Dermo-Seg: ResNet-UNet Architecture and Hybrid Loss Function for Detection of Differential Patterns to Diagnose Pigmented Skin Lesions
Abstract
:1. Introduction
- Dermo-Seg model is proposed consisting ResNet-50 as backbone architecture in the encoder part of UNet model to down-sample the input features of skin lesions. The purpose of down-sampling is to extract the high level feature maps of lesion’s area.
- These high-level feature maps contain very useful information about the attributes of skin lesions, which is then up-sampled in the decoder part of UNet and further concatenated with the high-level feature maps from the encoder part.
- Individual models are proposed for all five separate attributes of skin lesions.
- The hybrid loss function resolves the issue of class imbalance at the pixel level for each individual attribute.
- (1)
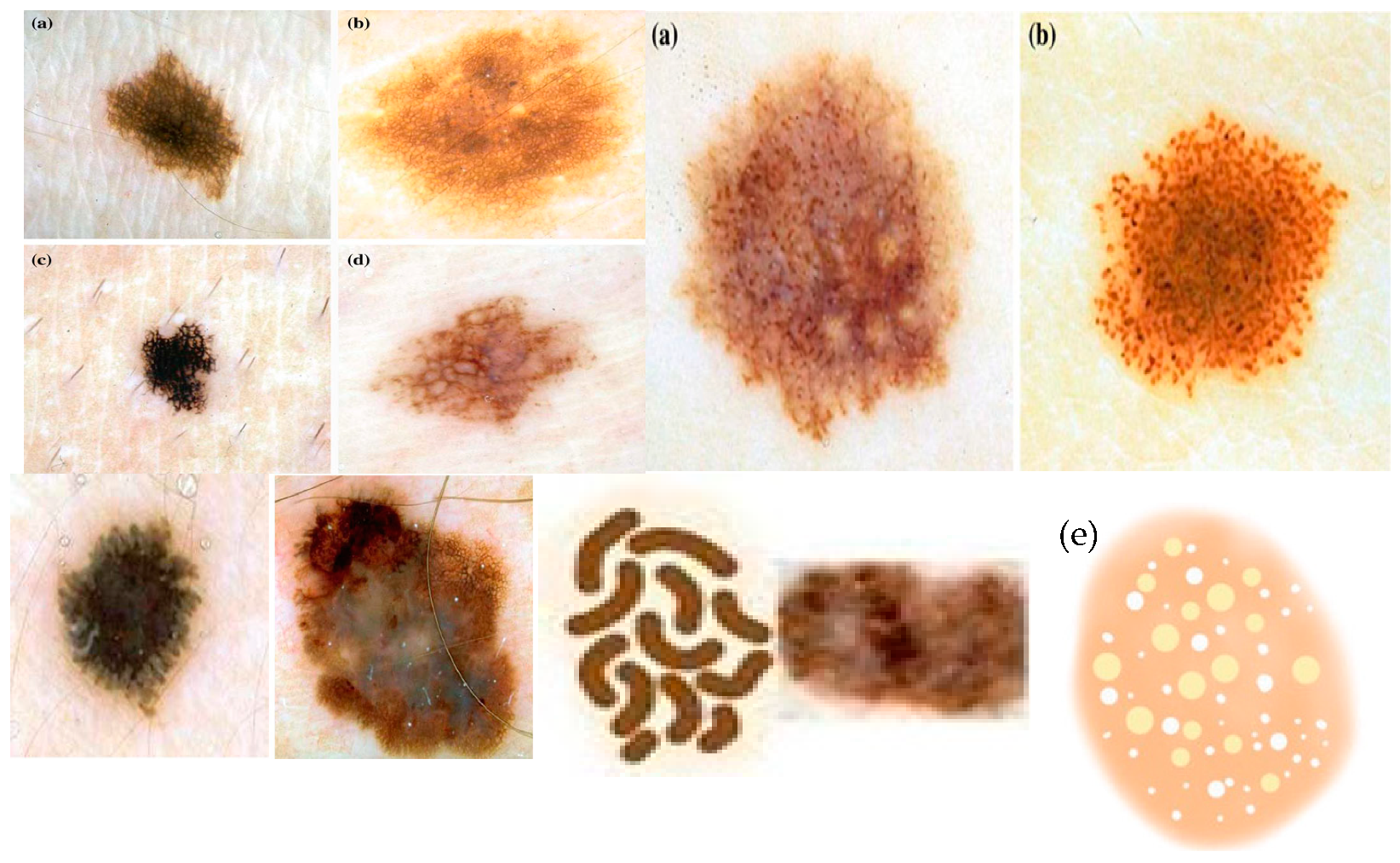
- Pigment Network: The pigment network is one of the fundamental dermoscopic structures observed in skin cancer lesions [2]. It refers to the regular or irregular distribution of pigmented lines and grids within the lesion. The evaluation of the pigment network can provide insights into the organization and pattern of pigmented cells, which aids in distinguishing benign lesions from malignant ones. Irregular or disturbed pigment network patterns are mostly an indication of the presence of malignancy. It is the most important structure in the diagnosis of melanoma. The assessment of the pigment network involves assessing its distribution, thickness, regularity, and overall arrangement. Differences from a regular, symmetrical pattern may give an indication of malignancy.
