GabROP: Gabor Wavelets-Based CAD for Retinopathy of Prematurity Diagnosis via Convolutional Neural Networks
Abstract
:1. Introduction
- Utilizing the GW texture analysis method to analyze fundus images and produce multiple sets of GW images to train DL models along with the original fundus images.
- Three cutting-edge pre-trained CNN models with various architectures are employed to construct GabROP.
- Integrating several textural features attained through training each CNN with a different set of GW images using DWT, resulting in textural-spectral-temporal interpretation. Moreover, a reduction in the dimension of integrated features is performed.
- Merging spatial information obtained using features extracted from each CNN constructed with the actual fundus photos with the textural-spectral-temporal representation of the combined features acquired via the same CNN constructed with the multiple sets of GW images.
- Incorporating fused features of the three CNNs, thus combining the benefits of their distinct structures.
2. Related CAD Systems for ROP Diagnosis
3. Materials and Methods
3.1. ROP Dataset Description
3.2. Proposed GabROP Tool
3.2.1. Gabor Wavelets Image Generation and Preprocessing
3.2.2. Multiple CNN Training and Extraction of Features
3.2.3. Triple Fusion
3.2.4. ROP Diagnosis
4. Performance Evaluation and Setup
5. Results
5.1. First Fusion Stage Results
5.2. Second Fusion Stage Results
5.3. Third Fusion Stage Results
6. Discussion
6.1. Comparisons
6.2. Limitations and Future Directions
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Flynn, J.T.; Bancalari, E.; Bachynski, B.N.; Buckley, E.B.; Bawol, R.; Goldberg, R.; Cassady, J.; Schiffman, J.; Feuer, W.; Gillings, D. Retinopathy of Prematurity: Diagnosis, Severity, and Natural History. Ophthalmology 1987, 94, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, H.; Lawn, J.E.; Vazquez, T.; Fielder, A.; Gilbert, C. Preterm-Associated Visual Impairment and Estimates of Retinopathy of Prematurity at Regional and Global Levels for 2010. Pediatr. Res. 2013, 74, 35–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Chen, Z.; Zhu, W.; Shi, F.; Wang, M.; Zhou, Y.; Xiang, D.; Chen, X.; Chen, F. ADS-Net: Attention-Awareness and Deep Supervision Based Network for Automatic Detection of Retinopathy of Prematurity. Biomed. Opt. Express 2022, 13, 4087–4101. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.F.; Wang, L.; Busuioc, M.; Du, Y.E.; Chan, P.; Kane, S.A.; Lee, T.C.; Weissgold, D.J.; Berrocal, A.M.; Coki, O. Telemedical Retinopathy of Prematurity Diagnosis: Accuracy, Reliability, and Image Quality. Arch. Ophthalmol. 2007, 125, 1531–1538. [Google Scholar] [CrossRef]
