The Nuclear Dense Fine Speckled (DFS) Immunofluorescence Pattern: Not All Roads Lead to DFS70/LEDGFp75
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Antibodies
2.3. Screening of Human Sera for Antibodies Producing the DFS IFA Pattern
2.4. Immunoblotting
3. Results
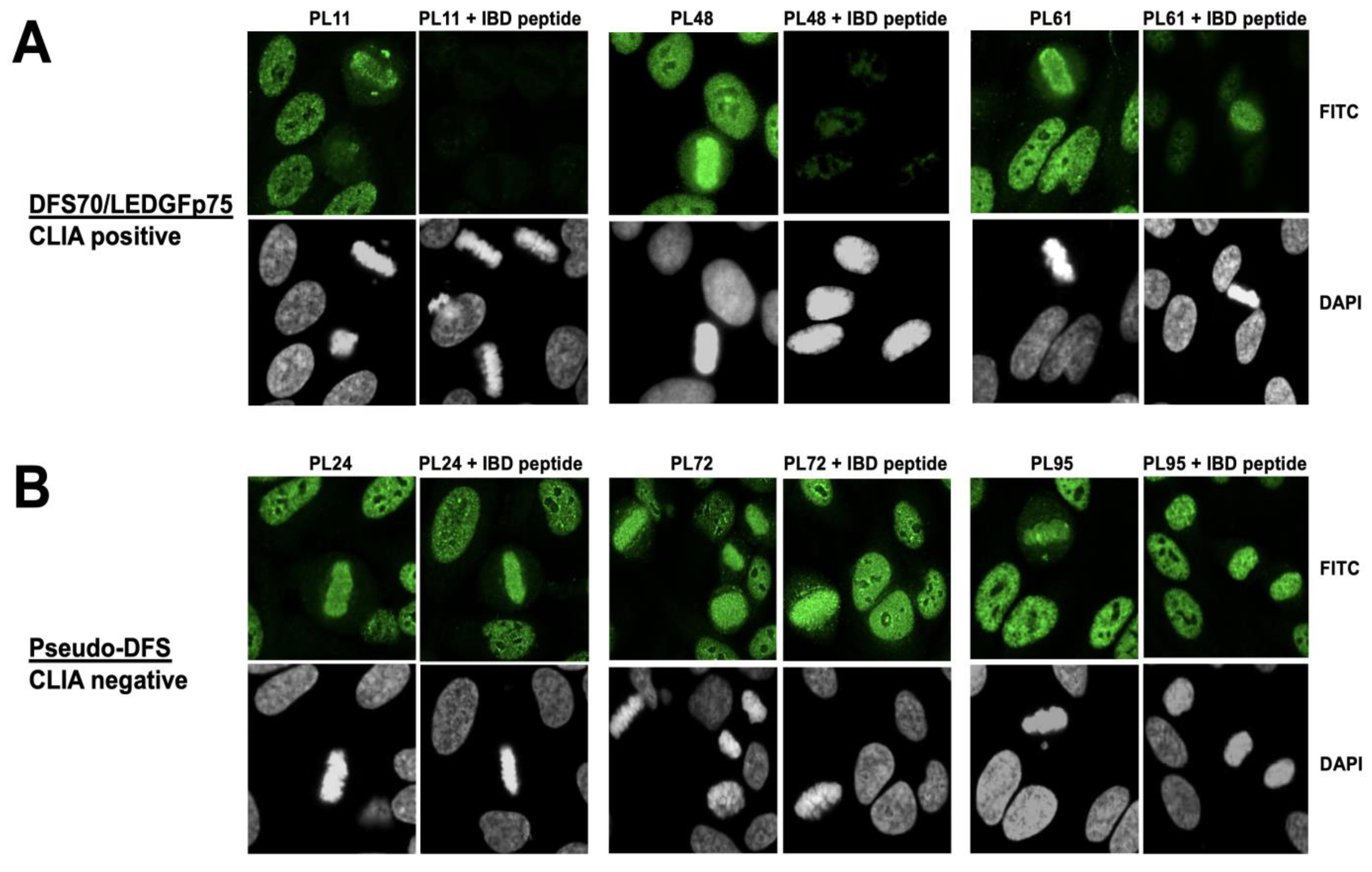
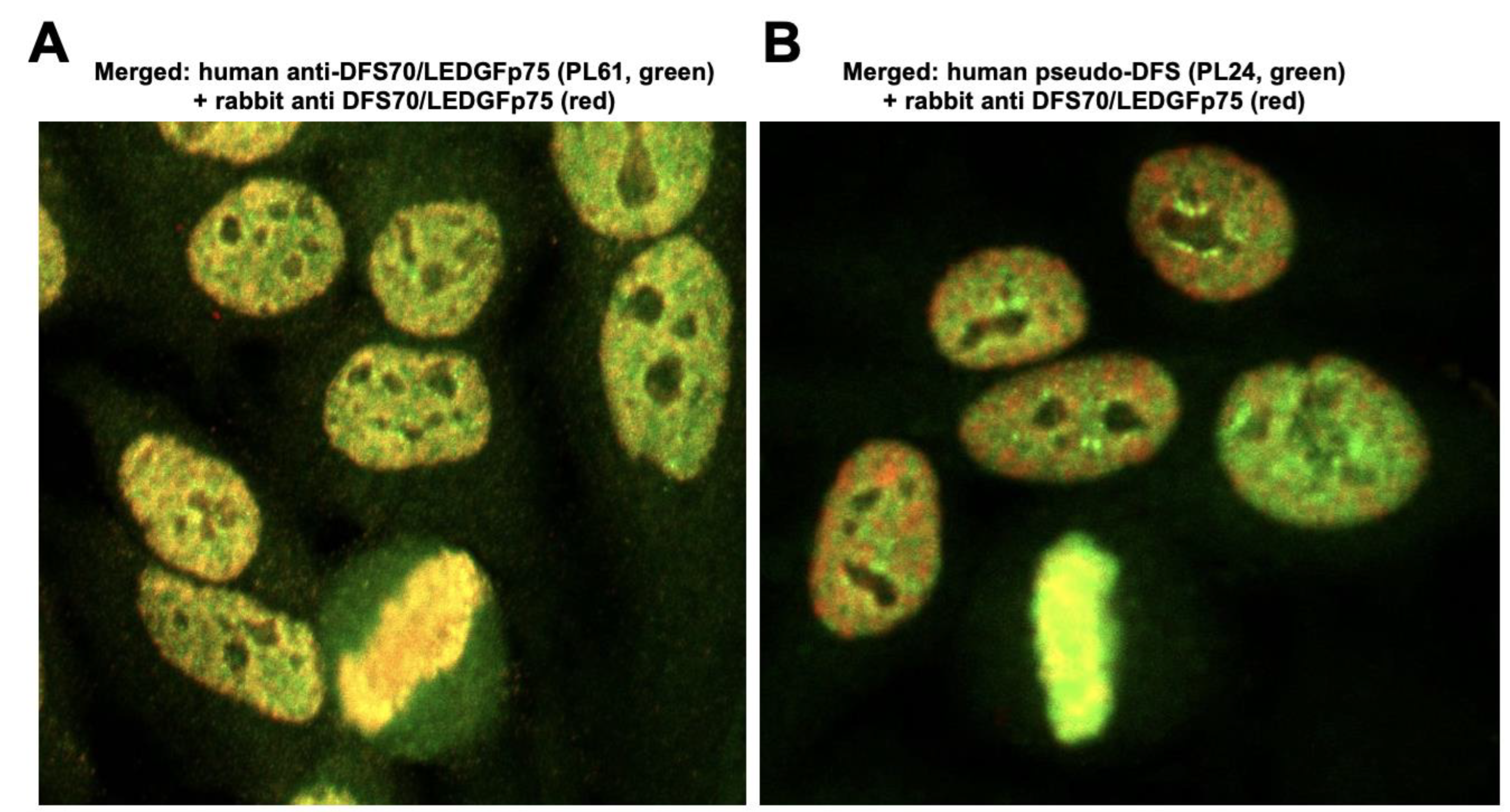
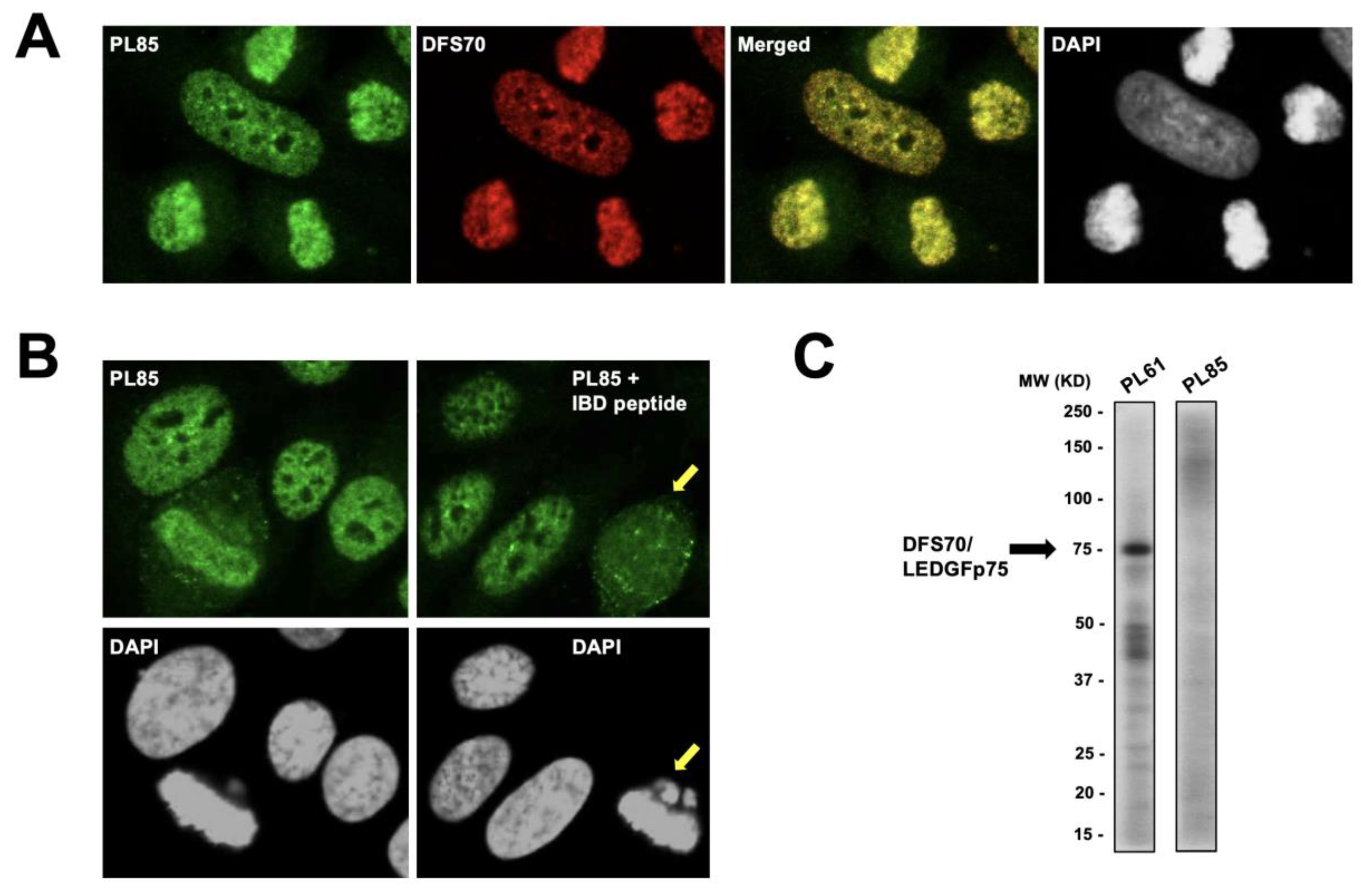
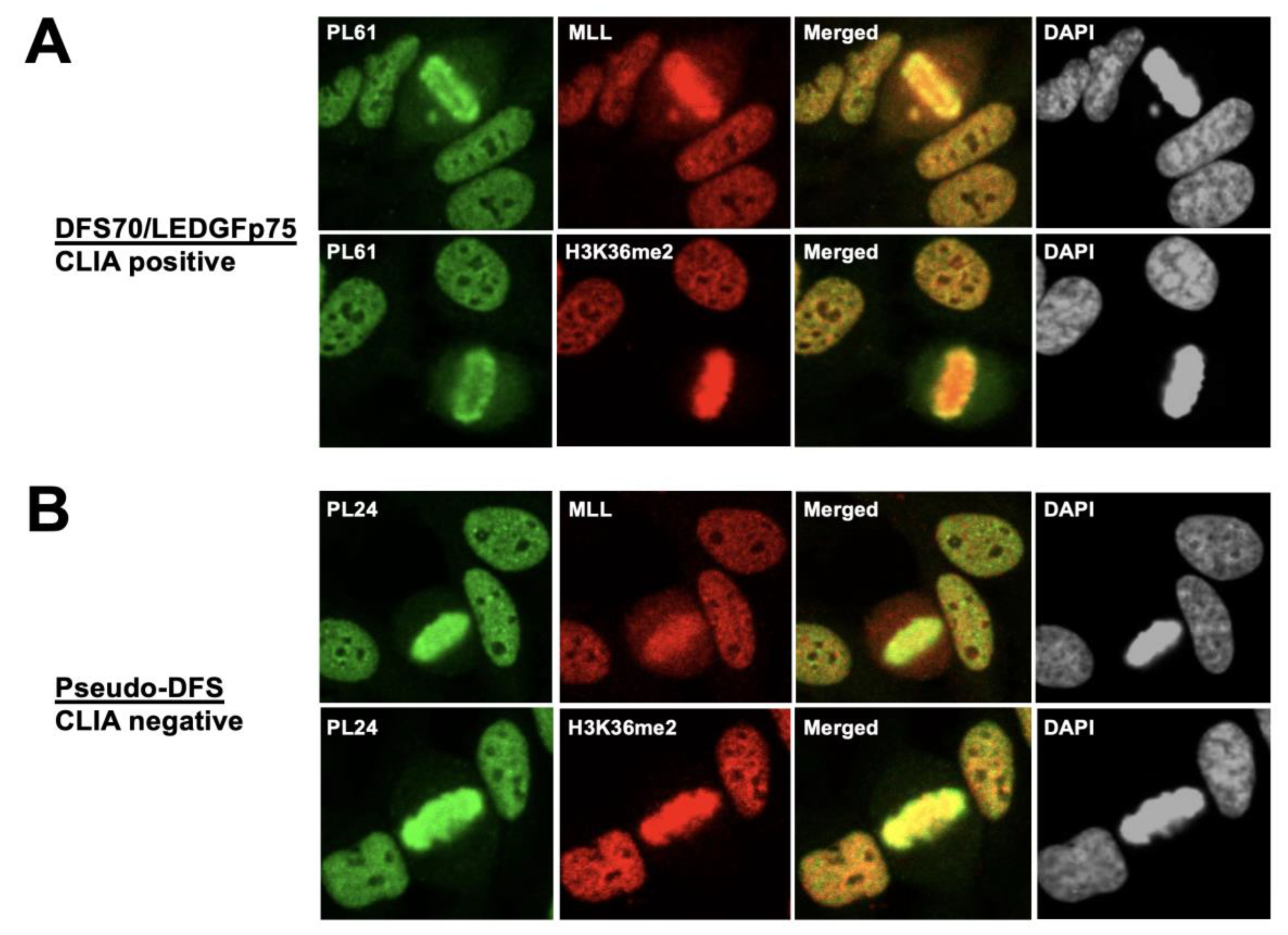
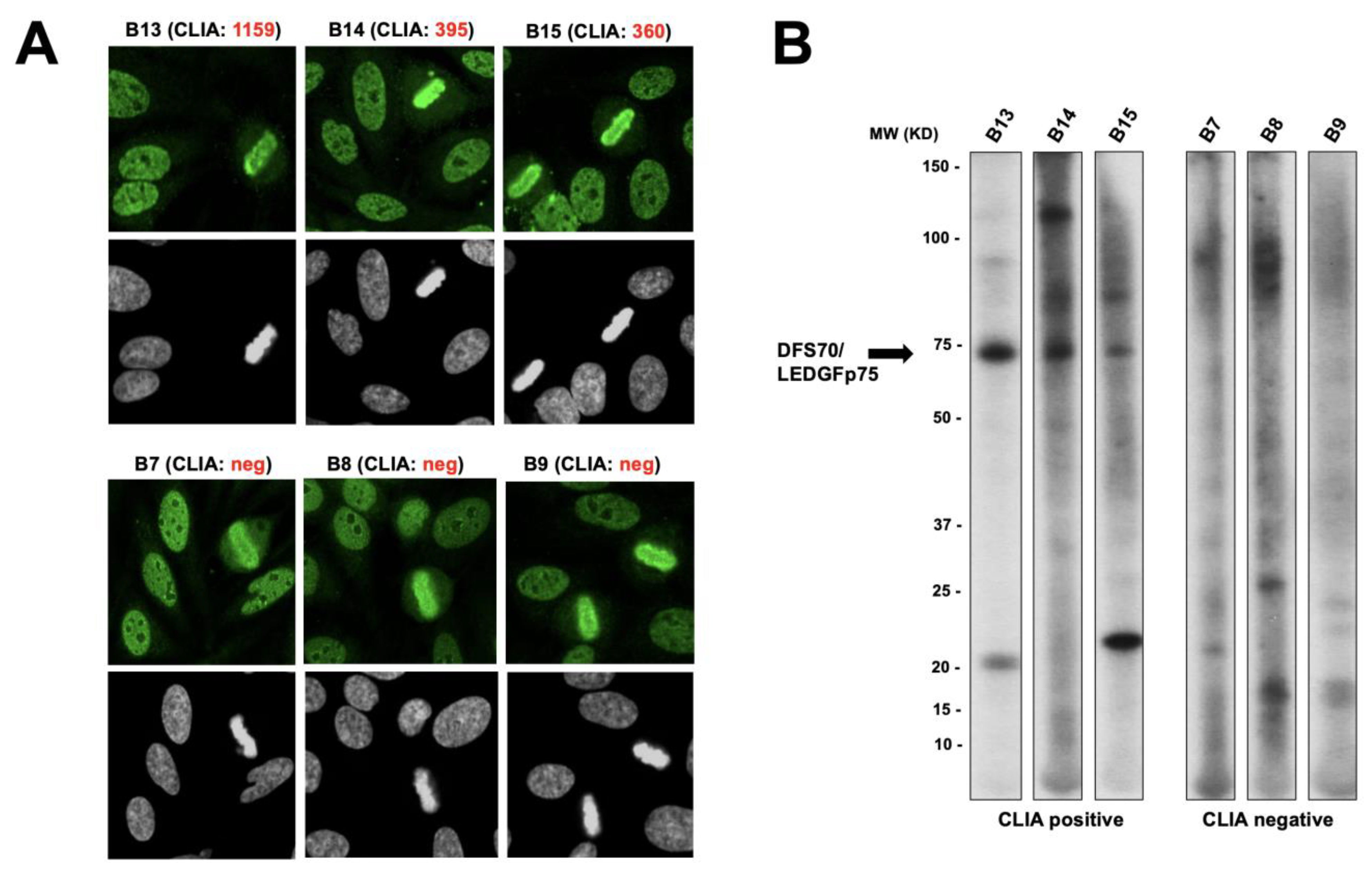
3.1. Selected CLIA Anti-DFS70-Negative Sera Produce a Pseudo-DFS IFA Pattern Resembling That of Anti-DFS70/LEDGFp75 Autoantibodies
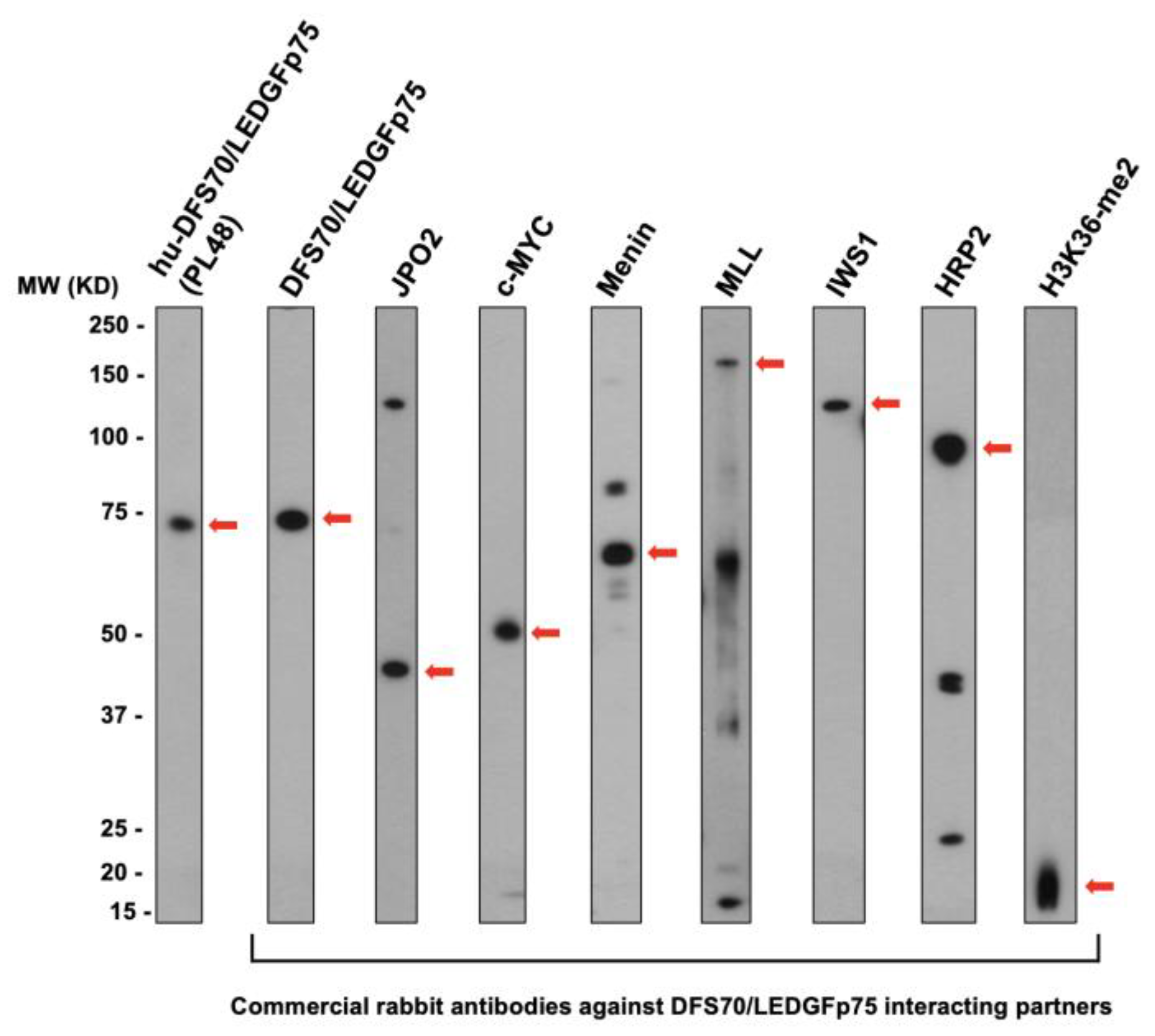
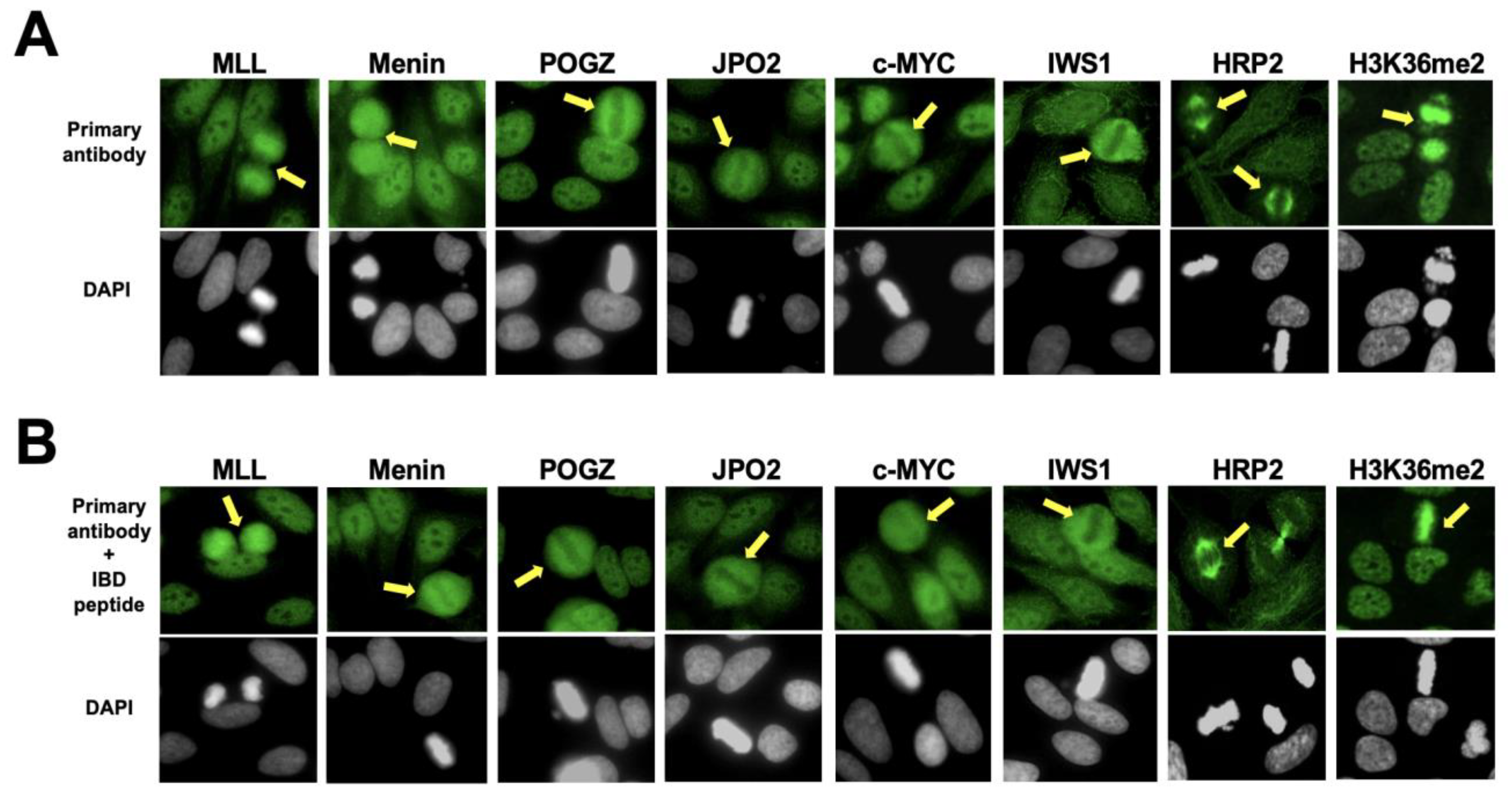
3.2. The Nuclear Localization Pattern of Selected Protein Interacting Partners of DFS70/LEDGFp75 Resembles the Classic DFS IFA Pattern
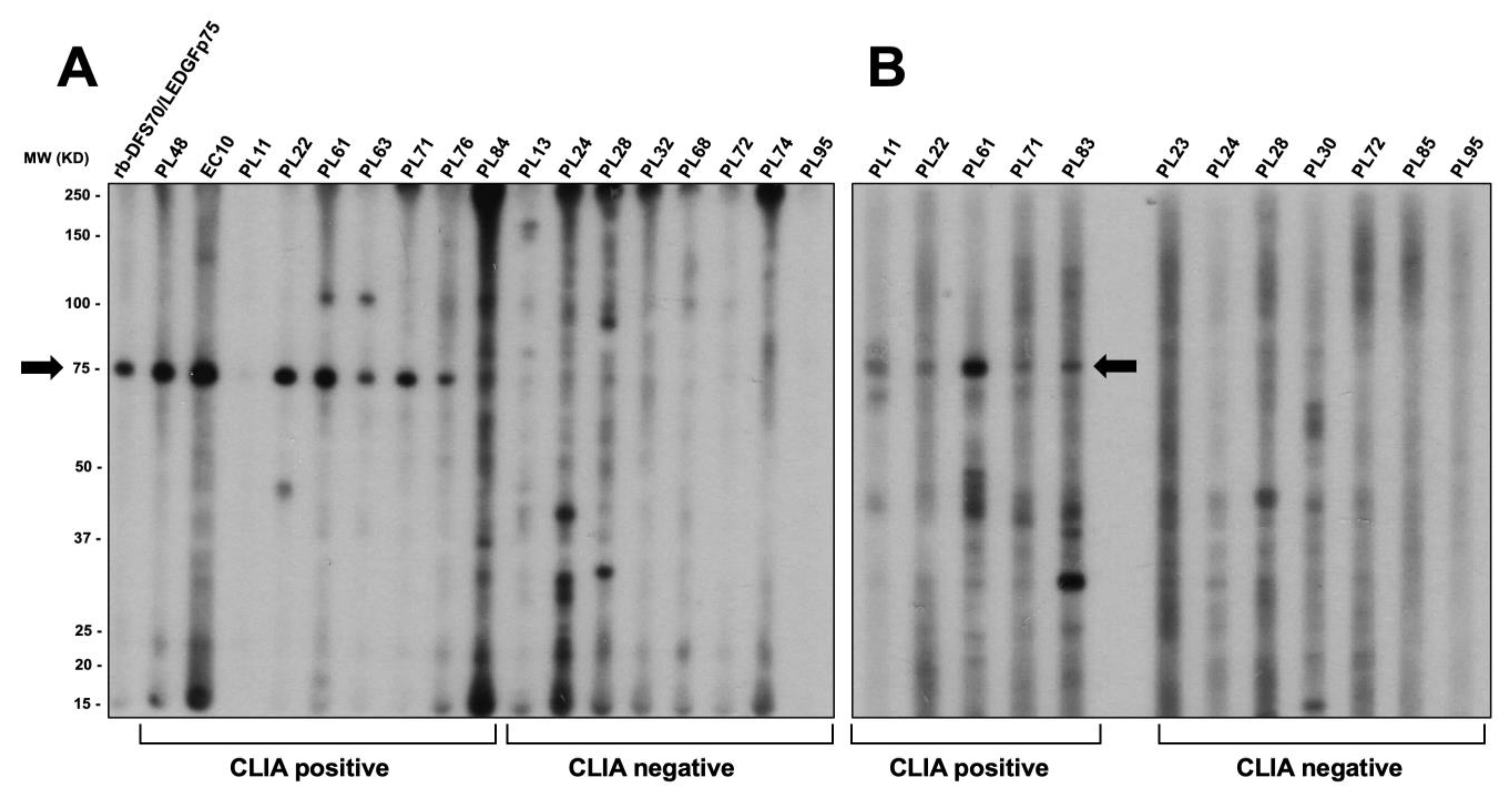
3.3. CLIA Anti-DFS70-Negative Sera Presenting the Pseudo-DFS IFA Pattern Do Not Recognize Common Protein Bands in Immunoblots
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tan, E.M. Autoantibodies and autoimmunity: A three-decade perspective. A tribute to Henry G. Kunkel. Ann. N. Y. Acad. Sci. 1997, 815, 1–14. [Google Scholar] [CrossRef]
- Bossuyt, X.; De Langhe, E.; Borghi, M.O.; Meroni, P.L. Understanding and interpreting antinuclear antibody tests in systemic rheumatic diseases. Nat. Rev. Rheumatol. 2020, 16, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Damoiseaux, J.; Andrade, L.E.C.; Carballo, O.G.; Conrad, K.; Francescantonio, P.L.C.; Fritzler, M.J.; Garcia de la Torre, I.; Herold, M.; Klotz, W.; Cruvinel, W.M.; et al. Clinical relevance of HEp-2 indirect immunofluorescent patterns: The International Consensus on ANA patterns (ICAP) perspective. Ann. Rheum. Dis. 2019, 78, 879–889. [Google Scholar] [CrossRef] [Green Version]
- von Mühlen, C.A.; Garcia-De La Torre, I.; Infantino, M.; Damoiseaux, J.; Andrade, L.E.C.; Carballo, O.G.; Conrad, K.; Francescantonio, P.L.C.; Fritzler, M.J.; Herold, M.; et al. How to report the antinuclear antibodies (anti-cell antibodies) test on HEp-2 cells: Guidelines from the ICAP initiative. Immunol. Res. 2021, 69, 594–608. [Google Scholar] [CrossRef] [PubMed]
- Andrade, L.E.C.; Klotz, W.; Herold, M.; Conrad, K.; Rönnelid, J.; Fritzler, M.J.; von Mühlen, C.A.; Satoh, M.; Damoiseaux, J.; de Melo Cruvinel, W.; et al. Executive Committee of ICAP. International consensus on antinuclear antibody patterns: Definition of the AC-29 pattern associated with antibodies to DNA topoisomerase I. Clin. Chem. Lab. Med. 2018, 56, 1783–1788. [Google Scholar] [CrossRef] [PubMed]