- (2)
- Globules: Globules are-round shaped structures seen within the skin lesion [2]. They may vary in size, color, and distribution. The deep assessment of globules assists in discriminating between benign and malignant lesions. This assessment involves assessing their size, shape, color uniformity, and arrangement. In most of the benign lesions, globules are usually regular, symmetric, and evenly distributed. Malignant lesions mostly exhibit irregular, asymmetrical, or clustered globules, indicating of malignancy. Globules attributes can be pigmented or non-pigmented or both and may provide valuable information for differentiating melanoma from benign lesions.
- (3)
- Streaks: Streaks are commonly linear structures mostly found within the lesion [35]. These attributes can give valuable diagnostic information. In benign lesions, streaks are uniform most of the time, thin, and evenly distributed. On the other hand, malignant lesions may show thick, irregular, and asymmetric streaks, which can indicate malignancy. Moreover, the presence of a radial streaking pattern, moving outward from the center of the lesion, may be a strong indicator of invasive melanoma.
- (4)
- Negative Network: The negative network, also called the hypopigmented network [35], appears as lighter or sometimes colorless lines or areas within the lesion. It shows the absence of pigmentation in various areas. The presence of a negative network may indicate various types of melanoma or other non-melanocytic skin lesions.
- (5)
- Milia-like Cysts: milia-like cysts are small and white cyst structures [35] found within the skin lesion. These structures resemble milia, which are tiny epidermal cysts commonly seen in healthy skin. In the context of skin cancer, the presence of milia-like cysts can be indicative of specific subtypes or serve as a diagnostic clue for differentiating benign and malignant lesions.
2. Methodological Background of U-Net and RseNet-50 Architectures
2.1. U-Net Architecture
2.2. ResNet Architecture
3. Proposed Methodology
3.1. Proposed Model Overview
3.2. Proposed ResNet-UNet Model
3.3. Hybrid Loss Function
4. Experiments and Results
4.1. Dataset
4.2. Experimental Setup
4.3. Experimental Results and Discussion
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Descriptive Form |
| ResNet | Residual Network |
| CNN | Convolutional Neural Network |
| ISIC | International Skin Imaging Collaboration |
| IOU | Intersection Over Union |
| CAD | Computer Aided Diagnostic |
| FTL | Focal Tversky loss |
| FFA | FireFly Algorithm |
| FCM | Fuzzy CMean |
| LVP | Local Vector Pattern |
| LBP | Local Binary Pattern |
| YOLO | You Only Look Once |
| BCC | Basal Cell Carcinoma |
| XOR | Exclusive OR |
| ADAM | Adaptive Momentum |
| TATL | Task Agnostic Transfer Learning |
| Grad-CAM | Gradient Class Activation Map |
| RSM | Random Shuffle Mechanism |
| SANet | Superpixel Attention Network |
| SAM | Superpixel Attention Module |
| ReLU | Rectified Linear Unit |
| BN_ACT | Batch Normalization_ Activation |
| ROC | Receiver Operating Curve |
References
- American Cancer Society. Cancer Facts & Figures 2019; American Cancer Society: Atlanta, GA, USA, 2019. [Google Scholar]
- Braun, K.K. Dermoscopedia R. Dermoscopic Structures. 2019. Available online: https://dermoscopedia.org/Dermoscopic_structures (accessed on 21 January 2019).
