Prognostic Value of BRAF, Programmed Cell Death 1 (PD1), and PD Ligand 1 (PDL1) Protein Expression in Colon Adenocarcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Histopathology
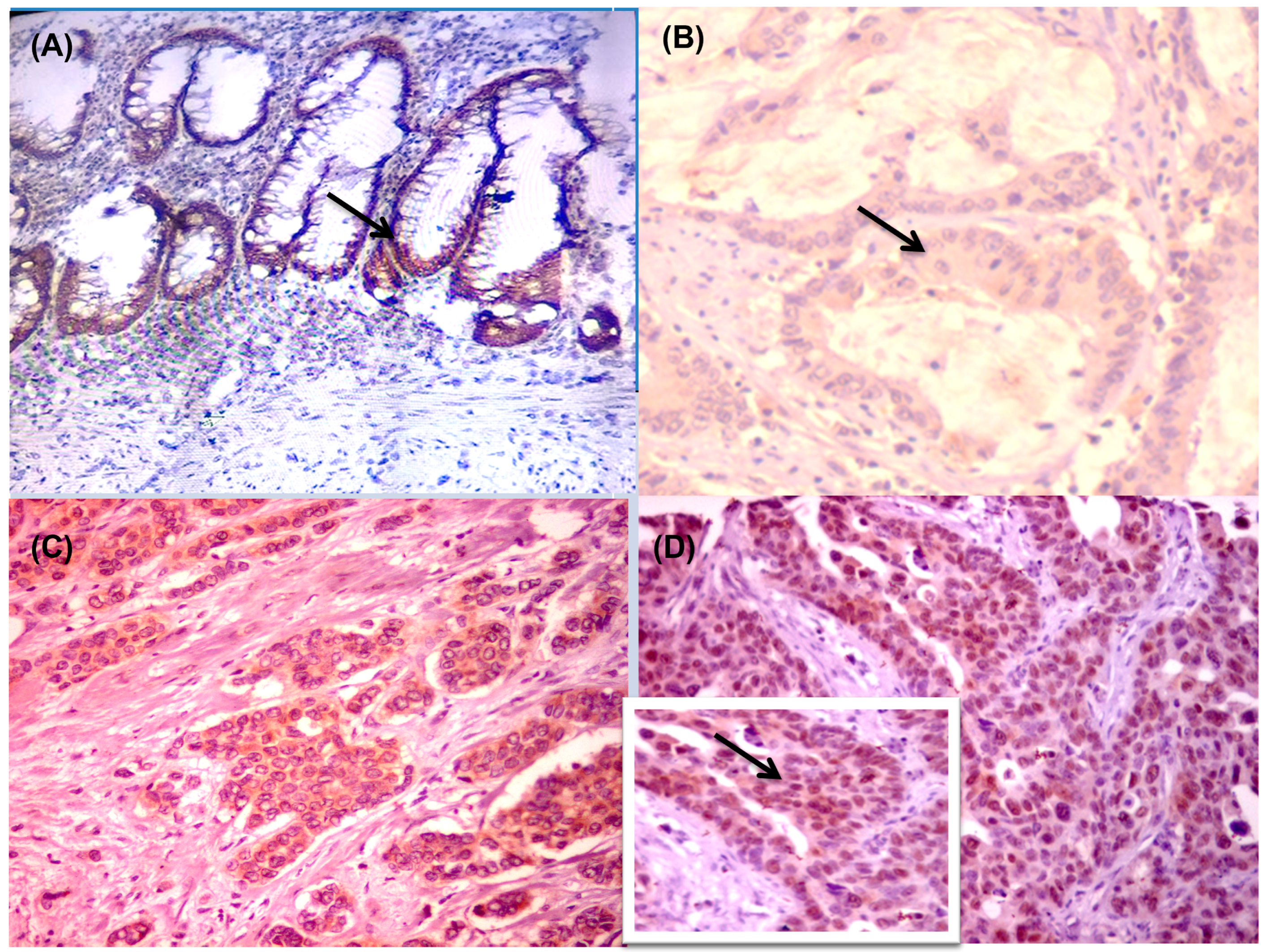
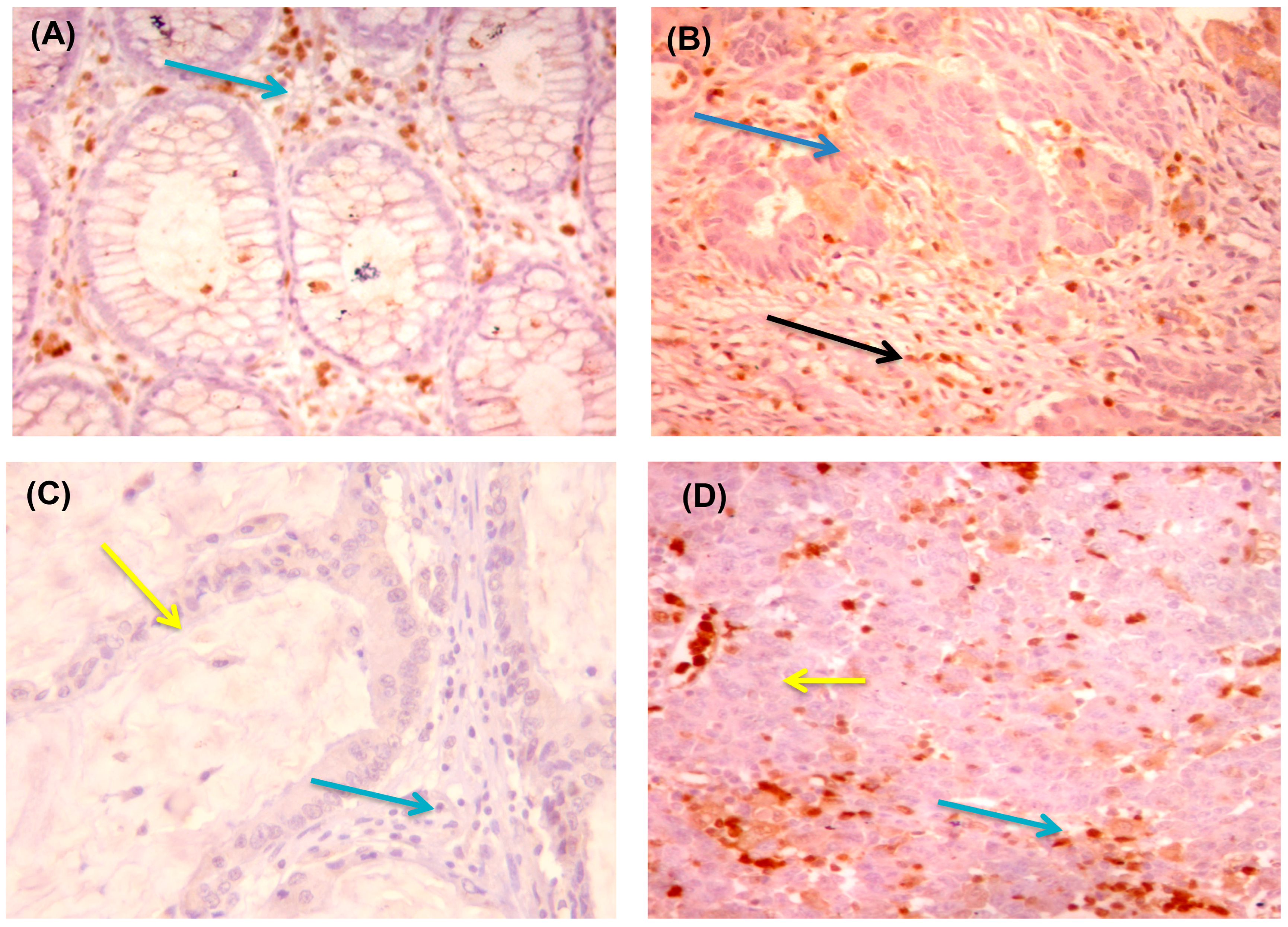
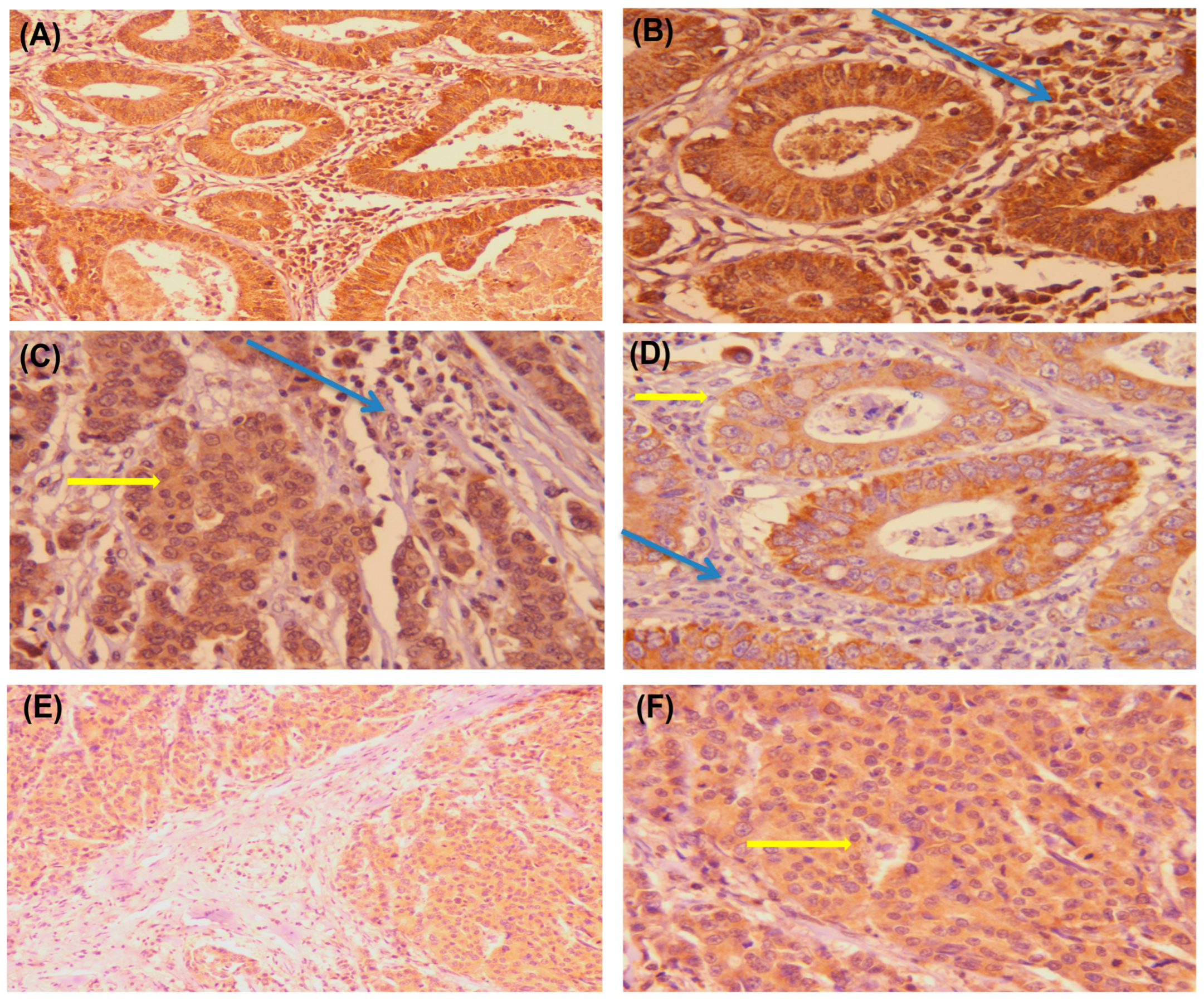
2.3. Immunohistochemical (IHC) Analysis and Interpretation
2.4. Combined Expression Patterns of PDL1 in Neoplastic Cells (NC) and Infiltrating Immune Cells (IIC)
2.5. Statistical Analysis
3. Results
3.1. The Clinicopathological Characteristics of the Studied Cases
3.2. BRAF Protein Expression and Association with Clinicopathological Prognostic Factors of Colon Adenocarcinoma
3.3. Expression of PD1 and PDL1 and Association with Clinicopathological Prognostic Factors of Colon Adenocarcinomas
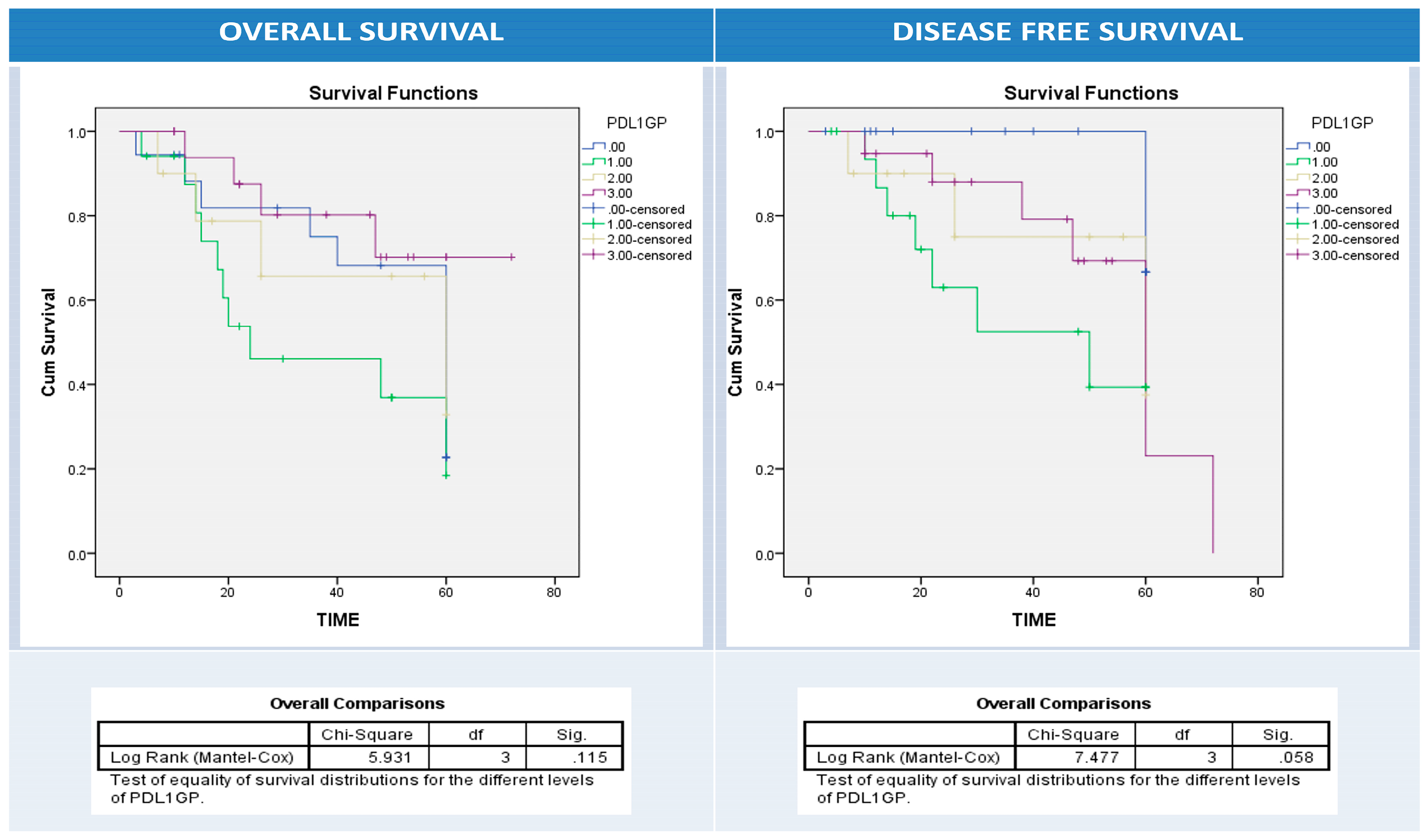
3.4. Combined Expression Patterns of PDL1 in Neoplastic and Infiltrating Immune Cells, and Association with the Clinicopathological Prognostic Factors of Colon Adenocarcinomas
3.5. Correlation between BRAF, PD1, and the PDL1 Protein Expressions
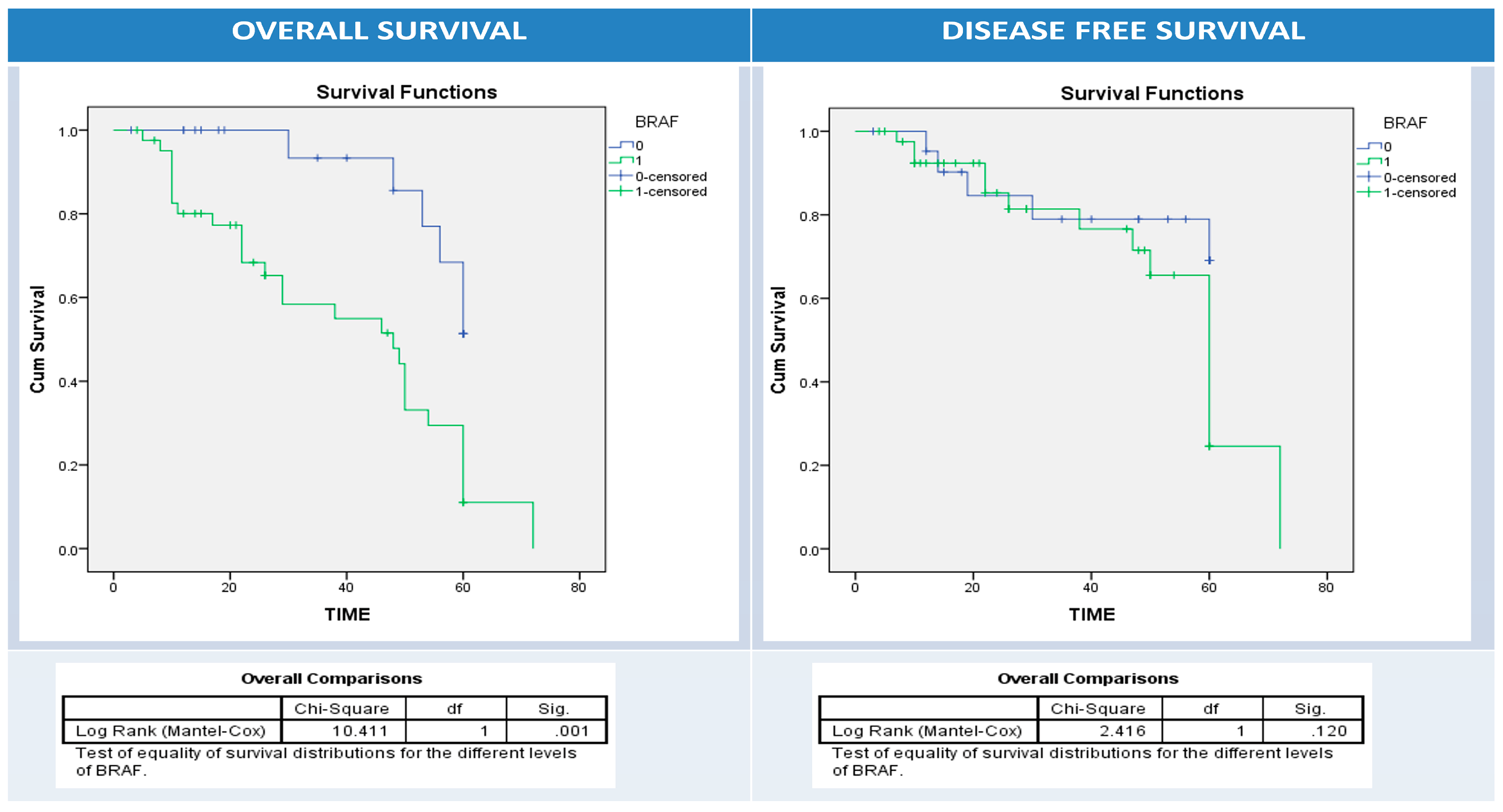
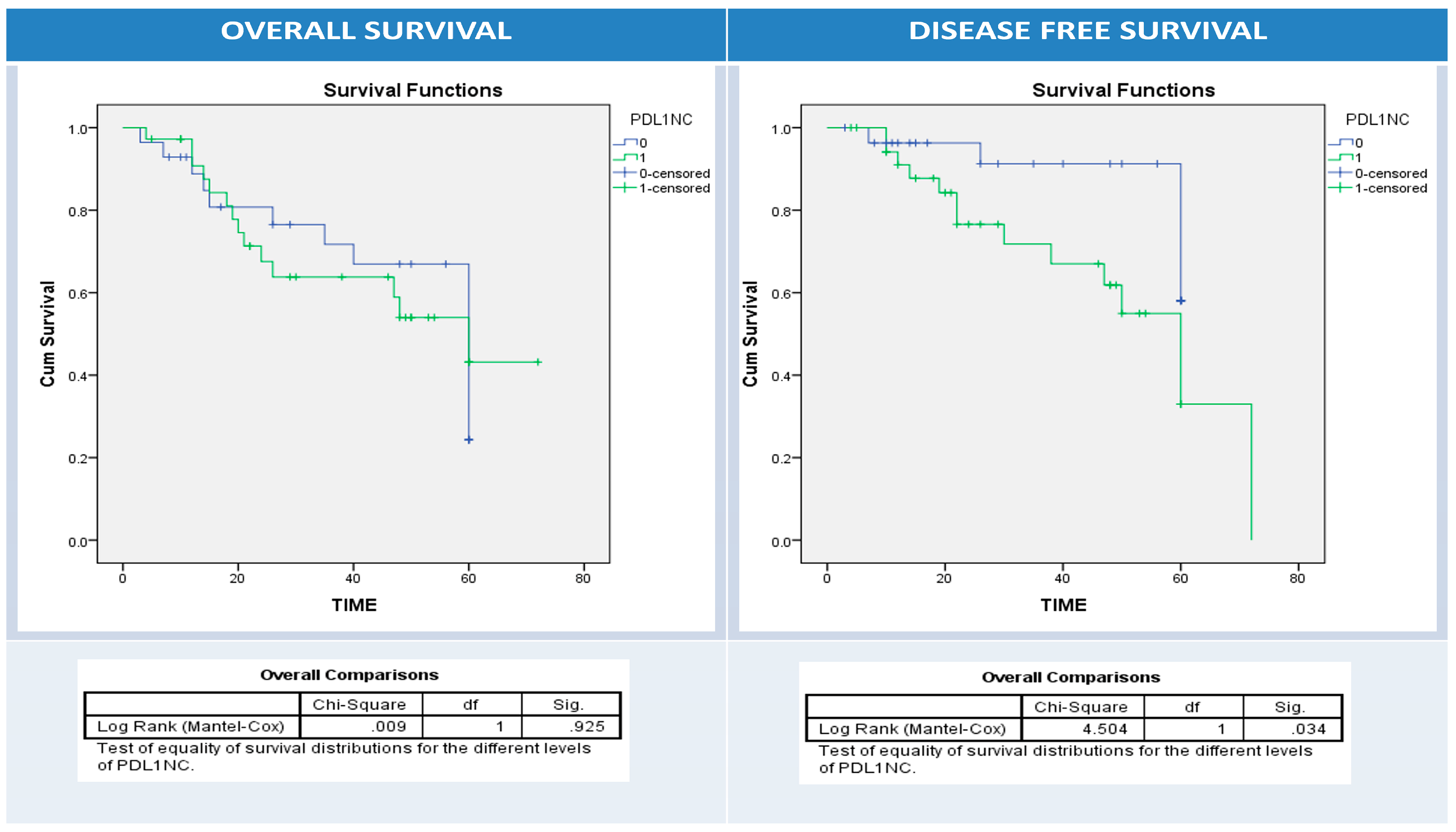
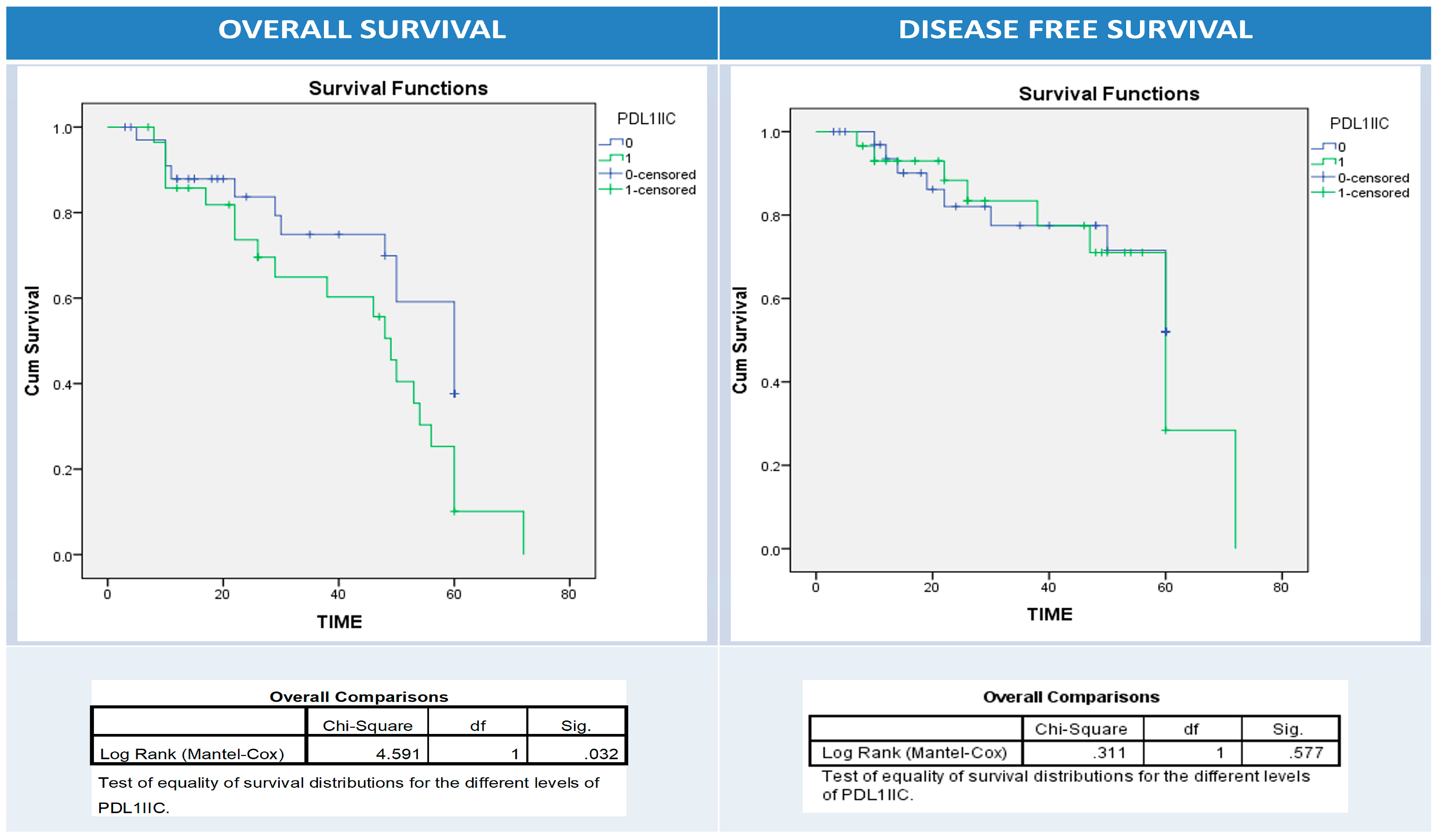
3.6. Univariate and Multivariate Analyses of Patients’ Clinical Outcomes (Relapse, Overall and Disease-Free Survival) and BRAF, PD1, and PDL1 Expressions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Welch, H.G.; Robertson, D.J. Colorectal Cancer on the Decline--Why Screening Can’t Explain It All. N. Engl. J. Med. 2016, 374, 1605–1607. