1. Introduction
Musculoskeletal ultrasound (MSK-US) imaging is a valuable diagnostic tool for the examination of the musculoskeletal system since it enables real-time and non-invasive investigation of muscles in both normal and pathological conditions. An important application in clinical practice is the measurement of the visible cross-sectional area (CSA) since it quantifies the muscle’s size and echogenicity. These two basic architectural parameters are closely correlated with the maximal force a muscle can produce and it has been proven that they can be measured using conventional transverse ultrasound scans [
1,
2,
3]. In addition, echogenicity is also important for detecting health disorders affecting the muscles; in certain diseases, intramuscular fat and fibrosis alter normal muscle composition causing a progressive increase in echogenicity [
4]. However, manually placing the region of interest (ROI) in each muscle to extract the echogenicity can be tedious, user-dependent, and time-consuming. Therefore, the automatic extraction of CSA via state-of-the-art deep learning algorithms is an important research field that has witnessed a growing interest in recent years. Its goal is to quantify the muscle size and echogenicity inside the muscle CSA since it can be experimentally determined that a maximum ROI size within a given muscle will produce the least measurement variation.
A few methods have been used to estimate muscle CSA in vivo. These include magnetic resonance imaging (MRI), computed tomography, and ultrasound imaging. It has been shown that ultrasound imaging produces reliable results compared to MRI, which is considered the golden standard [
5]. The extraction of the muscle’s anatomical cross-section area (ACSA) is commonly performed by positioning the transducer in the transversal plane of the muscle (i.e., across the largest muscle diameter) and using an extended field-of-view imaging technique which makes the recording of the larger portion of the muscle possible. Although this imaging modality contains more information about muscle characteristics, the acquisition process is complex. It requires high experience on the operator’s part, high accuracy of manual tracking, dedicated software tools, and is time-consuming. A simpler and more accurate way of performing this measurement is by extracting the visible muscle CSA using conventional transverse ultrasound scans. Specifically, in [
6,
7,
8], the authors evaluated the muscle size changes induced by musculoskeletal training and rehabilitation by analysing the visible muscle CSA. Furthermore, in other studies [
9,
10], it has been reported that a CSA variation >5% is an indicator of a clinically relevant change; therefore, the accurate extraction of this measurement is important.
However, delineating the CSA from the transverse MSK-US scans is a time-consuming and user-dependent task, thus making it error-prone. Therefore, recent studies have focused on developing automated ways to extract the CSA without manual intervention using image processing and deep learning techniques to address this issue. In particular, in [
11,
12], the authors introduced a gradient-based filtering approach to initially delineate the aponeurosis-like structures of the examined images and later used heuristic and morphological operations to identify the relevant deep and superficial aponeuroses. As a final step, the CSA area was obtained by connecting the endpoints of the aponeurosis. However, their approaches’ downsides are that the aponeuroses’ identification is based exclusively on gradient-based filtering, which can be error-prone in low-contrast images, a common situation in MSK-US scans of abnormal muscles.
To overcome this difficulty, in [
13] the authors investigated the performance of different deep learning architectures in automatically delineating the CSA in transverse MSK-US images in a relatively large-scale dataset of three different muscles. Additionally, they used the z-score of the CSA echogenicity to classify the muscles as normal and ab-normal based on reference values from healthy subjects. Their results indicate that it is feasible to automatically segment and perform quantitative grayscale analysis in the CSA with comparable performance to human experts. However, as the authors state, their study’s limitations are the small number of the examined skeletal muscles and the fact that all of their recordings were taken using a specific ultrasound machine. Similarly, in [
14], the authors proposed an automatic tracking method for the cross-sectional muscle area in ultrasound images of rectus femoris using a convolutional neural network during muscle contraction. Different approaches have also been used in analysing MSK-US images for the automatic extraction of ACSA. In particular, in [
15], a semi-automatic ImageJ script (named “ACSAuto”) for quantifying the ACSA of lower limb muscles was proposed, and in [
16], a UNet [
17] was trained to automatically segment ACSA in panoramic ultrasound images of the human lower limb muscles.