- Chen, L.L.; Dusza, S.W.; Jaimes, N.; Marghoob, A.A. Performance of the first step of the 2-step dermoscopy algorithm. JAMA Dermatol. 2015, 151, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Jamil, U.; Ann, Q.; Sajid, A.; Imran, M. Role of Digital Filters in Dermoscopic Image Enhancement. Tech. J. Univ. Eng. Technol. Taxila 2018, 23, 73–81. [Google Scholar]
- Garg, S.; Jindal, B. Skin lesion segmentation using k-mean and optimized fire fly algorithm. Multimed. Tools Appl. 2021, 80, 7397–7410. [Google Scholar] [CrossRef]
- Durgarao, N.; Sudhavani, G. Diagnosing skin cancer via C-means segmentation with enhanced fuzzy optimization. IET Image Process. 2021, 15, 2266–2280. [Google Scholar] [CrossRef]
- Kaur, R.; Gholam, H.; Sinha, R.; Lindén, M. Automatic lesion segmentation using atrous convolutional deep neural networks in dermoscopic skin cancer images. BMC Med. Imaging 2021, 22, 1–13. [Google Scholar] [CrossRef]
- Bagheri, F.; Tarokh, M.J.; Ziaratban, M. Skin lesion segmentation from dermoscopic images by using Mask R-CNN, Retina-Deeplab, and graph-based methods. Biomed. Signal Process. Control 2021, 67, 102533. [Google Scholar] [CrossRef]
- Qamar, S.; Ahmad, P.; Shen, L. Dense Encoder-Decoder–Based Architecture for Skin Lesion Segmentation. Cogn. Comput. 2021, 13, 583–594. [Google Scholar] [CrossRef]
- Wu, H.; Pan, J.; Li, Z.; Wen, Z.; Qin, J. Automated Skin Lesion Segmentation Via an Adaptive Dual Attention Module. IEEE Trans. Med. Imaging 2020, 40, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, S.; Özkaya, U. Skin Lesion Segmentation with Improved Convolutional Neural Network. J. Digit. Imaging 2020, 33, 958–970. [Google Scholar] [CrossRef]
- Shan, P.; Wang, Y.; Fu, C.; Song, W.; Chen, J. Automatic skin lesion segmentation based on FC-DPN. Comput. Biol. Med. 2020, 123, 103762. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Shi, F.; Song, H.; Ji, W.; Han, G. Attentive boundary aware network for multi-scale skin lesion segmentation with adversarial training. Multimed. Tools Appl. 2020, 79, 27115–27136. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Zhang, Y.-D.; Sharif, M. Attributes based skin lesion detection and recognition: A mask RCNN and transfer learning-based deep learning framework. Pattern Recognit. Lett. 2021, 143, 58–66. [Google Scholar] [CrossRef]
- Huang, C.; Yu, A.; Wang, Y.; He, H. Skin Lesion Segmentation Based on Mask R-CNN. In Proceedings of the 2020 International Conference on Virtual Reality and Visualization (ICVRV), Recife, Brazil, 13–14 November 2020. [Google Scholar]
- Shahin, A.H.; Amer, K.; Elattar, M.A. Deep Convolutional Encoder-Decoders with Aggregated Multi-Resolution Skip Connections for Skin Lesion Segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 451–454. [Google Scholar]
- Ünver, H.M.; Ayan, E. Skin Lesion Segmentation in Dermoscopic Images with Combination of YOLO and GrabCut Algorithm. Diagnostics 2019, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Goyal, M.; Oakley, A.; Bansal, P.; Dancey, D.; Yap, M.H. Skin Lesion Segmentation in Dermoscopic Images with Ensemble Deep Learning Methods. IEEE Access 2020, 8, 4171–4181. [Google Scholar] [CrossRef]
- Lameski, J.; Jovanov, A.; Zdravevski, E.; Lameski, P.; Gievska, S. Skin lesion segmentation with deep learning. In Proceedings of the IEEE EUROCON 2019-18th International Conference on Smart Technologies, Novi Sad, Serbia, 1–4 July 2019; pp. 1–5. [Google Scholar]