- Ochs, R.L.; Stein, T.W., Jr.; Peebles, C.L.; Gittes, R.F.; Tan, E.M. Autoantibodies in interstitial cystitis. J. Urol. 1994, 151, 587–592. [Google Scholar] [CrossRef]
- Ochs, R.L.; Muro, Y.; Si, Y.; Ge, H.; Chan, E.K.; Tan, E.M. Autoantibodies to DFS 70 kd/transcription coactivator p75 in atopic dermatitis and other conditions. J. Allergy Clin. Immunol. 2000, 105, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Mahler, M.; Hanly, J.G.; Fritzler, M.J. Importance of the dense fine speckled pattern on HEp-2 cells and anti-DFS70 antibodies for the diagnosis of systemic autoimmune diseases. Autoimmun. Rev. 2012, 11, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Hernandez, G.L.; Sanchez-Hernandez, E.S.; Casiano, C.A. Twenty years of research on the DFS70/LEDGF autoanti-body-autoantigen system: Many lessons learned but still many questions. Autoimmun. Highlights 2020, 11, 3. [Google Scholar] [CrossRef]
- Ochs, R.L.; Mahler, M.; Basu, A.; Rios-Colon, L.; Sanchez, T.W.; Andrade, L.E.; Fritzler, M.J.; Casiano, C.A. The significance of autoantibodies to DFS70/LEDGFp75 in health and disease: Integrating basic science with clinical understanding. Clin. Exp. Med. 2016, 6, 273–293. [Google Scholar] [CrossRef]
- Basu, A.; Sanchez, T.W.; Casiano, C.A. DFS70/LEDGFp75: An Enigmatic Autoantigen at the Interface between Autoimmunity, AIDS, and Cancer. Front. Immunol. 2015, 6, 116. [Google Scholar] [CrossRef] [Green Version]
- Shinohara, T.; Singh, D.P.; Fatma, N. LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res. 2002, 21, 341–358. [Google Scholar] [CrossRef]
- Debyser, Z.; Christ, F.; De Rijck, J.; Gijsbers, R. Host factors for retroviral integration site selection. Trends Biochem. Sci. 2015, 40, 108–116. [Google Scholar] [CrossRef]
- Engelman, A.N.; Singh, P.K. Cellular and molecular mechanisms of HIV-1 integration targeting. Cell. Mol. Life Sci. 2018, 75, 2491–2507. [Google Scholar] [CrossRef] [PubMed]
- Blokken, J.; De Rijck, J.; Christ, F.; Debyser, Z. Protein–protein and protein–chromatin interactions of LEDGF/p75 as novel drug targets. Drug Discov. Today Technol. 2017, 24, 25–31. [Google Scholar] [CrossRef]