Convolutional neural networks (CNNs) have not been used only for automating the CSA measurement. In recent studies, the effectiveness of deep learning methods has also been demonstrated in other medical image segmentation problems. In particular, in [
18], a deep learning approach was used to automatically extract the muscle thickness, fascicle length, and pennation angle from MSK-US longitudinal scans of three different muscles. The authors initially employed Attention-UNet [
19] to segment the deep and superficial aponeuroses, and then the measurements were extracted by incorporating a heuristic-based approach. Another notable recent development was the introduction of vision transformers [
20,
21], which attracted a growing interest in medical imaging for their ability to capture global context information compared to CNNs with local receptive fields. Specifically, in [
22], TMUNet is presented, which is a UNet variant with transformer capabilities. The authors integrated a contextual attention mechanism to adaptively aggregate pixel, object, and image-level features. Furthermore, they coupled a transformer module with the CNN encoder to model an object-level interaction. In addition, in [
23], a convolutional multilayer perceptron (MLP)-based network called UNeXt was proposed, which was evaluated in breast ultrasound image segmentation. Its advantage is that MLPs are less complicated than convolution or self-attention and transformers, so reducing the number of parameters and computational complexity was feasible.
Several studies have emerged in recent years regarding the textural analysis of musculoskeletal ultrasound images [
24,
25,
26,
27,
28]. The purpose of these studies is usually to categorise subjects’ biomarkers based on muscle texture, such as gender, the dominant side, the body mass index (BMI), or the pathological condition of the muscle, especially in specific musculoskeletal disorders. The methods mostly used for the categorisation belong to classical machine learning with hand-crafted feature extraction techniques and classical classifiers. However, deep learning methodologies have recently been used to characterise muscle texture. In [
27], the authors used three common deep learning architectures to classify a muscle region of interest (ROI) as sarcopenic based on its texture. In addition, the Grad-CAM [
29] analysis was applied to visualise the exact location inside the ROI that played the most significant role in the classifier decision. An accuracy ranging from 70.0% to 80.0% for predicting sarcopenia was reported in their experiments, showing that such a diagnostic tool is feasible.
This study presents a complete pipeline for automatically extracting the muscle CSA and its mean grey level value, representing echogenicity. Different state-of-the-art deep learning models and vision transformers have been incorporated along with customised procedures to extract this information automatically. Subsequently, comparative muscle size and echogenicity results are reported and discussed for patients of different age groups and health conditions, as well as the behaviour of these measurements in different muscle sections. Additionally, a textural analysis is performed in the muscle CSA of these subjects to investigate whether a classification into healthy and abnormal muscles is feasible by using higher order and more sophisticated information than echogenicity. In particular, a deep learning classifier trained in the muscle CSA texture of young and elderly subjects is used to automatically classify the ultrasound recordings into these categories. Finally, the performance of each muscle section in this classification was investigated to find which muscle was more fitted for this task.
This study’s main contribution is the improvement of the existing methods for a more efficient and reliable calculation of the muscle CSA, by introducing state-of-the art techniques such as vision transformers. Furthermore, future integration of this software in ultrasound machines would be of high value since this would allow ultrasonographers to calculate muscle size and echogenicity faster, more accurately, and in a more standardised fashion. Another contribution is automatically classifying the subject’s muscle condition solely from the CSA texture, again with a state-of-the-art deep learning classifier. To the best of our knowledge, this is the first time a complete pipeline is presented, beginning from the automatic extraction of the CSA and ending in classifying an abnormal muscle. Based on our results, we firmly believe that the proposed method can be applied in the clinical routine as an additional diagnostic tool to assist the experts. Finally, it is worth mentioning that all of the analyses have been performed on a novel, large, and diverse MSK-US dataset; therefore, the results presented in this study provide additional knowledge for the feasibility of the abovementioned tasks.