- Hasan, S.N.; Gezer, M.; Azeez, R.A.; Gulsecen, S. Skin Lesion Segmentation by using Deep Learning Techniques. In Proceedings of the 2019 Medical Technologies Congress (TIPTEKNO), Izmir, Turkey, 3–5 October 2019; pp. 1–4. [Google Scholar]
- Li, H.; He, X.; Zhou, F.; Yu, Z.; Ni, D.; Chen, S.; Wang, T.; Lei, B. Dense Deconvolutional Network for Skin Lesion Segmentation. IEEE J. Biomed. Health Inform. 2019, 23, 527–537. [Google Scholar] [CrossRef]
- Nguyen, D.M.H.; Nguyen, T.T.; Vu, H.; Pham, Q.; Nguyen, M.; Nguyen, B.T.; Sonntag, D. TATL: Task agnostic transfer learning for skin attributes detection. Med. Image Anal. 2022, 78, 102359. [Google Scholar] [CrossRef]
- Kadir, M.A.; Nunnari, F.; Sonntag, D. Fine-tuning of explainable CNNs for skin lesion classification based on dermatologists’ feedback towards increasing trust. arXiv 2023, arXiv:2304.01399. [Google Scholar]
- Nguyen, D.M.H.; Ezema, A.; Nunnari, F.; Sonntag, D. A visually explainable learning system for skin lesion detection using multiscale input with attention u-net. In Proceedings of the KI 2020: Advances in Artificial Intelligence: 43rd German Conference on AI, Bamberg, Germany, 21–25 September 2020; Volume 12325 LNAI, pp. 313–319. [Google Scholar] [CrossRef]
- Jahanifar, M.; Tajeddin, N.Z.; Koohbanani, N.A.; Gooya, A.; Rajpoot, N. Segmentation of Skin Lesions and their Attributes Using Multi-Scale Convolutional Neural Networks and Domain Specific Augmentations. arXiv 2018, arXiv:1809.10243. [Google Scholar]
- He, X.; Lei, B.; Wang, T. SANet: Superpixel Attention Network for Skin Lesion Attributes Detection. arXiv 2019, arXiv:1910.08995. [Google Scholar]
- Bissoto, A.; Perez, F.; Ribeiro, V.; Fornaciali, M.; Avila, S.; Valle, E. Deep-Learning Ensembles for Skin-Lesion Segmentation, Analysis, Classification: RECOD Titans at ISIC Challenge 2018. arXiv 2018, arXiv:1808.08480. [Google Scholar]
- Navarro, F.; Escudero-Viñolo, M.; Bescós, J. Accurate Segmentation and Registration of Skin Lesion Images to Evaluate Lesion Change. IEEE J. Biomed. Heal. Inform. 2019, 23, 501–508. [Google Scholar] [CrossRef]
- Saitov, I.; Polevaya, T.; Filchenkov, A. Dermoscopic attributes classification using deep learning and multi-Task learning. Procedia Comput. Sci. 2020, 178, 328–336. [Google Scholar] [CrossRef]
- Kawahara, J.; Hamarneh, G. Fully convolutional neural networks to detect clinical dermoscopic features. IEEE J. Biomed. Health Inform. 2018, 23, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Koohbanani, N.A.; Jahanifar, M.; Tajeddin, N.Z.; Gooya, A.; Rajpoot, N. Leveraging transfer learning for segmenting lesions and their attributes in dermoscopy images. arXiv 2018, arXiv:1809.10243. [Google Scholar]
- Nunnari, F.; Kadir, M.A.; Sonntag, D. On the overlap between grad-cam saliency maps and explainable visual features in skin cancer images. In International Cross-Domain Conference for Machine Learning and Knowledge Extraction; Springer: Cham, Switzerland, 2021; pp. 241–253. [Google Scholar]
- Gonzalez-Diaz, I. Dermaknet: Incorporating the knowledge of dermatologists to convolutional neural networks for skin lesion diagnosis. IEEE J. Biomed. Health Inform. 2018, 23, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Q.; Daadaa, Y.; Rashid, U.; Ibrahim, M.E.A. Assist-Dermo: A Lightweight Separable Vision Transformer Model for Multiclass Skin Lesion Classification. Diagnostics 2023, 13, 2531. [Google Scholar] [CrossRef]
