Deep Learning-Based Delayed PET Image Synthesis from Corresponding Early Scanned PET for Dosimetry Uptake Estimation
Abstract
:1. Introduction
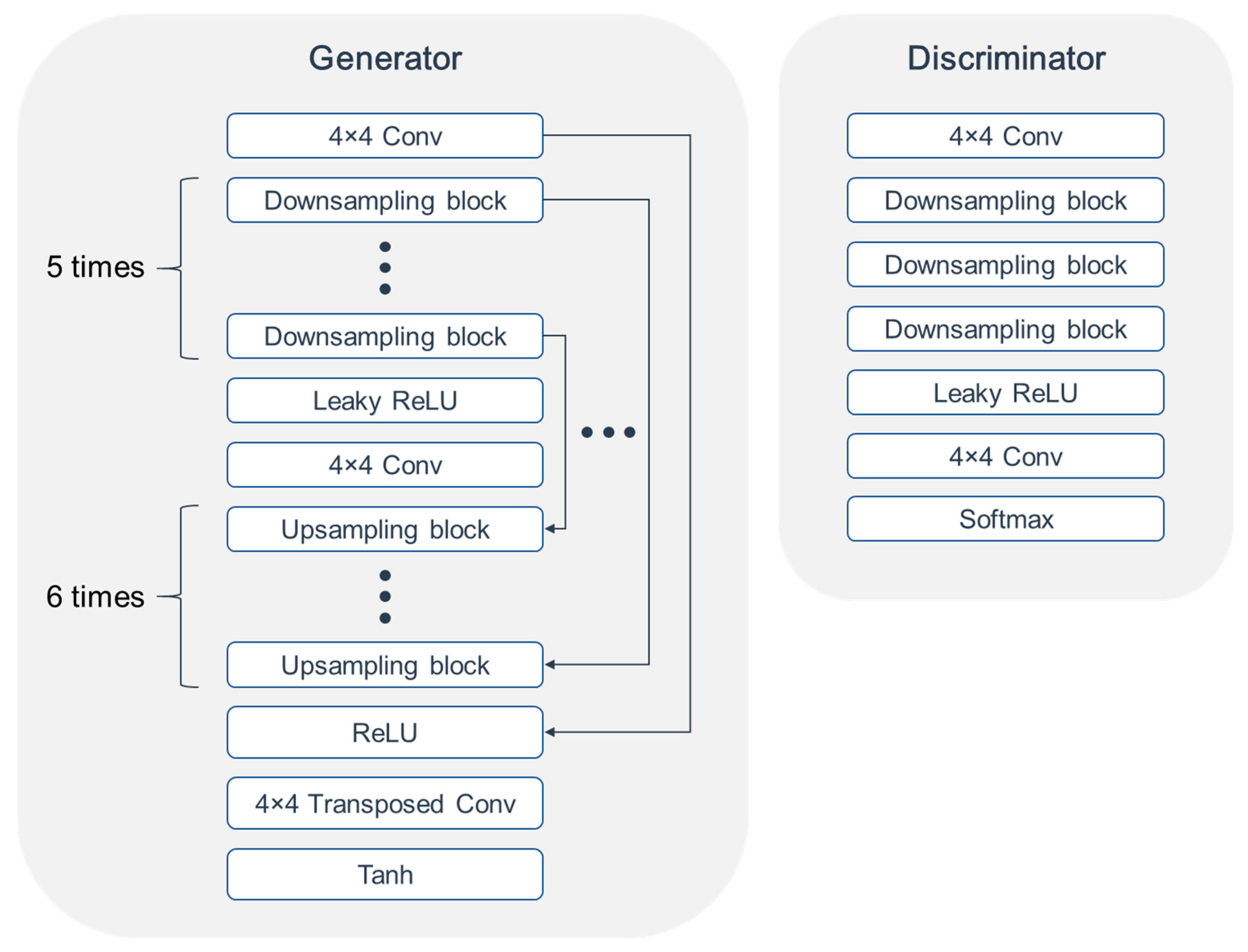
2. Materials and Methods
3. Results
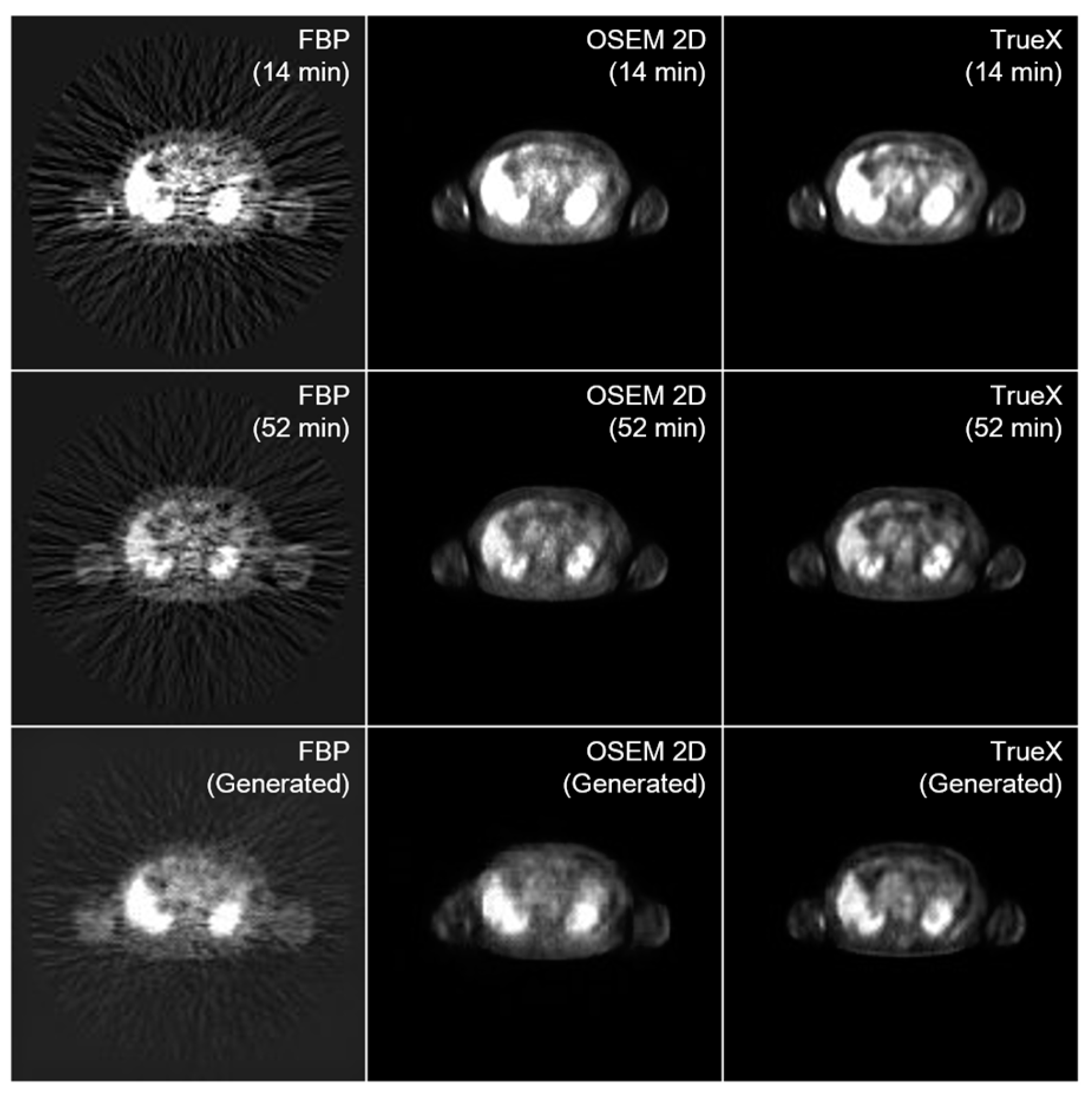
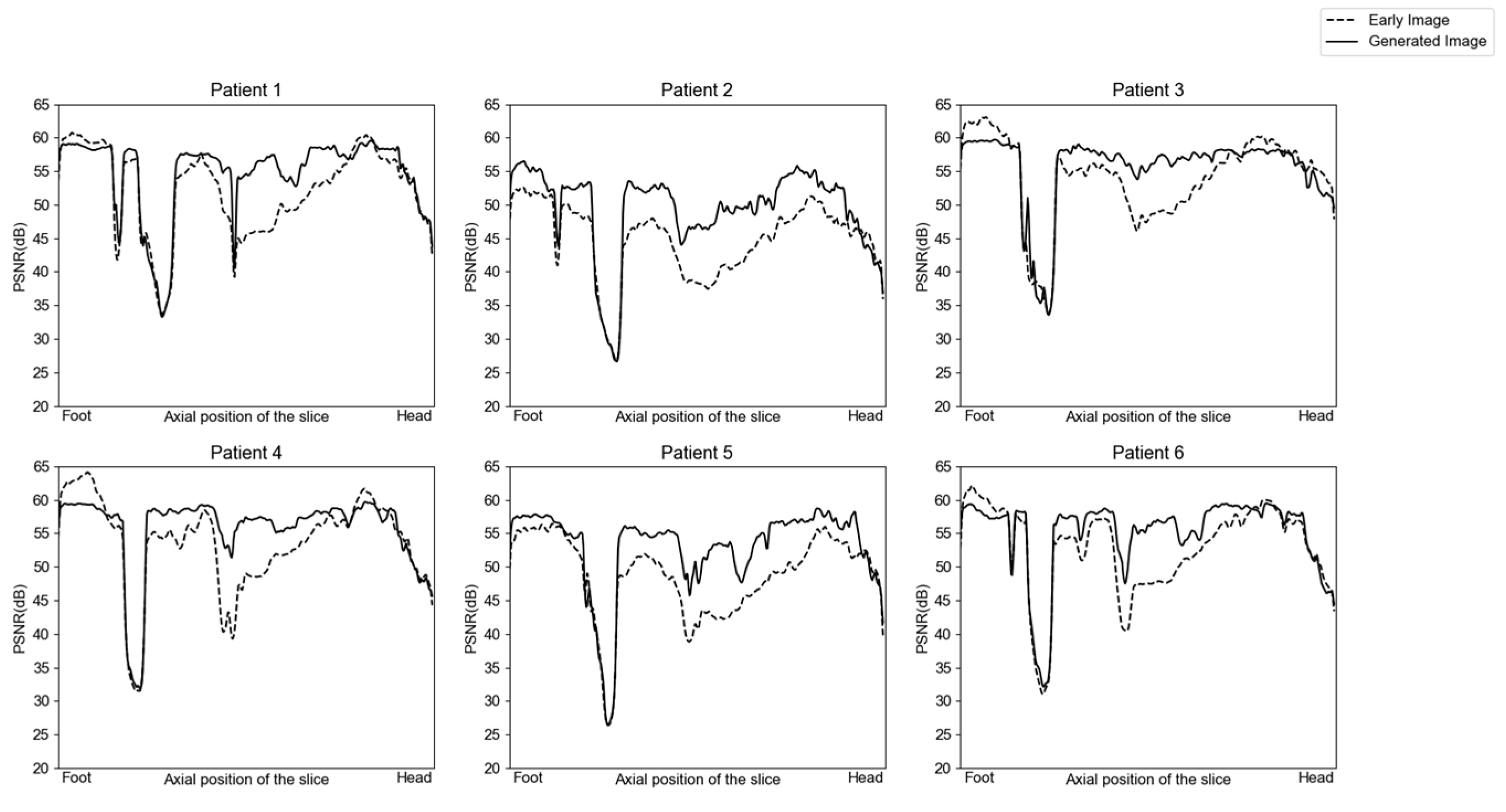
3.1. Image Generation
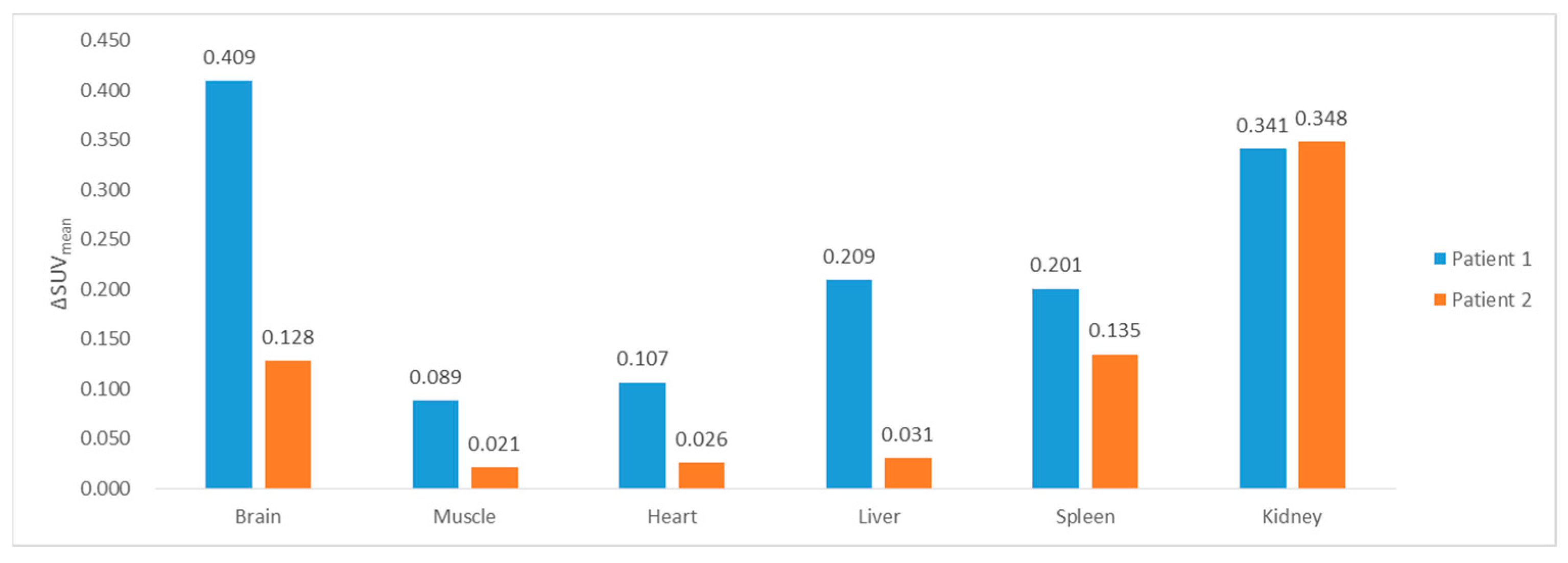
3.2. SUVmean Estimation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sgouros, G.; Hobbs, R.F. Dosimetry for Radiopharmaceutical Therapy. Semin. Nucl. Med. 2014, 44, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Graves, S.A.; Hobbs, R.F. Dosimetry for Optimized, Personalized Radiopharmaceutical Therapy. Semin. Radiat. Oncol. 2021, 31, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Bolch, W.E.; Eckerman, K.F.; Sgouros, G.; Thomas, S.R.; Brill, A.B.; Fisher, D.R.; Howell, R.W.; Meredith, R.; Wessels, B.W. MIRD Pamphlet No. 21: A Generalized Schema for Radiopharmaceutical Dosimetry—Standardization of Nomenclature. J. Nucl. Med. 2009, 50, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Hindorf, C.; Glatting, G.; Chiesa, C.; Lindén, O.; Flux, G. EANM Dosimetry Committee Guidelines for Bone Marrow and Whole-Body Dosimetry. Eur. J. Nucl. Med. Mol. Imaging 2010, 37, 1238–1250. [Google Scholar] [CrossRef] [PubMed]
- Danieli, R.; Milano, A.; Gallo, S.; Veronese, I.; Lascialfari, A.; Indovina, L.; Botta, F.; Ferrari, M.; Cicchetti, A.; Raspanti, D.; et al. Personalized Dosimetry in Targeted Radiation Therapy: A Look to Methods, Tools and Critical Aspects. J. Pers. Med. 2022, 12, 205. [Google Scholar] [CrossRef] [PubMed]