3. Results
3.1. CSA Extraction Results
Table 2 demonstrates the image segmentation results for the five examined network topologies and TRAMA algorithm. It must be noted that the implementation of the TRAMA algorithm is ours since there is no official one. However, we tried to follow as closely as possible the algorithm proposed in [
11]. For all of the networks, the average performance between the evaluation set of the five folds is reported. In all five metrics, the TMUNet exhibits superior performance in relevance with the other networks, and therefore all of the CSA segmentation results needed for the rest of the analyses of this study were extracted using it. In particular, the best-reported precision is 0.95, with the corresponding recall being 0.96 showing the network’s capability to localise the deep and superficial aponeuroses accurately. Furthermore, in terms of DSC and IoU, the reported results are equal to 0.96 and 0.92, respectively, with the corresponding accuracy in HD95 being only 10.09, another indicator of the excellent performance of the segmented masks of the validation set. Lastly, the small standard deviation of the results indicate that the performance of the deep learning models is relatively stable without severe or total failures.
It must be noted that all of the different deep learning architectures surpassed the TRAMA methodology by a large margin. This behaviour can be explained by the convolutional neural networks’ ability to filter the noise better than the traditional approaches. More specifically, from the visual inspection of the results, TRAMA performed better in the recordings belonging to the young and healthy subjects (Group 1) since, in these images, the aponeuroses are more distinct and have higher contrast to the muscle tissue in comparison with those of the elderly (Group 2 and Group 3). In particular, the average precision of the TRAMA algorithm in all of the samples equals 43%, which is significantly less than that of CNNs. Additionally, the high standard deviation of the evaluation metrics highlights that the TRAMA predictions are not stable and that there are a lot of cases with significant or even total failures. This is the reason we omitted to report the HD95 metric in this experiment, since due to these failures becomes invalid.
Continuing our analysis,
Table 3 compares the proposed method, which consists of the TMUNet and the post-processing refinement, to the manual measurements provided by a human operator for calculating the CSA size and their corresponding echogenicity in physical units. It presents the mean ± standard deviations and their discrepancy in RMSE and HD95, and their reliability as measurements in terms of the ICC metric and Pearson coefficient.
From the results in
Table 3, it can be noted that manual vs. automatic measurements have an extremely low RMSE and HD95, showing the high quality of the predicted cross-sectional area. In particular, the RMSE equals 38.15 mm
2 which equals 4% of the total CSA, a significant result that indicates that the proposed method can be used for future automation of this clinical task. Regarding the HD95, the average performance equals 2.20 mm, which indicates the high quality of the produced masks. Regarding the statistical analysis of our results, the ICC and Pearson coefficient are close to 1, showing the reliability of the two measurements. Furthermore, in terms of echogenicity, the RMSE difference between the two readings is only 0.88, a discrepancy near 1% that shows that these two measurements can be used interchangeably. Finally, in
Figure 8, qualitative results of the proposed method are provided in different samples of each examined muscle section. From the qualitative results of
Figure 8, it is clear that the predicted masks after the post-processing refinement are very accurate and managed in most cases to capture the entire muscle CSA in all of the different muscle sections.
Another useful analysis presented in
Figure 9 is the Bland–Altman plot of the CSA and echogenicity measurements. Regarding the CSA measurements in
Figure 9A, the plot shows negligible additive bias and that most differences fall between the 95% limits of agreement. Finally, there are no distinguishable patterns depicted in the plot. Considering the echogenicity measurements in
Figure 9B, once again in this Bland–Altman plot, there are no distinguishable patterns and neither systematic error is presented.
Continuing our analysis,
Table 4 compares the automatic vs. the manual CSA measurements and their corresponding echogenicity in all of the examined muscles. For each muscle, the average discrepancy is presented in terms of RMSE.