- Tesina, P.; Čermáková, K.; Hořejší, M.; Procházková, K.; Fábry, M.; Sharma, S.; Christ, F.; Demeulemeester, J.; Debyser, Z.; De Rijck, J.; et al. Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif. Nat. Commun. 2015, 6, 7968. [Google Scholar] [CrossRef] [Green Version]
- Leoh, L.S.; Van Heertum, B.; De Rijck, J.; Filippova, M.; Rios-Colon, L.; Basu, A.; Martinez, S.; Tungteakkhun, S.S.; Filippov, V.; Christ, F.; et al. The stress oncoprotein LEDGF/p75 interacts with the methyl CpG binding Protein MeCP2 and influences its transcriptional activity. Mol. Cancer Res. 2012, 10, 378–391. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, A.; Cleary, M.L. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 2008, 14, 36–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liedtke, V.; Schröder, C.; Roggenbuck, D.; Weiss, R.; Stohwasser, R.; Schierack, P.; Rödiger, S.; Schenk, L. LEDGF/p75 is required for an efficient DNA damage response. Int. J. Mol. Sci. 2021, 22, 5866. [Google Scholar] [CrossRef] [PubMed]
- Canella, A.; Van Belle, S.; Brouns, T.; Nigita, G.; Carlon, M.S.; Christ, F.; Debyser, Z. LEDGF/p75-mediated chemoresistance of mixed-lineage leukemia involves cell survival pathways and super enhancer activators. Cancer Gene Ther. 2022, 29, 133–140. [Google Scholar] [CrossRef]
- Singh, P.K.; Plumb, M.R.; Ferris, A.L.; Iben, J.R.; Wu, X.; Fadel, H.J.; Luke, B.T.; Esnault, C.; Poeschla, E.M.; Hughes, S.H.; et al. LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev. 2015, 29, 2287–2297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundgren, M.C.; Sapkota, S.; Peterson, D.J.; Crosson, J.T. The antinuclear antibody dense fine speckled pattern and possible clinical associations: An indication of a proinflammatory microenvironment. J. Immunol. Methods 2021, 488, 112904. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Kodera, M.; Sugiura, K.; Usuda, T.; Tan, E.M.; Takasaki, Y.; Tomita, Y.; Muro, Y. Anti-DFS70 antibodies in 597 healthy hospital workers. Arthritis Rheum. 2004, 50, 892–900. [Google Scholar] [CrossRef]
- Dellavance, A.; Viana, V.S.; Leon, E.P.; Bonfa, E.S.; Andrade, L.E.; Leser, P.G. The clinical spectrum of antinuclear antibodies associated with the nuclear dense fine speckled immunofluorescence pattern. J. Rheumatol. 2005, 32, 2144–2149. [Google Scholar]