- Bao, Y.; Ming, W.-K.; Mou, Z.-W.; Kong, Q.-H.; Li, A.; Yuan, T.-F.; Mi, X.-S. Current Application of Digital Diagnosing Systems for Retinopathy of Prematurity. Comput. Methods Programs Biomed. 2020, 200, 105871. [Google Scholar] [CrossRef]
- Attallah, O. A Computer-Aided Diagnostic Framework for Coronavirus Diagnosis Using Texture-Based Radiomics Images. Digit. Health 2022, 8, 20552076221092544. [Google Scholar] [CrossRef]
- Attallah, O.; Sharkas, M. GASTRO-CADx: A Three Stages Framework for Diagnosing Gastrointestinal Diseases. PeerJ Comput. Sci. 2021, 7, e423. [Google Scholar] [CrossRef]
- Fujita, H. AI-Based Computer-Aided Diagnosis (AI-CAD): The Latest Review to Read First. Radiol. Phys. Technol. 2020, 13, 6–19. [Google Scholar] [CrossRef]
- Attallah, O.; Anwar, F.; Ghanem, N.M.; Ismail, M.A. Histo-CADx: Duo Cascaded Fusion Stages for Breast Cancer Diagnosis from Histopathological Images. PeerJ Comput. Sci. 2021, 7, e493. [Google Scholar] [CrossRef]
- Attallah, O.; Ragab, D.A. Auto-MyIn: Automatic Diagnosis of Myocardial Infarction via Multiple GLCMs, CNNs, and SVMs. Biomed. Signal Process. Control 2023, 80, 104273. [Google Scholar] [CrossRef]
- Attallah, O.; Ma, X. Bayesian Neural Network Approach for Determining the Risk of Re-Intervention after Endovascular Aortic Aneurysm Repair. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2014, 228, 857–866. [Google Scholar] [CrossRef]
- Bakator, M.; Radosav, D. Deep Learning and Medical Diagnosis: A Review of Literature. Multimodal Technol. Interact. 2018, 2, 47. [Google Scholar] [CrossRef] [Green Version]
- Attallah, O.; Sharkas, M. Intelligent Dermatologist Tool for Classifying Multiple Skin Cancer Subtypes by Incorporating Manifold Radiomics Features Categories. Contrast Media Mol. Imaging 2021, 2021, 7192016. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O.; Zaghlool, S. AI-Based Pipeline for Classifying Pediatric Medulloblastoma Using Histopathological and Textural Images. Life 2022, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O.; Sharkas, M.A.; Gadelkarim, H. Deep Learning Techniques for Automatic Detection of Embryonic Neurodevelopmental Disorders. Diagnostics 2020, 10, 27–49. [Google Scholar] [CrossRef] [Green Version]
- Ragab, D.A.; Attallah, O. FUSI-CAD: Coronavirus (COVID-19) Diagnosis Based on the Fusion of CNNs and Handcrafted Features. PeerJ Comput. Sci. 2020, 6, e306. [Google Scholar] [CrossRef]