From the above results, the B.B. has the largest CSA in all of the examined muscles in both readings. This result is expected since the B.B. consists of two muscles (biceps brachii and brachialis anterior) commonly functionally considered as one unit. Regarding the average discrepancy between the two readings, R.F. has the lowest RMSE, equal to only 32.86 mm2. In terms of percentage, the B.B. and GCM have a 5% difference between the manual and automatic measurements, the TA has a 6% difference, and the best result was reported in R.F. with only a 4% difference between the two readings. Regarding the echogenicity, R.F. again shows the lowest absolute value at 44.78, and T.A. presents the largest at 70.48. Finally, regarding the average differences, B.B. demonstrates the best result, equalling only 0.73, with all the other muscles following nearby.
Additionally, in
Figure 10, the Bland–Altman plots depict the performance of the four examined muscles. These plots show negligible additive bias and no systematic errors since most differences fall between the 95% limits of agreement. Finally, there are no distinguishable patterns depicted in the plots.
Furthermore,
Table 5 presents an analysis of the three groups that participated in this study, defined in
Table 1. Specifically, the CSA and its corresponding echogenicity are reported along with its average discrepancy in RMSE.
From the results above, it is clear that Group 1, consisting of young and healthy subjects, has the largest muscle cross-sectional area and the lowest echogenicity. This result is expected since the young subjects are anticipated to have the best muscle architecture and characteristics. In particular, the average automatic CSA measurement between all of the recordings was found to be 841.06 mm2, differing by only 34.90 in terms of RMSE from the manual readings. The corresponding average echogenicity was reported to be 52.42, once more very close to the manual measurements. Additionally, the CSA measurements of Group 2 and Group 3 were nearby in average magnitude. Therefore, this can be explained by the fact that both groups include the elderly population. The difference between them is that the subjects of Group 3 are sarcopenic; hence, the lower CSA and the larger echogenicity reported in this group’s measurements are compatible with the clinical diagnosis.
Another useful analysis presented in
Figure 11 is the Bland–Altman plots of the CSA measurements per study group. It is clear from the plots that most of the samples belong to the Group 1 category, and the smallest number of the samples are in Group 3. Furthermore, the plots show negligible additive bias and no systematic errors since most differences fall between the 95% limits of agreement. Lastly, once more there are no distinguishable patterns depicted in the plots.
Finally, for assessing the normality of the precision and recall distributions for each examined group, the box plot diagram is presented in
Figure 12. It is observable from this diagram that the distributions failed to match a normal distribution since skew is presented in all of them. Furthermore, it must be noted that the box plots of Group 1 are comparatively shorter than those of the other groups, indicating that these measurements have a higher level of agreement with each other.
3.2. CSA Textural Analysis Results
This section presents the textural analysis of each group’s CSA.
Table 6 presents the classification results for the manually and automatically extracted CSA image textures. Each method’s performance is reported in terms of the classification metrics described in
Section 2.5.
From the results above, it can be highlighted that the classifier labelled most of the samples correctly, over 84% of the time. Furthermore, the manually extracted CSA and the image segmentation network predictions have almost identical performance in the three group categories’ classification problems. Although, since the dataset suffers from imbalance, apart from the weighted average performance depicted in
Table 6, the confusion matrices are also extracted in
Figure 13 for analysing the per-group accuracy.
From the results in
Figure 13, the texture of the muscle CSAs of Group 1 is more distinguishable than the other two groups. In particular, the images of this group achieve more than 93.1% accuracy in this task. The images of Group 2 also performed well, with an average precision of 78.4%. In contrast, we observe that the results of Group 3 deteriorate to nearly 44.2% and it is common for the classifier to mislabel them as Group 2 and vice versa. The above can be explained by the fact that in both Group 2 and Group 3, many muscle CSAs have high echogenicity and similar textural characteristics that cause this confusion in the classifier. Additionally, the same pattern in the results is observed in the automatically extracted CSAs. In the textural analysis of the predicted CSAs, the classification task in Group 1 achieved 94.0% accuracy, followed by Group 2 with 73.2% and Group 3 with 37.8%. Once more, the same confusion in the classifier occurs between the samples of Group 3 and Group 2.