- Mahler, M.; Parker, T.; Peebles, C.L.; Andrade, L.E.; Swart, A.; Carbone, Y.; Ferguson, D.J.; Villalta, D.; Bizzaro, N.; Hanly, J.G.; et al. Anti-DFS70/LEDGF antibodies are more prevalent in healthy individuals compared to patients with systemic autoimmune rheumatic diseases. J. Rheumatol. 2012, 39, 2104–2110. [Google Scholar] [CrossRef]
- Muro, Y.; Sugiura, K.; Nakashima, R.; Mimori, T.; Akiyama, M. Low prevalence of anti-DFS70/LEDGF antibodies in patients with dermatomyositis and other systemic autoimmune rheumatic diseases. J. Rheumatol. 2013, 40, 92–93. [Google Scholar] [CrossRef] [Green Version]
- Bonroy, C.; Schouwers, S.; Berth, M.; Stubbe, M.; Piette, Y.; Hoffman, I.; Devreese, K.; Van Hoovels, L. The importance of detecting anti-DFS70 in routine clinical practice: Comparison of different care settings. Clin. Chem. Lab. Med. 2018, 56, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Miyara, M.; Albesa, R.; Charuel, J.L. Clinical phenotypes of patients with anti-DFS70/LEDGF antibodies in a routine ANA referral cohort. Clin. Dev. Immunol. 2013, 2013, 703759. [Google Scholar] [CrossRef]
- Fitch-Rogalsky, C.; Steber, W.; Mahler, M.; Lupton, T.; Martin, L.; Barr, S.G.; Mosher, D.P.; Wick, J.; Fritzler, M.J. Clinical and serological features of patients referred through a rheumatology triage system because of positive antinuclear antibodies. PLoS ONE 2014, 9, e93812. [Google Scholar] [CrossRef]
- Conrad, K.; Röber, N.; Andrade, L.E.; Mahler, M. The Clinical Relevance of Anti-DFS70 Autoantibodies. Clin. Rev. Allergy Immunol. 2017, 52, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Shovman, O.; Gilburd, B.; Chayat, C.; Amital, H.; Langevitz, P.; Watad, A.; Guy, A.; Perez, D.; Azoulay, D.; Blank, M.; et al. Prevalence of anti-DFS70 antibodies in patients with and without systemic autoimmune rheumatic diseases. Clin. Exp. Rheumatol. 2018, 36, 121–126. [Google Scholar] [PubMed]
- Infantino, M.; Pregnolato, F.; Bentow, C.; Mahler, M.; Benucci, M.; Li Gobbi, F.; Damiani, A.; Grossi, V.; Franceschini, F.; Bodio, C.; et al. Only monospecific anti-DFS70 antibodies aid in the exclusion of antinuclear antibody associated rheumatic diseases: An Italian experience. Clin. Chem. Lab. Med. 2019, 57, 1764–1769. [Google Scholar] [CrossRef]
- Mariz, H.A.; Sato, E.I.; Barbosa, S.H.; Rodrigues, S.H.; Dellavance, A.; Andrade, L.E. Pattern on the antinuclear antibody-HEp-2 test is a critical parameter for discriminating antinuclear antibody-positive healthy individuals and patients with autoimmune rheumatic diseases. Arthritis Rheum. 2011, 63, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Seelig, C.A.; Bauer, O.; Seelig, H.P. Autoantibodies Against DFS70/LEDGF Exclusion Markers for Systemic Autoimmune Rheumatic Diseases (SARD). Clin. Lab. 2016, 62, 499–517. [Google Scholar] [CrossRef]