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Gu, L.; Mao, D.; Chen, M.; Jin, R. Clinicopathological and prognostic significance of PD-L1 expression in colorectal cancer: A systematic review and meta-analysis. World J. Surg. Oncol. 2019, 17, 4. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, R.; Kommoss, S.; Winterhoff, B.J.; Kipp, B.R.; Garcia, J.J.; Voss, J.; Halling, K.; Karnezis, A.; Senz, J.; Yang, W.; et al. Targeted deep sequencing of mucinous ovarian tumors reveals multiple overlapping RAS-pathway activating mutations in borderline and cancerous neoplasms. BMC Cancer 2015, 15, 415. [Google Scholar] [CrossRef]
- Isomoto, K.; Haratani, K.; Hayashi, H.; Shimizu, S.; Tomida, S.; Niwa, T.; Yokoyama, T.; Fukuda, Y.; Chiba, Y.; Kato, R.; et al. Impact of EGFR-TKI Treatment on the Tumor Immune Microenvironment in EGFR Mutation–Positive Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 2037–2046. [Google Scholar] [CrossRef]
- Salem, M.E.; Puccini, A.; Tie, J. Redefining Colorectal Cancer by Tumor Biology. Am. Soc. Clin. Oncol. Educ. Book 2020, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Claps, F.; Mir, M.C.; Zargar, H. Molecular markers of systemic therapy response in urothelial carcinoma. Asian J. Urol. 2021, 8, 376–390. [Google Scholar] [CrossRef]
- Mertens, L.S.; Claps, F.; Mayr, R.; Bostrom, P.J.; Shariat, S.F.; Zwarthoff, E.C.; Boormans, J.L.; Abas, C.; van Leenders, G.J.L.H.; Götz, S.; et al. Prognostic markers in invasive bladder cancer: FGFR3 mutation status versus P53 and KI-67 expression: A multi-center, multi-laboratory analysis in 1058 radical cystectomy patients. Urol. Oncol. 2022, 40, 110.e1–110.e9. [Google Scholar] [CrossRef]