Furthermore, it must be noted that the number of individuals in Group 3 is much less than in the other two categories, an important limitation to consider. Additionally, even though the patients of Group 3 are sarcopenic, not all muscles are equally affected. Therefore, it is possible that in several muscles, the effects of this disease are more severe than others, as already presented in [
30], which can lead to a performance reduction. Finally, some of the muscle sections of the elderly subjects in Group 2 can suffer from other pathological conditions, such as obesity, since sarcopenia is not the only condition than can cause deterioration in muscle architecture.
Figure 14 presents the confusion matrices for each examined muscle for the manually extracted CSA.
From the results in
Figure 14, it must be noted that all of the examined muscles showed similar performance. Specifically, the average accuracy of the T.A. was 86%, that of the R.F. was 84%, that of the GCM was 85%, and finally that of the B.B. was 81%. In all muscles, the most significant accuracy reduction was due to confusion of the classifier in the samples of Group 3 as Group 2 and vice versa. Lastly, it is worth mentioning that the GCM presented the best results concerning the classification accuracy of Group 3 which indicates that this muscle is more suitable for distinguishing sarcopenic patients from healthy individuals.
Finally, the Grad-Cam [
29] analysis was performed to investigate which areas of the activation maps were triggered when the classifier made a decision. This analysis will help us to interpret the results better and avoid any possible bias in the decision model. In
Figure 15, the Grad-Cam results are presented. From these, it can be observed that the areas that have the most important role in the final decision are mostly texture intensive since they contain a lot of muscle tissue, indicating that the classifier concentrates on the CSA texture. Hence, this is an important result for creating a diagnostic tool that uses the image texture to make decisions about the pathological state of the muscle. However, it is also possible that the GCM and the B.B. shape of the CSA are other factors that affect the classification performance. In some cases, this can be derived from the fact that the most intense (red) activation is near an irregular contour of the CSA and not in a more central area. Further experimentation will provide better insights into the true capabilities of such a system.
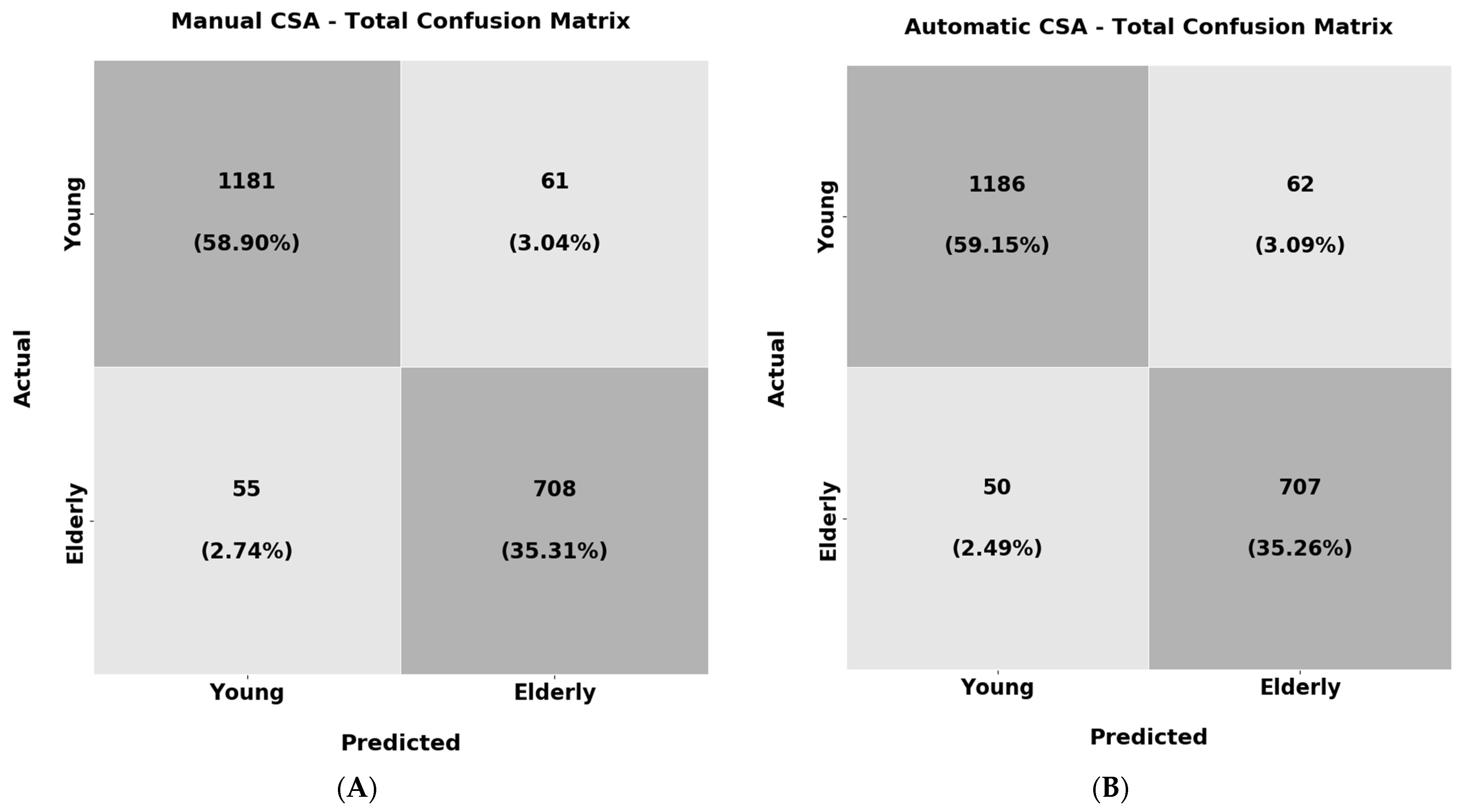
In the last experiment of the texture analysis, the objective was to train a ResNet18 classifier to recognise the young individuals (Group 1) and the elderly subjects (Group 2 and Group 3 combined) solely from the muscle CSA texture. As before, the classification results for the manually extracted and automatic CSA are reported in