- Cheng, C.F.; Shih, M.C.; Lan, T.Y.; Li, K.J. Anti-DFS70 Antibodies for Differentiating Systemic Autoimmune Rheumatic Disease in Patients with Positive ANA Tests: A Systematic Review and Meta-Analysis. Diagnostics 2021, 11, 1592. [Google Scholar] [CrossRef]
- Mahler, M.; Andrade, L.E.; Casiano, C.A.; Malyavantham, K.; Fritzler, M.J. Anti-DFS70 antibodies: An update on our current understanding and their clinical usefulness. Expert Rev. Clin. Immunol. 2019, 15, 241–250. [Google Scholar] [CrossRef]
- Mahler, M.; Fritzler, M.J. The clinical significance of the dense fine speckled immunofluorescence pattern on HEp-2 cells for the diagnosis of systemic autoimmune diseases. Clin. Dev. Immunol. 2012, 2012, 494356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gundín, S.; Irure-Ventura, J.; Asensio, E.; Ramos, D.; Mahler, M.; Martínez-Taboada, V.; López-Hoyos, M. Measurement of anti-DFS70 antibodies in patients with ANA-associated autoimmune rheumatic diseases suspicion is cost-effective. Autoimmun. Highlights 2016, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Moroni, L.; Restovic, G.; Cervera, R.; Espinosa, G.; Viñas, O.; García, M.; Sampietro-Colom, L. Economic Impact Analysis of the Use of Anti-DFS70 Antibody Test in Patients with Undifferentiated Systemic Autoimmune Disease Symptoms. J. Rheumatol. 2020, 47, 1275–1284. [Google Scholar] [CrossRef]
- Bizzaro, N.; Tonutti, E.; Villalta, D. Recognizing the dense fine speckled/lens epithelium-derived growth factor/p75 pattern on HEP-2 cells: Not an easy task! Comment on the article by Mariz et al. Arthritis Rheum. 2011, 63, 4036–4037. [Google Scholar] [CrossRef]
- Bizzaro, N.; Tonutti, E.; Tampoia, M.; Infantino, M.; Cucchiaro, F.; Pesente, F.; Morozzi, G.; Fabris, M.; Villalta, D. Specific chemoluminescence and immunoasdorption tests for anti-DFS70 antibodies avoid false positive results by indirect immunofluorescence. Clin. Chim. Acta 2015, 451, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Bizzaro, N.; Pesente, F.; Cucchiaro, F.; Infantino, M.; Tampoia, M.; Villalta, D.; Fabris, M.; Tonutti, E. Anti-DFS70 antibodies detected by immunoblot methods: A reliable tool to