- Dewey, M.; Schlattmann, P. Deep Learning and Medical Diagnosis. Lancet 2019, 394, 1710–1711. [Google Scholar] [CrossRef] [Green Version]
- Attallah, O.; Aslan, M.F.; Sabanci, K. A Framework for Lung and Colon Cancer Diagnosis via Lightweight Deep Learning Models and Transformation Methods. Diagnostics 2022, 12, 2926. [Google Scholar] [CrossRef]
- Razzak, M.I.; Naz, S.; Zaib, A. Deep Learning for Medical Image Processing: Overview, Challenges and the Future. In Classification in BioApps; Springer: Cham, Switzerland, 2018; pp. 323–350. [Google Scholar]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis Using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [Green Version]
- Attallah, O. A Deep Learning-Based Diagnostic Tool for Identifying Various Diseases via Facial Images. Digit. Health 2022, 8, 20552076221124432. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Mitsuhashi, T.; Matsuo, T. Accuracy of Deep Learning Algorithms for the Diagnosis of Retinopathy of Prematurity by Fundus Images: A Systematic Review and Meta-Analysis. J. Ophthalmol. 2021, 2021, 8883946. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.P.; Singh, P.; Redd, T.K.; Brown, J.M.; Shah, P.K.; Subramanian, P.; Rajan, R.; Valikodath, N.; Cole, E.; Ostmo, S. Applications of Artificial Intelligence for Retinopathy of Prematurity Screening. Pediatrics 2021, 147, e2020016618. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, M.F.; Danford, I.D.; Shahrawat, M.; Ostmo, S.; Brown, J.; Kalpathy-Cramer, J.; Bradshaw, K.; Schelonka, R.; Cohen, H.S.; Chan, R.P. Evaluation of Artificial Intelligence-Based Telemedicine Screening for Retinopathy of Prematurity. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2020, 24, 160–162. [Google Scholar] [CrossRef] [PubMed]
- Castellano, G.; Bonilha, L.; Li, L.M.; Cendes, F. Texture Analysis of Medical Images. Clin. Radiol. 2004, 59, 1061–1069. [Google Scholar] [CrossRef]
- Attallah, O.; Sharkas, M.A.; Gadelkarim, H. Fetal Brain Abnormality Classification from MRI Images of Different Gestational Age. Brain Sci. 2019, 9, 231–252. [Google Scholar] [CrossRef] [Green Version]
- Parekh, R. Using Texture Analysis for Medical Diagnosis. IEEE MultiMedia 2012, 19, 28–37. [Google Scholar] [CrossRef]
- Buciu, I.; Gacsadi, A. Gabor Wavelet Based Features for Medical Image Analysis and Classification. In Proceedings of the 2009 2nd International Symposium on Applied Sciences in Biomedical and Communication Technologies, Bratislava, Slovakia, 24–27 November 2009; IEEE Press: New York, NY, USA, 2009; pp. 1–4. [Google Scholar]