- Chen, E.Z.; Dong, X.; Wu, J.; Jiang, H.; Li, X.; Rong, R. Lesion attributes segmentation for melanoma detection with deep learning. bioRxiv 2018, 381855. [Google Scholar] [CrossRef]
- López-Labraca, J.; Fernández-Torres M, Á.; González-Díaz, I.; Diaz-de-Maria, F.; Pizarro, Á. Enriched dermoscopicstructure- based cad system for melanoma diagnosis. Multimed. Tools Appl. 2018, 77, 12171–12202. [Google Scholar] [CrossRef]
- Akram, T.; Khan, M.A.; Sharif, M.; Yasmin, M. Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features. J. Ambient Intell. Humaniz. Comput. 2018, 34, 1–20. [Google Scholar] [CrossRef]
- Kaur, R.; Leander, R.; Mishra, N.K.; Hagerty, J.R.; Kasmi, R.; Stanley, R.J.; Celebi, M.E.; Stoecker, W.V. Thresholding methods for lesion segmentation of basal cell carcinoma in dermoscopy images. Ski. Res. Technol. 2016, 23, 416–428. [Google Scholar] [CrossRef]
- Kasmi, R.; Mokrani, K.; Rader, R.K.; Cole, J.G.; Stoecker, W.V. Biologically inspired skin lesion segmentation using a geodesic active contour technique. Ski. Res. Technol. 2016, 22, 208–222. [Google Scholar] [CrossRef]
- Yu, C.Y.; Zhang, W.S.; Yu, Y.Y.; Li, Y. A novel active contour model for image segmentation using distance regularization term. Comput. Math. Appl. 2013, 65, 1746–1759. [Google Scholar] [CrossRef]
- Ashour, A.S.; Nagieb, R.M.; El-Khobby, H.A.; Elnaby, M.M.A.; Dey, N. Genetic algorithm-based initial contour optimization for skin lesion border detection. Multimed. Tools Appl. 2021, 80, 2583–2597. [Google Scholar] [CrossRef]
- Mohakud, R.; Dash, R. Skin cancer image segmentation utilizing a novel EN-GWO based hyper-parameter optimized FCEDN. J. King Saud Univ. Comput. Inf. Sci 2022, 34, 9889–9904. [Google Scholar] [CrossRef]
- LeLee-Thorp, J.; Ainslie, J.; Eckstein, I.; Ontanon, S. Fnet: Mixing tokens with fourier transforms. arXiv 2021, arXiv:2105.03824. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer International Publishing: Berlin, Germany, 2015; pp. 234–241. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; IEEE Computer Society: Washington, DC, USA, 2017; pp. 2261–2269. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. arXiv 2017, arXiv:1610.02357. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-ResNet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; pp. 4278–4284. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef]
- Baig, A.R.; Abbas, Q.; Almakki, R.; Ibrahim, M.E.A.; AlSuwaidan, L.; Ahmed, A.E.S. Light-Dermo: A Lightweight Pretrained Convolution Neural Network for the Diagnosis of Multiclass Skin Lesions. Diagnostics 2023, 13, 385. [Google Scholar] [CrossRef]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2019, arXiv:1902.03368. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef]
- Abraham, N.; Khan, N.M. A Novel Focal Tversky Loss Function with Improved Attention U-Net for Lesion Segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019. [Google Scholar]
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-IoU Loss: Faster and Better Learning for Bounding Box Regression. arXiv 2019, arXiv:1911.08287v1. [Google Scholar] [CrossRef]
- James, C.; Bezdek, C.; Robert Ehrlich, W.F. FCM: The fuzzy c-means clustering algorithm. Comput. Geosci. 1984, 10, 191–203. [Google Scholar]
- Fan, K.C.; Hung, T.Y. A Novel Local Pattern Descriptor—Local Vector Pattern in High-Order Derivative Space for Face Recognition. IEEE Trans. Image Process. 2014, 23, 2877–2891. [Google Scholar] [CrossRef]