confirm the dense fine speckles ANA pattern. J. Immunol. Methods 2016, 436, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Mahler, M.; Meroni, P.L.; Andrade, L.E.; Khamashta, M.; Bizzaro, N.; Casiano, C.A.; Fritzler, M.J. Towards a better understanding of the clinical association of anti-DFS70 autoantibodies. Autoimmun. Rev. 2016, 15, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Infantino, M.; Shovman, O.; Pérez, D.; Grossi, V.; Manfredi, M.; Benucci, M.; Damiani, A.; Gilburd, B.; Azoulay, D.; Serrano, A.; et al. A better definition of the anti-DFS70 antibody screening by IIF methods. J. Immunol. Methods 2018, 461, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Infantino, M.; Shovman, O.; Gilburd, B.; Manfredi, M.; Grossi, V.; Benucci, M.; Damiani, A.; Chimenti, D.; Malyavantham, K.; Shoenfeld, Y. Improved accuracy in DFS pattern interpretation using a novel HEp-2 ELITE system. Clin. Rheumatol 2019, 38, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- Infantino, M.; Carbone, T.; Manfredi, M.; Grossi, V.; Benucci, M.; Casiano, C.A.; Bizzaro, N. Dense fine speckled (DFS) immunofluorescence pattern and anti-DFS70 antibodies: Cleaning up the current concepts. Clin. Chim. Acta 2020, 510, 157–159. [Google Scholar] [CrossRef]
- Zheng, B.; Wang, Z.; Mora, R.A.; Liu, A.; Li, C.; Liu, D.; Zhai, F.; Liu, H.; Gong, H.; Zhou, J.; et al. Anti-DFS70 Antibodies Among Patient and Healthy Population Cohorts in China: Results From a Multicenter Training Program Showing Spontaneous Abortion and Pediatric Systemic Autoimmune Rheumatic Diseases Are Common in Anti-DFS70 Positive Patients. Front. Immunol. 2020, 11, 562138. [Google Scholar] [CrossRef]
- Bentow, C.; Fritzler, M.J.; Mummert, E.; Mahler, M. Recognition of the dense fine speckled (DFS) pattern remains challenging: Results from an international internet-based survey. Autoimmun. Highlights 2016, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Basu, A.; Woods-Burnham, L.; Ortiz, G.; Rios-Colon, L.; Figueroa, J.; Albesa, R.; Andrade, L.E.; Mahler, M.; Casiano, C.A. Specificity of Antinuclear Autoantibodies Recognizing the Dense Fine Speckled Nuclear Pattern: Preferential Targeting of DFS70/LEDGF Over its Interacting Partner MeCP2. Clin. Immunol. 2015, 161, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Hernandez, G.L.; Sanchez-Hernandez, E.S.; Ochoa, P.T.; Elix, C.C.; Alkashgari, H.R.; McMullen, J.R.W.; Soto, U.; Martinez, S.R.; Diaz Osterman, C.J.; Mahler, M.; et al. The LEDGF/p75 Integrase Binding Domain Interactome Contributes to the Survival, Clonogenicity, and Tumorsphere Formation of Docetaxel-Resistant Prostate Cancer Cells. Cells 2021, 10, 2723. [Google Scholar] [CrossRef] [PubMed]