Table 7, and their confusion matrices are shown in
Figure 16.
From the results above, it must be highlighted that the classifier trained both in the manually extracted CSA textures as well as the automatic correctly labelled the majority of the samples over 94% of the time. Furthermore, the recall and F1 scores also present very high performance (94%) in both of the experiments. These results are an additional indicator that the muscle CSA texture can be used to distinguish the young subjects from the elderly ones. This could lead to diagnostic tools solely from the muscle texture that could be useful in clinical practice. Additionally, apart from the weighted average performance depicted in
Table 7, the confusion matrices are also presented in
Figure 16 for analysing the per-class accuracy.
From the results above, it is observable that the classifier shows almost identical performance in both experiments. Regarding the per-class accuracy for the manually extracted CSAs texture, the classifier reached 95.0% in the samples belonging to the young population and 92.7% for the elderly. For the texture of the automatically extracted CSAs, the corresponding precisions were 95.0% for the samples belonging to the young category and 93.3% for the elderly. One conclusion that can be made is that the young subjects again have more distinguishable muscle texture in comparison with the elderly subjects. Nevertheless, in the elderly subjects, the classifier could still recognise the vast majority of the samples correctly.
4. Discussion
This study employed different state-of-the-art deep learning architectures and vision transformers to segment the cross-sectional area in a new, large, and diverse musculoskeletal ultrasound database. After delineating the CSA, the muscle echogenicity was calculated using this area’s mean grey level intensity. The presented preliminary results regarding four very informative muscles for investigating neuromuscular disorders and sarcopenia [
13,
30], indicate that such an automated approach can be used successfully in clinical practice. Additionally, in a supplementary analysis, the texture of the muscle CSA (extracted manually and automatically) was investigated for its ability to recognise the subject’s group. Our results indicate that for the younger population, the differences in muscle texture in comparison with the elderly subjects were significant and therefore the classifier was capable of correctly categorising the majority of the samples. Regarding the recognition results of the elderly non-sarcopenic and sarcopenic patients, the classifier presented significant confusion between these two categories leading to performance reduction. However, larger studies with more balanced datasets are required to assess the feasibility of this task conclusively.