- Song, K.-C.; Yan, Y.-H.; Chen, W.-H. Research and Perspective on Local Binary Pattern. Acta Autom. Sin. 2013, 39, 730–744. [Google Scholar] [CrossRef]
- Wang, Z.; Jun, L. A review of object detection based on convolutional neural network. In Proceedings of the 2017 36th Chinese control conference (CCC), Dalian, China, 26–28 July 2017; pp. 11104–11109. [Google Scholar]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. Faceb. AI Res. (FAIR). arXiv 2018, arXiv:1703.06870v3. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. arXiv 2017, arXiv:1606.00915v2. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Vladimir Iglovikov, A.S. TernausNet: U-Net with VGG11 Encoder Pre-Trained on ImageNet for Image Segmentation. arXiv 2018, arXiv:1801.05746. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Zou, J.; Ma, X.; Zhong, C.; Zhang, Y. Dermoscopic image analysis for ISIC challenge 2018. arXiv 2018, arXiv:1807.08948. [Google Scholar]
- Diao, Q.; Dai, Y.; Zhang, C.; Wu, Y.; Feng, X.; Pan, F. Superpixel-Based Attention Graph Neural Network for Semantic Segmentation in Aerial Images. J. Remote Sens. 2022, 14, 305. [Google Scholar] [CrossRef]
- Weng, W.; Zhu, X. INet: Convolutional Networks for Biomedical Image Segmentation. IEEE Access 2021, 9, 16591–16603. [Google Scholar] [CrossRef]
- Marghoob, N.G.; Liopyris, K.; Jaimes, N. Dermoscopy: A review of the structures that facilitate melanoma detection. J. Am. Osteopath. Assoc. 2019, 119, 380–390. [Google Scholar] [CrossRef] [PubMed]

















| Cited | Methodology | Dataset | Results | Limitations |
|---|---|---|---|---|
| [29] | Task-Agnostic Transfer Learning (TATL) using U-Shape with b0-EfficientNet; L-Shape with b0-EfficientNet | ISIC 2018 Task 2 | Mean IOU (U-Shape with b0-EfficientNet) Pigment network: 0.565 Globules: 0.373 Milia: 0.157 Negative Network: 0.268 Streaks: 0.243 Mean IOU (L-Shape with b0-EfficientNet) Pigment network: 0.562 Globules: 0.356 Milia: 0.168 Negative Network: 0.292 Streaks: 0.252 | Heavy computational resources required; A lot of pre-processing required; Obtained results are very low. i.e., less than 50% of the except pigment network; A very lengthy process followed by the use of various models that consumes resources. |
| [31] | Attention-UNet model using transfer learning | ISIC 2018 Task 2 | Mean IOU Pigment network: 0.535 Globules: 0.312 Milia: 0.162 Negative Network: 0.187 Streaks: 0.197 | A lot of pre-processing required. |
| [32] | Transfer learning-based UNet model with multi-scale convolution (MSC) block pyramid pooling paradigm | ISIC 2018 Task 2 | Mean IOU Pigment network: 0.563 Globules: 0.341 Milia: 0.171 Negative Network: 0.228 Streaks: 0.156 | A lot of image processing applied including contrast and sharpness adjustment, shrinking and stretching contrast, hair occlusion; Data imbalance handled with augmentation. |
| [37] | UNet with pyramid pooling paradigm | ISIC 2018 Task 2 | Mean IOU Pigment network: 0.544 Globules: 0.252 Milia: 0.165 Negative Network: 0.285 Streaks: 0.123 | At test time, various augmentations are applied, and their outputs are merged to predict the final output, which shows dependency and reduces the efficiency of the model. |
| [66] | Modified pyramid scene-parsing network (modified PSPNet) | ISIC 2018 Task 2 | Mean IOU (training) Pigment network: 0.482 Globules: 0.239 Milia: 0.132 Negative Network: 0.225 Streaks: 0.145 | They did not show the test data results. Only given training results. |