- Infantino, M.; Bizzaro, N.; Grossi, V.; Manfredi, M. The long-awaited ‘pseudo-DFS pattern’. Expert Rev. Clin. Immunol. 2019, 15, 445. [Google Scholar] [CrossRef] [PubMed]
- Mahler, M.; Andrade, L.E.; Casiano, C.A.; Malyavantham, K.; Fritzler, M.J. Implications for redefining the dense fine speckled and related indirect immunofluorescence patterns. Expert Rev. Clin. Immunol. 2019, 15, 447–448. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, Y.; Sugiura, K.; Watanabe, A.; Kunimatsu, M.; Mishima, M.; Tomita, Y.; Muro, Y. Autoantigenicity of DFS70 is restricted to the conformational epitope of C-terminal alpha-helical domain. J. Autoimmun. 2004, 23, 221–231. [Google Scholar] [CrossRef]
- Wei, X.; Chen, R.; Yang, C.; Nguyen, K.; Wakefield, D. Improved performance of confirmatory assays for detecting dense fine speckled (DFS) 70 antibodies. Pathology 2022, 54, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Malyavantham, K.; Suresh, L. Analysis of DFS70 pattern and impact on ANA screening using a novel HEp-2 ELITE/DFS70 knockout substrate. Autoimmun. Highlights 2017, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez-Del Mercado, M.; Gómez-Bañuelos, E.; Navarro-Hernández, R.E.; Pizano-Martinez, O.; Saldaña-Millán, A.; Chavarria-Avila, E.; Gonzalez-Rosas, L.; Andrade-Ortega, L.; Saavedra, M.A.; Vera-Lastra, O.L.; et al. Detection of autoantibodies to DSF70/LEDGFp75 in Mexican Hispanics using multiple complementary assay platforms. Autoimmun. Highlights 2017, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Bizzaro, N.; Pesce, G.; Trevisan, M.T.; Marchiano, M.; Cinquanta, L.; Infantino, M.; Paura, G.; Tampoia, M.; Alessio, M.G.; Previtali, G.; et al. Anti-DFS70 antibodies detected by specific methods in patients with thrombosis or recurrent pregnancy loss: No evidence of an association. Sci. Rep. 2020, 10, 7748. [Google Scholar] [CrossRef]