A significant advancement of this study in comparison with other recent works presented in [
11,
12] is the use of deep learning models to address this problem. Filter-based approaches are hard to design, more prone to overfitting, and have difficulties in handling the noise in real-world conditions. In particular, we demonstrated that the TRAMA algorithm [
11] severely underperforms compared to all of the examined convolutional neural networks. The reason for this is that in ultrasound images with abnormal echogenicity, the differentiation in the muscle aponeuroses from the inner muscle tissues is limited due to their low contrast. Hence, this leads to severe algorithm failure since muscle aponeuroses cannot be adequately localised, causing significant errors in CSA estimation and corresponding echogenicity.
Another similar study has been recently presented in [
13]. In this study, the authors also used deep learning techniques to improve the performance of the existing automated methods. Our additional contribution in relevance to their work is, firstly, the introduction for the first time of vision transformer technology for the automatic segmentation of CSA, which is the most recent advancement in the deep learning field. Secondly, our analysis was performed in a new, large, and diverse database; therefore, the results presented here provide additional knowledge for fully automating the CSA measurement. Furthermore, this database consists of ultrasound recordings acquired with a highly reproducible examination protocol since minimal pressure was applied in order to not deform the inner muscle tissues, hence adding reliability to the measurements. This is not true for the datasets studied in [
11,
12,
13], since it is clear from the images that a non-measurable pressure (highly user-dependent) was applied by the transducer during the examination. This variable degree of pressure can lead to muscle tissue deformation with subsequent changes in the thickness and echogenicity of the studied muscle, with eventual erroneous measurements. Finally, it must be noted that the k-fold evaluation protocol (k = 5) was followed which is more immune to overfitting and selection bias, leading to a better estimation of the final performance since it considers all of the available samples.
For the problem of the automatic CSA delineation, the best convolutional neural network was found to be TMUNet, a recently proposed visual transformer [
22]. In particular, the automatic measurements achieved over 95% precision and recall compared with a human expert’s manual annotation of the CSA. The other examined architectures have achieved similar performance, another indicator of the capability of the deep learning models of providing an automated solution to this problem. Furthermore, the route mean square error of the two readings in all of the 2005 ultrasound transverse recordings was found to be 38.15 mm
2, a minimal error that indicates the success of the proposed methodology. Regarding the discrepancy in the mean grey level value of CSA representing echogenicity, the manual and the predicted values are almost identical, with the average difference being only 0.88, hence near to 1% in percentage. Additionally, from the Bland–Altman plots in all automated measurements, it was demonstrated that no systematic error appeared, and most of the measurements fell within the limits of agreement. Finally, regarding the statistical analysis of the results, ICC (2,1) reached 0.97, showing the level of agreement between the two readings. Furthermore, the Pearson coefficient reached 0.99, demonstrating that these measurements are highly correlated.
Additionally, different supplementary analyses were performed in the newly annotated database to obtain significant insights into the sample distribution. Initially, the measurements of the ultrasound recordings of the different muscles were extracted and compared. From there, it was found that the B.B. possesses, on average, the largest CSA in the examined dataset and T.A. the smallest. Regarding muscle echogenicity, the smallest was observed in the R.F. and the largest was observed in the T.A. Additionally, the smallest error between the two readings was achieved in the R.F. This result can be explained by the muscle shape; in most cases, R.F. was composed of a smaller pixel area compared with the other examined muscles. Furthermore, the group analysis highlighted that the subjects who were elderly (Groups 2 and 3) had significantly less CSA and more echogenicity than their younger counterparts (Group 1). The above-mentioned is expected since ageing leads to a reduction in muscle mass and an increase in muscle fat which causes higher echogenicity. Furthermore, the lowest muscle CSA was found in elderly patients suffering from sarcopenia (Group 3), a disease that further deteriorates muscle architecture. Moreover, in this group, the highest echogenicity was also found, another indicator of muscle loss. Finally, in Group 1, the automatic measurements achieved the best result (young and healthy). The explanation for this is that the recordings in this group have more distinguishable boundaries between the aponeuroses and the inner tissue due to healthier muscles than the other study groups. Regarding the clinical application of the presented results, in [
10] it was reported that a CSA variation >5% indicates a clinically relevant change and a variation above 10% can be a sign of muscle atrophy; therefore, the proposed method presented in this study which achieved an average 4% discrepancy between the manual and automatic measurements can be considered to be reliable for the future integration of this software in ultrasound machines.