| Layer Name | Output Size | 18-Layer | 34-Layer | 50-Layer | 101-Layer | 152-Layer |
|---|---|---|---|---|---|---|
| conv1 | 112 × 112 | 7 × 7, 64, stride 2 | ||||
| conv2_x | 56 × 56 | 3 × 3 max pool, stride 2 | ||||
| Conv3_x | 28 × 28 | |||||
| Conv4_x | 14 × 14 | |||||
| Conv5_x | 7 × 7 | |||||
| 1 × 1 | Average pool, 100-d fc, softmax | |||||
| FLOPs | ||||||
| Attributes | No. of Images | % of Images |
|---|---|---|
| Streaks | 100 | 2.9% |
| Pigment Network | 1522 | 58.7% |
| Globules | 602 | 23.2% |
| Negative Network | 189 | 7.3% |
| Milia-like-cysts | 681 | 26.3% |
| Total images | 2594 | 100% |
| Model Parameters | Values |
|---|---|
| Image Resolution | 512 × 512 × 3 |
| Batch size | 8 |
| No. of epochs | 60 |
| Learning Rate | 0.001 |
| Patience | 20 |
| Optimizer | Adam |
| Loss function | Hybrid |
| Early Stopping at (Automatic) | |
| Globules | 36 |
| Pigment Network | 49 |
| Negative Network | 59 |
| Streaks | 50 |
| Milia | 30 |
| Class/Attribute | Mean IoU |
|---|---|
| Streaks | 0.53 |
| Pigment Network | 0.67 |
| Globules | 0.66 |
| Negative Network | 0.58 |
| Milia-like-cysts | 0.53 |
| Factors | Values |
|---|---|
| Execution Time | 02 h 40 min |
| Total No. of parameters | 32,561,114 |
| Trainable parameters | 9,058,644 |
| Non-trainable parameters | 23,502,470 |
| Space required | 16 GB RAM |
| Method | Pigment Network | Globules | Milia-like Cyst | Negative Network | Streaks |
|---|---|---|---|---|---|
| ResNet-151 [25] | 0.527 | 0.304 | 0.144 | 0.149 | 0.125 |
| ResNet-v2 [25] | 0.539 | 0.310 | 0.159 | 0.189 | 0.121 |
| DenseNet-169 [25] | 0.538 | 0.324 | 0.158 | 0.213 | 0.134 |
| b0-EfficientNet [22] | 0.554 | 0.324 | 0.157 | 0.213 | 0.139 |
| U-Eff (TATL) [22] | 0.565 | 0.373 | 0.157 | 0.268 | 0.243 |
| L-Eff (TATL) [22] | 0.562 | 0.356 | 0.168 | 0.292 | 0.252 |
| Ensemble [25] | 0.563 | 0.341 | 0.171 | 0.228 | 0.156 |
| Attention UNet [24] | 0.535 | 0.312 | 0.162 | 0.187 | 0.197 |
| LeHealth method (Second ranked ISIC 2018 challenge) [66] | 0.482 | 0.239 | 0.132 | 0.225 | 0.145 |
| NMN’s method [31] | 0.544 | 0.252 | 0.165 | 0.285 | 0.123 |
| SANet [26] | 0.576 | 0.346 | 0.251 | 0.286 | 0.248 |
| Dermo-Seg | 0.67 | 0.66 | 0.53 | 0.58 | 0.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arshad, S.; Amjad, T.; Hussain, A.; Qureshi, I.; Abbas, Q. Dermo-Seg: ResNet-UNet Architecture and Hybrid Loss Function for Detection of Differential Patterns to Diagnose Pigmented Skin Lesions. Diagnostics 2023, 13, 2924. https://doi.org/10.3390/diagnostics13182924
Arshad S, Amjad T, Hussain A, Qureshi I, Abbas Q. Dermo-Seg: ResNet-UNet Architecture and Hybrid Loss Function for Detection of Differential Patterns to Diagnose Pigmented Skin Lesions. Diagnostics. 2023; 13(18):2924. https://doi.org/10.3390/diagnostics13182924
Chicago/Turabian StyleArshad, Sannia, Tehmina Amjad, Ayyaz Hussain, Imran Qureshi, and Qaisar Abbas. 2023. "Dermo-Seg: ResNet-UNet Architecture and Hybrid Loss Function for Detection of Differential Patterns to Diagnose Pigmented Skin Lesions" Diagnostics 13, no. 18: 2924. https://doi.org/10.3390/diagnostics13182924
APA StyleArshad, S., Amjad, T., Hussain, A., Qureshi, I., & Abbas, Q. (2023). Dermo-Seg: ResNet-UNet Architecture and Hybrid Loss Function for Detection of Differential Patterns to Diagnose Pigmented Skin Lesions. Diagnostics, 13(18), 2924. https://doi.org/10.3390/diagnostics13182924