- Siegel, J.A.; Thomas, S.R.; Stubbs, J.B.; Stabin, M.G.; Hays, M.T.; Koral, K.F.; Robertson, J.S.; Howell, R.W.; Wessels, B.W.; Fisher, D.R.; et al. MIRD Pamphlet No. 16: Techniques for Quantitative Radiopharmaceutical Biodistribution Data Acquisition and Analysis for Use in Human Radiation Dose Estimates. J. Nucl. Med. 1999, 40, 37–61. [Google Scholar]
- Yi, X.; Walia, E.; Babyn, P. Generative Adversarial Network in Medical Imaging: A Review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, A. Deep Generative Adversarial Networks for Image-to-Image Translation: A Review. Symmetry 2020, 12, 1705. [Google Scholar] [CrossRef]
- Jin, D.; Xu, Z.; Tang, Y.; Harrison, A.P.; Mollura, D.J. CT-Realistic Lung Nodule Simulation from 3D Conditional Generative Adversarial Networks for Robust Lung Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; pp. 732–740. [Google Scholar] [CrossRef]
- Pang, S.; Du, A.; Orgun, M.A.; Yu, Z.; Wang, Y.; Wang, Y.; Liu, G. CTumorGAN: A Unified Framework for Automatic Computed Tomography Tumor Segmentation. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2248–2268. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, M.D.; Abramian, D.; Eklund, A. Vox2Vox: 3D-GAN for Brain Tumour Segmentation. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020; pp. 274–284. [Google Scholar] [CrossRef]
- Nie, D.; Trullo, R.; Lian, J.; Petitjean, C.; Ruan, S.; Wang, Q.; Shen, D. Medical Image Synthesis with Context-Aware Generative Adversarial Networks. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; pp. 417–425. [Google Scholar] [CrossRef]