- Mir, M.C.; Campi, R.; Loriot, Y.; Puente, J.; Giannarini, G.; Necchi, A.; Rouprêt, M.; on behalf of the EAU Section of Oncological Urology ESOU Board. Adjuvant Systemic Therapy for High-risk Muscle-invasive Bladder Cancer After Radical Cystectomy: Current Options and Future Opportunities. Eur. Urol. Oncol. 2022, 5, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Högner, A.; Moehler, M. Immunotherapy in Gastric Cancer. Curr. Oncol. 2022, 29, 1559–1574. [Google Scholar] [CrossRef]
- Ralli, M.; Botticelli, A.; Visconti, I.C.; Angeletti, D.; Fiore, M.; Marchetti, P.; Lambiase, A.; de Vincentiis, M.; Greco, A. Immunotherapy in the Treatment of Metastatic Melanoma: Current Knowledge and Future Directions. J. Immunol. Res. 2020, 2020, 9235638. [Google Scholar] [CrossRef]
- Punekar, S.R.; Shum, E.; Grello, C.M.; Lau, S.C.; Velcheti, V. Immunotherapy in non-small cell lung cancer: Past, present, and future directions. Front. Oncol. 2022, 12, 877594. [Google Scholar] [CrossRef] [PubMed]
- Deleuze, A.; Saout, J.; Dugay, F.; Peyronnet, B.; Mathieu, R.; Verhoest, G.; Bensalah, K.; Crouzet, L.; Laguerre, B.; Belaud-Rotureau, M.A.; et al. Immunotherapy in Renal Cell Carcinoma: The Future Is Now. Int. J. Mol. Sci. 2020, 21, 2532. [Google Scholar] [CrossRef] [PubMed]
- Rhea, L.P.; Mendez-Marti, S.; Kim, D.; Aragon-Ching, J.B. Role of immunotherapy in bladder cancer. Cancer Treat. Res. Commun. 2021, 26, 100296. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Xu, Y.; Ye, W.; Xie, Q.; Gao, H.; Xu, B.; Zhang, D.; Jiang, J. Expression of PD-L1 in ovarian cancer and its synergistic antitumor effect with PARP inhibitor. Gynecol. Oncol. 2020, 157, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.L. Robbins Basic Pathology. Arch. Pathol. Lab. Med. 2003, 127, 1532. [Google Scholar] [CrossRef]
- Yu, Y.; Sitaraman, S.; Gewirtz, A.T. Intestinal epithelial cell regulation of mucosal inflammation. Immunol. Res. 2004, 29, 55–68. [Google Scholar] [CrossRef]
- Gianchecchi, E.; Delfino, D.V.; Fierabracci, A. Recent insights into the role of the PD-1/PD-L1 pathway in immunological tolerance and autoimmunity. Autoimmun. Rev. 2013, 12, 1091–1100. [Google Scholar] [CrossRef]
- Lim, S.O.; Li, C.W.; Xia, W.; Cha, J.H.; Chan, L.C.; Wu, Y.; Chang, S.S.; Lin, W.C.; Hsu, J.M.; Hsu, Y.H.; et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell 2016, 30, 925–939. [Google Scholar] [CrossRef]
- Juneja, V.R.; McGuire, K.A.; Manguso, R.T.; LaFleur, M.W.; Collins, N.; Haining, W.N.; Freeman, G.J.; Sharpe, A.H. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J. Exp. Med. 2017, 214, 895–904. [Google Scholar] [CrossRef]
- Zuazo, M.; Gato-Cañas, M.; Llorente, N.; Ibañez-Vea, M.; Arasanz, H.; Kochan, G.; Escors, D. Molecular mechanisms of programmed cell death-1 dependent T cell suppression: Relevance for immunotherapy. Ann. Transl. Med. 2017, 5, 385. [Google Scholar] [CrossRef]
- Kamphorst, A.O.; Wieland, A.; Nasti, T.; Yang, S.; Zhang, R.; Barber, D.L.; Konieczny, B.T.; Daugherty, C.Z.; Koenig, L.; Yu, K.; et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 2017, 355, 1423–1427. [Google Scholar] [CrossRef]