- Pour, E.K.; Pourreza, H.; Zamani, K.A.; Mahmoudi, A.; Sadeghi, A.M.M.; Shadravan, M.; Karkhaneh, R.; Pour, R.R.; Esfahani, M.R. Retinopathy of Prematurity-Assist: Novel Software for Detecting plus Disease. Korean J. Ophthalmol. KJO 2017, 31, 524–532. [Google Scholar] [CrossRef] [Green Version]
- Oloumi, F.; Rangayyan, R.M.; Ells, A.L. Computer-Aided Diagnosis of Plus Disease in Retinal Fundus Images of Preterm Infants via Measurement of Vessel Tortuosity. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milano, Italy, 25–29 August 2015; IEEE Press: New York, NY, USA, 2015; pp. 4338–4342. [Google Scholar]
- Oloumi, F.; Rangayyan, R.M.; Casti, P.; Ells, A.L. Computer-Aided Diagnosis of plus Disease via Measurement of Vessel Thickness in Retinal Fundus Images of Preterm Infants. Comput. Biol. Med. 2015, 66, 316–329. [Google Scholar] [CrossRef]
- Nasrazadani, D.A.; Wallace, D.K.; Freedman, S.F.; Prakalapakorn, S.G. Development of a Scale for Grading Pre-plus and plus Disease Using Retinal Images: A Pilot Study. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2018, 22, 316–319. [Google Scholar] [CrossRef]
- Ataer-Cansizoglu, E.; Bolon-Canedo, V.; Campbell, J.P.; Bozkurt, A.; Erdogmus, D.; Kalpathy-Cramer, J.; Patel, S.; Jonas, K.; Chan, R.P.; Ostmo, S. Computer-Based Image Analysis for plus Disease Diagnosis in Retinopathy of Prematurity: Performance of the “i-ROP” System and Image Features Associated with Expert Diagnosis. Transl. Vis. Sci. Technol. 2015, 4, 5. [Google Scholar] [CrossRef]
- Han, D.; Liu, Q.; Fan, W. A New Image Classification Method Using CNN Transfer Learning and Web Data Augmentation. Expert Syst. Appl. 2018, 95, 43–56. [Google Scholar] [CrossRef]
- Attallah, O. An Intelligent ECG-Based Tool for Diagnosing COVID-19 via Ensemble Deep Learning Techniques. Biosensors 2022, 12, 299. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. CoMB-Deep: Composite Deep Learning-Based Pipeline for Classifying Childhood Medulloblastoma and Its Classes. Front. Neuroinformatics 2021, 15, 663592. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ju, R.; Chen, Y.; Zhang, L.; Hu, J.; Wu, Y.; Dong, W.; Zhong, J.; Yi, Z. Automated Retinopathy of Prematurity Screening Using Deep Neural Networks. EBioMedicine 2018, 35, 361–368. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Liu, J. Early Diagnosis and Quantitative Analysis of Stages in Retinopathy of Prematurity Based on Deep Convolutional Neural Networks. Transl. Vis. Sci. Technol. 2022, 11, 17. [Google Scholar] [CrossRef]