- Armanious, K.; Jiang, C.; Fischer, M.; Küstner, T.; Hepp, T.; Nikolaou, K.; Gatidis, S.; Yang, B. MedGAN: Medical Image Translation Using GANs. Comput. Med. Imaging Graph. 2020, 79, 101684. [Google Scholar] [CrossRef] [PubMed]
- Abu-Srhan, A.; Almallahi, I.; Abushariah, M.A.M.; Mahafza, W.; Al-Kadi, O.S. Paired-Unpaired Unsupervised Attention Guided GAN with Transfer Learning for Bidirectional Brain MR-CT Synthesis. Comput. Biol. Med. 2021, 136, 104763. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Zhang, H.; Wang, N.; Gao, X.; Shen, D. Auto-GAN: Self-Supervised Collaborative Learning for Medical Image Synthesis. Proc. AAAI Conf. Artif. Intell. 2020, 34, 10486–10493. [Google Scholar] [CrossRef]
- Lin, W.; Lin, W.; Chen, G.; Zhang, H.; Gao, Q.; Huang, Y.; Tong, T.; Du, M. Bidirectional Mapping of Brain MRI and PET With 3D Reversible GAN for the Diagnosis of Alzheimer’s Disease. Front. Neurosci. 2021, 15, 357. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Advances in Neural Information Processing Systems; NeurIPS: La Jolla, CA, USA, 2014; Volume 27. [Google Scholar]
- Islam, J.; Zhang, Y. GAN-Based Synthetic Brain PET Image Generation. Brain Inform. 2020, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Abazari, M.A.; Soltani, M.; Moradi Kashkooli, F.; Raahemifar, K. Synthetic 18F-FDG PET Image Generation Using a Combination of Biomathematical Modeling and Machine Learning. Cancers 2022, 14, 2786. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar] [CrossRef]