- Ibrahiem, A.T.; Fawzy, M.S.; Abu AlSel, B.T.; Toraih, E.A. Prognostic value of BRAF/MIR-17 signature and B-Raf protein expression in patients with colorectal cancer: A pilot study. J. Clin. Lab. Anal. 2021, 35, e23679. [Google Scholar] [CrossRef]
- Di Nicolantonio, F.; Martini, M.; Molinari, F.; Sartore-Bianchi, A.; Arena, S.; Saletti, P.; De Dosso, S.; Mazzucchelli, L.; Frattini, M.; Siena, S.; et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 5705–5712. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Guo, T.; Huang, X.; Wu, Q.; Niu, D.; Ji, X.; Feng, Q.; Li, Z.; Kakudo, K. In papillary thyroid carcinoma, expression by immunohistochemistry of BRAF V600E, PD-L1, and PD-1 is closely related. Virchows Arch. 2018, 472, 779–787. [Google Scholar] [CrossRef]
- Atefi, M.; Avramis, E.; Lassen, A.; Wong, D.J.; Robert, L.; Foulad, D.; Cerniglia, M.; Titz, B.; Chodon, T.; Graeber, T.G.; et al. Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin. Cancer Res. 2014, 20, 3446–3457. [Google Scholar] [CrossRef] [PubMed]
- Schmiegel, W.; Pox, C.; Reinacher-Schick, A.; Adler, G.; Arnold, D.; Fleig, W.; Fölsch, U.R.; Frühmorgen, P.; Graeven, U.; Heinemann, V.; et al. S3 guidelines for colorectal carcinoma: Results of an evidence-based consensus conference on February 6/7, 2004 and June 8/9, 2007 (for the topics IV, VI and VII). Z. Gastroenterol. 2010, 48, 65–136. [Google Scholar] [CrossRef]