- Brown, J.M.; Campbell, J.P.; Beers, A.; Chang, K.; Ostmo, S.; Chan, R.P.; Dy, J.; Erdogmus, D.; Ioannidis, S.; Kalpathy-Cramer, J. Automated Diagnosis of plus Disease in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2018, 136, 803–810. [Google Scholar] [CrossRef]
- Yildiz, V.M.; Tian, P.; Yildiz, I.; Brown, J.M.; Kalpathy-Cramer, J.; Dy, J.; Ioannidis, S.; Erdogmus, D.; Ostmo, S.; Kim, S.J. Plus Disease in Retinopathy of Prematurity: Convolutional Neural Network Performance Using a Combined Neural Network and Feature Extraction Approach. Transl. Vis. Sci. Technol. 2020, 9, 10. [Google Scholar] [CrossRef] [Green Version]
- Rani, P.; Elagiri Ramalingam, R.; Rajamani, K.T.; Kandemir, M.; Singh, D. Multiple Instance Learning: Robust Validation on Retinopathy of Prematurity. Int. J. Ctrl Theory Appl. 2016, 9, 451–459. [Google Scholar]
- Tan, Z.; Simkin, S.; Lai, C.; Dai, S. Deep Learning Algorithm for Automated Diagnosis of Retinopathy of Prematurity plus Disease. Transl. Vis. Sci. Technol. 2019, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Lei, B.; Zeng, X.; Huang, S.; Zhang, R.; Chen, G.; Zhao, J.; Wang, T.; Wang, J.; Zhang, G. Automated Detection of Retinopathy of Prematurity by Deep Attention Network. Multimed. Tools Appl. 2021, 80, 36341–36360. [Google Scholar] [CrossRef]
- Zhang, R.; Zhao, J.; Xie, H.; Wang, T.; Chen, G.; Zhang, G.; Lei, B. Automatic Diagnosis for Aggressive Posterior Retinopathy of Prematurity via Deep Attentive Convolutional Neural Network. Expert Syst. Appl. 2021, 187, 115843. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Vadloori, S.; Chu, H.-C.; Kang, E.Y.-C.; Wu, W.-C.; Kusaka, S.; Fukushima, Y. Deep Learning Models for Automated Diagnosis of Retinopathy of Prematurity in Preterm Infants. Electronics 2020, 9, 1444. [Google Scholar] [CrossRef]
- Attallah, O. DIAROP: Automated Deep Learning-Based Diagnostic Tool for Retinopathy of Prematurity. Diagnostics 2021, 11, 2034. [Google Scholar] [CrossRef]
- Hu, J.; Chen, Y.; Zhong, J.; Ju, R.; Yi, Z. Automated Analysis for Retinopathy of Prematurity by Deep Neural Networks. IEEE Trans. Med. Imaging 2018, 38, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhong, J.; Yang, S.; Gao, Z.; Hu, J.; Chen, Y.; Yi, Z. Automated Identification and Grading System of Diabetic Retinopathy Using Deep Neural Networks. Knowl. Based Syst. 2019, 175, 12–25. [Google Scholar] [CrossRef]