- Isola, P.; Zhu, J.-Y.; Zhou, T.; Efros, A.A.; Research, B.A. Image-To-Image Translation With Conditional Adversarial Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Ledig, C.; Theis, L.; Huszár, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-Realistic Single Image Super-Resolution Using a Generative Adversarial Network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4681–4690. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv 2015, arXiv:1511.06434. [Google Scholar] [CrossRef]
- Li, C.; Wand, M. Precomputed Real-Time Texture Synthesis with Markovian Generative Adversarial Networks. In Proceedings of the Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; pp. 702–716. [Google Scholar] [CrossRef]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein Generative Adversarial Networks. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 214–223. [Google Scholar]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A.C. Improved Training of Wasserstein GANs. In Advances in Neural Information Processing Systems; NeurIPS: La Jolla, CA, USA, 2017; Volume 30. [Google Scholar]
- Mao, X.; Li, Q.; Xie, H.; Lau, R.Y.K.; Wang, Z.; Paul Smolley, S. Least Squares Generative Adversarial Networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2794–2802. [Google Scholar]
- Gatys, L.A.; Ecker, A.S.; Bethge, M. A Neural Algorithm of Artistic Style. J. Vis. 2015, 16, 326. [Google Scholar] [CrossRef]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs Trained by a Two Time-Scale Update Rule Converge to a Local Nash Equilibrium. In Advances in Neural Information Processing Systems; NeurIPS: La Jolla, CA, USA, 2017; Volume 30. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury Google, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems; NeurIPS: La Jolla, CA, USA, 2019; Volume 32. [Google Scholar]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A.; Research, B.A. Unpaired Image-To-Image Translation Using Cycle-Consistent Adversarial Networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Park, T.; Efros, A.A.; Zhang, R.; Zhu, J.Y. Contrastive Learning for Unpaired Image-to-Image Translation. In Proceedings of the Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020; pp. 319–345. [Google Scholar] [CrossRef]
- Lei, Y.; Harms, J.; Wang, T.; Liu, Y.; Shu, H.K.; Jani, A.B.; Curran, W.J.; Mao, H.; Liu, T.; Yang, X. MRI-Only Based Synthetic CT Generation Using Dense Cycle Consistent Generative Adversarial Networks. Med. Phys. 2019, 46, 3565–3581. [Google Scholar] [CrossRef]
- Klages, P.; Benslimane, I.; Riyahi, S.; Jiang, J.; Hunt, M.; Deasy, J.O.; Veeraraghavan, H.; Tyagi, N. Patch-Based Generative Adversarial Neural Network Models for Head and Neck MR-Only Planning. Med. Phys. 2020, 47, 626–642. [Google Scholar] [CrossRef]


| Loss Function | Uptake Time (Min) | Reconstruction Method | FID | PSNR (dB) |
|---|---|---|---|---|
| GAN + L1 loss | 14 | OSEM 3D | 45.71 | 52.29 |
| WGAN + L1 loss | 14 | OSEM 3D | 18.45 | 52.54 |
| LSGAN + L1 loss | 14 | OSEM 3D | 19.02 | 53.06 |
| GAN + perceptual loss | 14 | OSEM 3D | 31.05 | 51.65 |
| WGAN + perceptual loss | 14 | OSEM 3D | 18.69 | 52.86 |
