Diagnostics 13, no. 16: 2629. https://doi.org/10.3390/diagnostics13162629
APA StyleAbdullah, S. A. A., Goa, P., Vandenberghe, E., & Flavin, R. (2023). Update on the Pathogenesis of Enteropathy-Associated T-Cell Lymphoma. Diagnostics, 13(16), 2629. https://doi.org/10.3390/diagnostics13162629