Another important contribution of this study is the textural analysis of the CSA using a well-established deep learning model. This analysis consisted of two important experiments. In the second experiment, we investigated whether there are differences in the muscle texture of the young (Group 1) vs. the elderly population (Group 2 and Group 3 combined). The preliminary results indicate that the texture of the muscle CSA of the elderly is very distinguishable compared to that of the young population. In particular, the classifier achieved over 94% overall accuracy between the two populations, showing that a diagnostic tool that can predict the condition of the muscle solely from its texture is feasible. Regarding the first experiment of the textural analysis, in which we tried to classify the muscle texture in the three examined groups of this study, the results was not conclusive. Although the classifier achieved over 84% correct categorisation on average, from the confusion matrix of these results we demonstrated that much of the accuracy was due to the imbalance of the dataset. In particular, the classifier properly categorised the vast majority of the samples belonging to Group 1 and Group 2 (93.1% and 78.4%, respectively) but severely underperformed in the samples belonging to Group 3 (44.2%). We attribute this result mainly to three reasons. Firstly, there is a high dataset imbalance in the number of samples per category and the sex distribution. The number of samples in Group 3 is significantly less than in the other two groups; most samples come from female patients. Secondly, the muscles are not affected uniformly by sarcopenia since, as presented in [
30], in some, the muscle loss is more severe than in others. Lastly, the subjects belonging to Group 2 are elderly, and their muscle architecture and quality are also affected negatively by their age. For these reasons, larger studies are required for more conclusive results about the feasibility of this task.
It is also worth mentioning that this study has some general limitations. First, a limitation is that the proposed method was assessed in only four muscles from over 200 that the human body possesses; hence, further studies with a larger population and with more muscle sections are required to confirm the accuracy of our results. Nevertheless, since the proposed method is scalable, we are confident that these challenges can be overcome with adequate training data. Another limiting factor is that all of the ultrasound recordings were acquired from the same ultrasound machine, with standardisation in the software settings. The variation in this setup in real-world scenarios could alter the final performance. However, the examination protocol for data collection and the imaging settings used were chosen to be as generic as possible. Specifically, since the transducer’s pressure on the skin during an examination is difficult to measure and standardise, the ultrasonographer avoided any pressure that could cause muscle tissue deformation by using a generous quantity of gel, thus making the examination methodology more reproducible for other operators. Regarding the imaging settings of the ultrasound device, all image optimisations were turned off to avoid emitting noise and deformation in the final representation from these settings. Again, we believe this examination setup is reproducible and can lead to some standardisation. Lastly, another limitation is the small number of sarcopenic patients participating in this study. More patients in this group could yield more reliable results, especially in the texture analysis of their muscle CSA.
Finally, in future work, we plan to investigate the automatic extraction of CSA measurement in more muscle sections and use a larger dataset of ultrasound images. Furthermore, we plan to analyse the image texture of the longitudinal ultrasound recordings to investigate whether specific patterns exist in patients suffering from sarcopenia or other neuromuscular disorders. Lastly, we plan to combine clinical data, such as gender, BMI, and the dominant side of a patient, with the features extracted from the muscle texture to fuse this information to develop a more optimised solution. This solution, which belongs to the multimodal learning field [
34], will combine heterogeneous data that provide different views of the same patient to better support various clinical decisions.