- Fabris, M.; Zago, S.; Tosolini, R.; Melli, P.; Bizzaro, N.; Tonutti, E. Anti-DFS70 antibodies: A useful biomarker in a pediatric case with suspected autoimmune disease. Pediatrics 2014, 134, e1706–e1708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Jurado, K.A.; Wu, X.; Shun, M.-C.; Li, X.; Ferris, A.L.; Smith, S.J.; Patel, P.A.; Fuchs, J.R.; Cherepanov, P.; et al. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res. 2012, 40, 11518–11530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Belle, S.; El Ashkar, S.; Čermáková, K.; Matthijssens, F.; Goossens, S.; Canella, A.; Hodges, C.H.; Christ, F.; De Rijck, J.; Van Vlierberghe, P.; et al. Unlike its paralog LEDGF/p75, HRP-2 is dispensable for MLL-R leukemogenesis but important for leukemic cell survival. Cells 2021, 10, 192. [Google Scholar] [CrossRef]
- LeRoy, G.; Oksuz, O.; Descostes, N.; Aoi, Y.; Ganai, R.A.; Kara, H.O.; Yu, J.-R.; Lee, C.-H.; Stafford, J.; Shilatifard, A.; et al. LEDGF and HDGF2 relieve the nucleosome-induced barrier to transcription in differentiated cells. Sci. Adv. 2019, 5, eaay3068. [Google Scholar] [CrossRef] [PubMed]
- Thakar, K.; Votteler, I.; Kelkar, D.; Shidore, T.; Gupta, S.; Kelm, S.; Dietz, F. Interaction of HRP-2 isoforms with HDGF: Chromatin binding of a specific heteromer. FEBS J. 2012, 279, 737–751. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez-Hernandez, E.S.; Ortiz-Hernandez, G.L.; Ochoa, P.T.; Reeves, M.; Bizzaro, N.; Andrade, L.E.C.; Mahler, M.; Casiano, C.A. The Nuclear Dense Fine Speckled (DFS) Immunofluorescence Pattern: Not All Roads Lead to DFS70/LEDGFp75. Diagnostics 2023, 13, 222. https://doi.org/10.3390/diagnostics13020222
Sanchez-Hernandez ES, Ortiz-Hernandez GL, Ochoa PT, Reeves M, Bizzaro N, Andrade LEC, Mahler M, Casiano CA. The Nuclear Dense Fine Speckled (DFS) Immunofluorescence Pattern: Not All Roads Lead to DFS70/LEDGFp75. Diagnostics. 2023; 13(2):222. https://doi.org/10.3390/diagnostics13020222
Chicago/Turabian StyleSanchez-Hernandez, Evelyn S., Greisha L. Ortiz-Hernandez, Pedro T. Ochoa, Michael Reeves, Nicola Bizzaro, Luis E. C. Andrade, Michael Mahler, and Carlos A. Casiano. 2023. "The Nuclear Dense Fine Speckled (DFS) Immunofluorescence Pattern: Not All Roads Lead to DFS70/LEDGFp75" Diagnostics 13, no. 2: 222. https://doi.org/10.3390/diagnostics13020222
APA StyleSanchez-Hernandez, E. S., Ortiz-Hernandez, G. L., Ochoa, P. T., Reeves, M., Bizzaro, N., Andrade, L. E. C., Mahler, M., & Casiano, C. A. (2023). The Nuclear Dense Fine Speckled (DFS) Immunofluorescence Pattern: Not All Roads Lead to DFS70/LEDGFp75. Diagnostics, 13(2), 222. https://doi.org/10.3390/diagnostics13020222