- Attallah, O.; Samir, A. A Wavelet-Based Deep Learning Pipeline for Efficient COVID-19 Diagnosis via CT Slices. Appl. Soft Comput. 2022, 128, 109401. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. Deep Learning-Based CAD System for COVID-19 Diagnosis via Spectral-Temporal Images. In Proceedings of the 2022 The 12th International Conference on Information Communication and Management, London, UK, 13–15 July 2022; pp. 25–33. [Google Scholar]
- Ali, H.; Sharif, M.; Yasmin, M.; Rehmani, M.H.; Riaz, F. A Survey of Feature Extraction and Fusion of Deep Learning for Detection of Abnormalities in Video Endoscopy of Gastrointestinal-Tract. Artif. Intell. Rev. 2019, 53, 2635–2707. [Google Scholar] [CrossRef]
- Cheng, Y.; Feng, J.; Jia, K. A Lung Disease Classification Based on Feature Fusion Convolutional Neural Network with X-Ray Image Enhancement. In Proceedings of the 2018 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA ASC), Honolulu, HI, USA, 12–15 November 2018; pp. 2032–2035. [Google Scholar]
- Attallah, O. MB-AI-His: Histopathological Diagnosis of Pediatric Medulloblastoma and Its Subtypes via AI. Diagnostics 2021, 11, 359–384. [Google Scholar] [CrossRef]
- Attallah, O. ECG-BiCoNet: An ECG-Based Pipeline for COVID-19 Diagnosis Using Bi-Layers of Deep Features Integration. Comput. Biol. Med. 2022, 142, 105210. [Google Scholar] [CrossRef]
- Fang, H.; Li, F.; Fu, H.; Sun, X.; Cao, X.; Lin, F.; Son, J.; Kim, S.; Quellec, G.; Matta, S. ADAM Challenge: Detecting Age-Related Macular Degeneration from Fundus Images. IEEE Trans. Med. Imaging 2022, 41, 2828–2847. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.; Wu, Z.; Zeng, J.; Chen, Y.; Tian, R.; Zhao, J.; Zhang, G. Development of an Automated Screening System for Retinopathy of Prematurity Using a Deep Neural Network for Wide-Angle Retinal Images. IEEE Access 2018, 7, 10232–10241. [Google Scholar] [CrossRef]
- Moon, K.; Jetawat, A. Medical Image Enhancement Technique Using Multiresolution Gabor Wavelet Transform. In Innovations in Computer Science and Engineering; Springer: Singapore, 2021; pp. 385–392. [Google Scholar]
- Serte, S.; Demirel, H. Gabor Wavelet-Based Deep Learning for Skin Lesion Classification. Comput. Biol. Med. 2019, 113, 103423. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Zhu, Z.; Zou, Y.; Wang, X. Facial Expression Recognition Based on Gabor Wavelet Transform and 2-Channel CNN. Int. J. Wavelets Multiresol. Inf. Process. 2020, 18, 2050003. [Google Scholar] [CrossRef]
- Haghighat, M.; Zonouz, S.; Abdel-Mottaleb, M. CloudID: Trustworthy Cloud-Based and Cross-Enterprise Biometric Identification. Expert Syst. Appl. 2015, 42, 7905–7916. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Ponnusamy, R.; Sathiamoorthy, S. Bleeding and Z-Line Classification by DWT Based SIFT Using KNN and SVM. In Proceedings of the International Conference On Computational Vision and Bio Inspired Computing, Coimbatore, India, 18–19 November 2022; Springer: Cham, Switzerland, 2019; pp. 679–688. [Google Scholar]