| LSGAN + perceptual loss | 14 | OSEM 3D | 14.49 | 53.25 |
| LSGAN + perceptual loss | 5 | OSEM 3D | 21.36 | 53.29 |
| LSGAN + perceptual loss | 14 | OSEM 3D | 14.49 | 53.25 |
| LSGAN + perceptual loss | 31 | OSEM 3D | 11.61 | 53.75 |
| LSGAN + perceptual loss | 14 | FBP | 85.93 | 40.46 |
| LSGAN + perceptual loss | 14 | OSEM 2D | 15.05 | 52.71 |
| LSGAN + perceptual loss | 14 | OSEM 3D | 14.49 | 53.25 |
| LSGAN + perceptual loss | 14 | TrueX | 10.43 | 56.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Byun, B.H.; Lim, I.; Lim, S.M.; Woo, S.-K. Deep Learning-Based Delayed PET Image Synthesis from Corresponding Early Scanned PET for Dosimetry Uptake Estimation. Diagnostics 2023, 13, 3045. https://doi.org/10.3390/diagnostics13193045
Kim K, Byun BH, Lim I, Lim SM, Woo S-K. Deep Learning-Based Delayed PET Image Synthesis from Corresponding Early Scanned PET for Dosimetry Uptake Estimation. Diagnostics. 2023; 13(19):3045. https://doi.org/10.3390/diagnostics13193045
Chicago/Turabian StyleKim, Kangsan, Byung Hyun Byun, Ilhan Lim, Sang Moo Lim, and Sang-Keun Woo. 2023. "Deep Learning-Based Delayed PET Image Synthesis from Corresponding Early Scanned PET for Dosimetry Uptake Estimation" Diagnostics 13, no. 19: 3045. https://doi.org/10.3390/diagnostics13193045
APA StyleKim, K., Byun, B. H., Lim, I., Lim, S. M., & Woo, S.-K. (2023). Deep Learning-Based Delayed PET Image Synthesis from Corresponding Early Scanned PET for Dosimetry Uptake Estimation. Diagnostics, 13(19), 3045. https://doi.org/10.3390/diagnostics13193045