- Miri, A.; Sharifian, S.; Rashidi, S.; Ghods, M. Medical Image Denoising Based on 2D Discrete Cosine Transform via Ant Colony Optimization. Optik 2018, 156, 938–948. [Google Scholar] [CrossRef]
- Colquhoun, D. An Investigation of the False Discovery Rate and the Misinterpretation of P-Values. R. Soc. Open Sci. 2014, 1, 140216. [Google Scholar] [CrossRef] [Green Version]
- Attallah, O. An Effective Mental Stress State Detection and Evaluation System Using Minimum Number of Frontal Brain Electrodes. Diagnostics 2020, 10, 292–327. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; IEEE Press: New York, NY, USA, 2017; pp. 7263–7271. [Google Scholar]
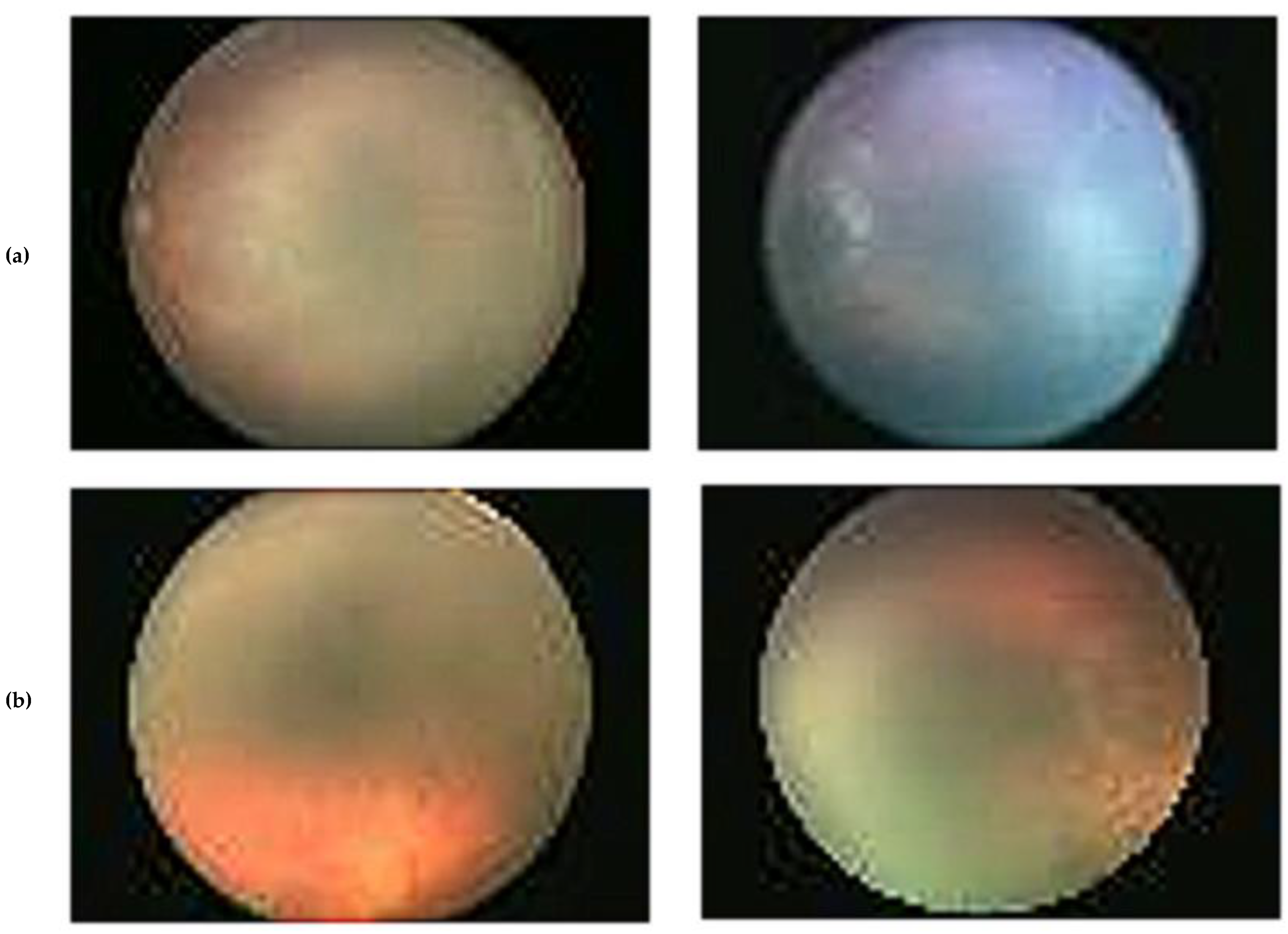

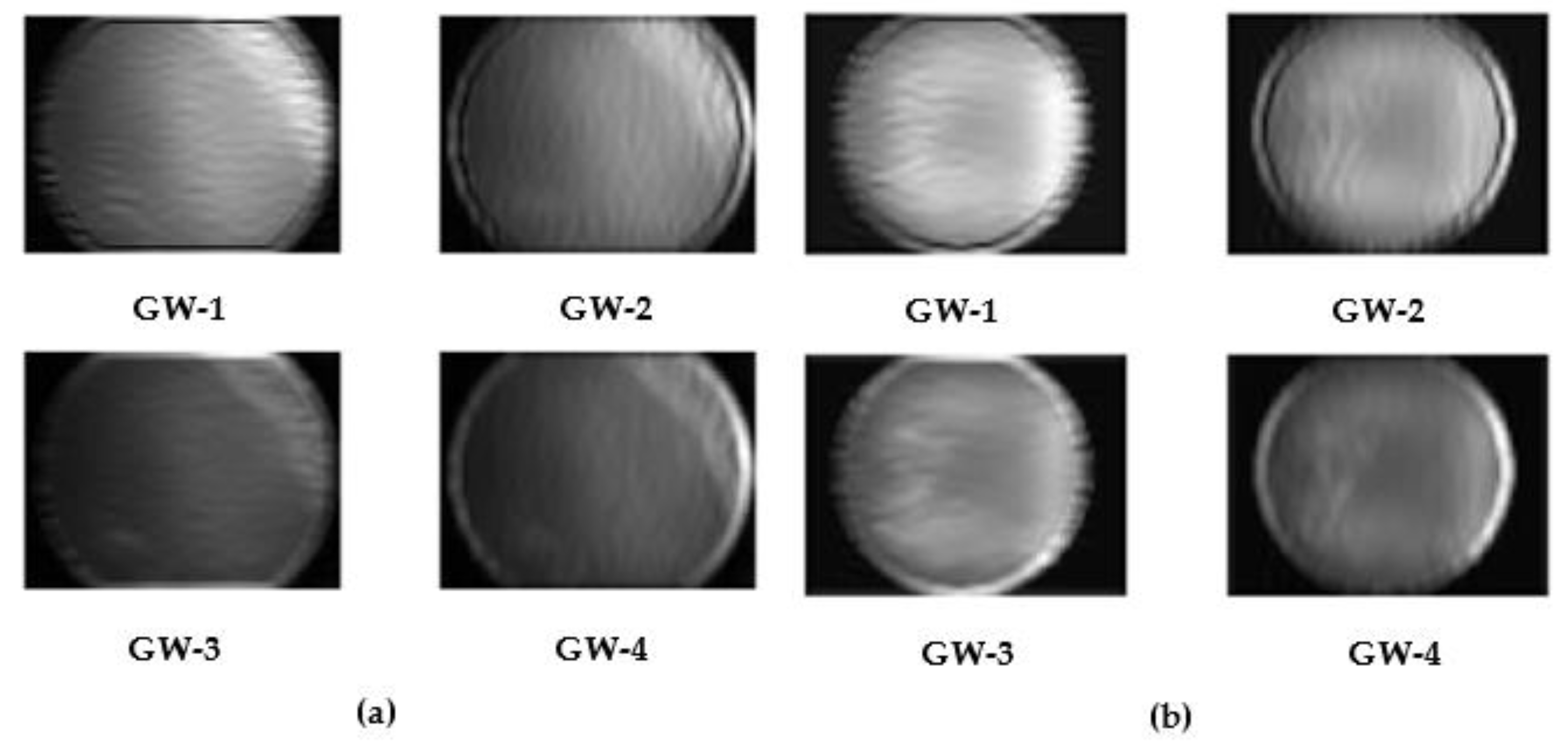









| Attribute | Set 1 | Set 2 |
|---|---|---|
| Gender | ||
| Female | 7726 | 754 |
| Male | 10,075 | 988 |
| Age | 31.9 (24–36.4) | 32.0 (25–36.2) |
| Birth Weight | 1.49 (0.63–2.00) | 1.50 (0.78–2.00) |
| Classification of Images | ||
| ROP Images | 8090 | 155 |
| Not ROP Images | 9711 | 1587 |
| Classifier | Precision | Specificity | Sensitivity | F1-score |
|---|---|---|---|---|
| LDA | 95.57 | 96.54 | 89.65 | 92.52 |
| SVM | 96.11 | 96.96 | 90.12 | 93.02 |
| ESD | 96.01 | 96.90 | 89.59 | 92.69 |
| Article | Model | Accuracy (%) |
|---|---|---|
| [56] | GoogleNet | 73.9 |
| [56] | AlexNet | 77.9 |
| [56] | VGG-16 | 80.4 |
| [46] | ResNet + Inception + InceptionResNet | 93.2 |
| [48] | Inception-ResNet V2 | 90.9 |
| [48] | Xception | 86.95 |
| [44] | ResNet-50 | 91.48 |
| [47] | Inception V3 | 91.42 |
| [66] | MobileNet | 87.94 |
| [67] | DarkNet-53 | 91.0 |
| GabROP | 93.9% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attallah, O. GabROP: Gabor Wavelets-Based CAD for Retinopathy of Prematurity Diagnosis via Convolutional Neural Networks. Diagnostics 2023, 13, 171. https://doi.org/10.3390/diagnostics13020171
Attallah O. GabROP: Gabor Wavelets-Based CAD for Retinopathy of Prematurity Diagnosis via Convolutional Neural Networks. Diagnostics. 2023; 13(2):171. https://doi.org/10.3390/diagnostics13020171
Chicago/Turabian StyleAttallah, Omneya. 2023. "GabROP: Gabor Wavelets-Based CAD for Retinopathy of Prematurity Diagnosis via Convolutional Neural Networks" Diagnostics 13, no. 2: 171. https://doi.org/10.3390/diagnostics13020171