- Bosman, F.T.; Carneiro, F.; Hruban, R.H.; Theise, N.D. WHO Classification of Tumours of the Digestive System; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Akkoca, A.N.; Yanık, S.; Ozdemir, Z.T.; Cihan, F.G.; Sayar, S.; Cincin, T.G.; Cam, A.; Ozer, C. TNM and Modified Dukes staging along with the demographic characteristics of patients with colorectal carcinoma. Int. J. Clin. Exp. Med. 2014, 7, 2828–2835. [Google Scholar]
- Shan, T.; Chen, S.; Wu, T.; Yang, Y.; Li, S.; Chen, X. PD-L1 expression in colon cancer and its relationship with clinical prognosis. Int. J. Clin. Exp. Pathol. 2019, 12, 1764–1769. [Google Scholar] [PubMed]
- Tolba, M.F. Revolutionizing the landscape of colorectal cancer treatment: The potential role of immune checkpoint inhibitors. Int. J. Cancer 2020, 147, 2996–3006. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, T.S.; Hu-Lieskovan, S.; Ribas, A. Mechanisms of Resistance to PD-1 and PD-L1 Blockade. Cancer J. 2018, 24, 47–53. [Google Scholar] [CrossRef]
- Boni, A.; Cogdill, A.P.; Dang, P.; Udayakumar, D.; Njauw, C.N.; Sloss, C.M.; Ferrone, C.R.; Flaherty, K.T.; Lawrence, D.P.; Fisher, D.E.; et al. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 2010, 70, 5213–5219. [Google Scholar] [CrossRef] [PubMed]
- van Brummelen, E.M.J.; de Boer, A.; Beijnen, J.H.; Schellens, J.H.M. Mutations as Predictive Biomarker for Response to Anti-EGFR Monoclonal Antibodies. Oncologist 2017, 22, 864–872. [Google Scholar] [CrossRef]
- Rössle, M.; Sigg, M.; Rüschoff, J.H.; Wild, P.J.; Moch, H.; Weber, A.; Rechsteiner, M.P. Ultra-deep sequencing confirms immunohistochemistry as a highly sensitive and specific method for detecting BRAF V600E mutations in colorectal carcinoma. Virchows Arch. 2013, 463, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Sinicrope, F.A.; Smyrk, T.C.; Tougeron, D.; Thibodeau, S.N.; Singh, S.; Muranyi, A.; Shanmugam, K.; Grogan, T.M.; Alberts, S.R.; Shi, Q. Mutation-specific antibody detects mutant BRAFV600E protein expression in human colon carcinomas. Cancer 2013, 119, 2765–2770. [Google Scholar] [CrossRef]
- Hang, J.F.; Li, A.F.; Chang, S.C.; Liang, W.Y. Immunohistochemical detection of the BRAF V600E mutant protein in colorectal cancers in Taiwan is highly concordant with the molecular test. Histopathology 2016, 69, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Gavin, P.G.; Colangelo, L.H.; Fumagalli, D.; Tanaka, N.; Remillard, M.Y.; Yothers, G.; Kim, C.; Taniyama, Y.; Kim, S.I.; Choi, H.J.; et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: An assessment of their prognostic and oxaliplatin predictive value. Clin. Cancer Res. 2012, 18, 6531–6541. [Google Scholar] [CrossRef]
- Brennan, D.F.; Dar, A.C.; Hertz, N.T.; Chao, W.C.; Burlingame, A.L.; Shokat, K.M.; Barford, D. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 2011, 472, 366–369. [Google Scholar] [CrossRef]
- Capper, D.; Berghoff, A.S.; Magerle, M.; Ilhan, A.; Wöhrer, A.; Hackl, M.; Pichler, J.; Pusch, S.; Meyer, J.; Habel, A.; et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol. 2012, 123, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Berntsson, J.; Eberhard, J.; Nodin, B.; Leandersson, K.; Larsson, A.H.; Jirström, K. Expression of programmed cell death protein 1 (PD-1) and its ligand PD-L1 in colorectal cancer: Relationship with sidedness and prognosis. Oncoimmunology 2018, 7, e1465165. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, G.N.; Dal Pozzo, C.A.; Depetris, I.; Schirripa, M.; Brignola, S.; Biason, P.; Balistreri, M.; Dal Santo, L.; Lonardi, S.; Munari, G.; et al. The heterogeneous clinical and pathological landscapes of metastatic. Cancer Cell Int. 2020, 20, 30. [Google Scholar] [CrossRef]
- Wang, S.; Yuan, B.; Wang, Y.; Li, M.; Liu, X.; Cao, J.; Li, C.; Hu, J. Clinicopathological and prognostic significance of PD-L1 expression in colorectal cancer: A meta-analysis. Int. J. Color. Dis. 2021, 36, 117–130. [Google Scholar] [CrossRef]
- Baran, B.; Mert Ozupek, N.; Yerli Tetik, N.; Acar, E.; Bekcioglu, O.; Baskin, Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterol. Res. 2018, 11, 264–273. [Google Scholar] [CrossRef] [PubMed]


| Number | 64 (100) | |
|---|---|---|
| Age (years) | Mean ± SD | 57.3 ± 12.5 |
| ≤55 | 29 (45.3) | |
| >55 | 35 (54.7) | |
| Sex | F | 29 (45.3) |
| M | 35 (54.7) | |
| Laterality | RT | 52 (81.3) |
| LT | 12 (18.8) | |
| Grade | G1 | 9 (14.1) |
| G2 | 38 (59.4) | |
| G3 | 17 (26.5) | |
| T stage | T1 | 5 (7.8) |
| T2 | 15 (23.4) | |
| T3 | 40 (62.5) | |
| T4 | 4 (6.3) | |
| N stage | N0 | 37 (57.8) |
| N1 | 24 (37.5) | |
| N2 | 3 (4.7) | |
| M stage | M0 | 59 (92.2) |
| M1 | 5 (7.8) | |
| LVI | No | 35 (45.3) |
| Yes | 29 (54.7) | |
| Dukes | A | 10 (15.6) |
| B | 26 (40.6) | |
| C | 23 (35.9) | |
| D | 5 (7.8) | |
| A+B | 35 (54.7) | |
| C+D | 29 (45.3) | |
| Relapse | No | 44 (60.9) |
| Yes | 20 (39.1) | |
| Alive | Dead | 35 (45.3) |
| Survived | 29 (54.7) | |
| BRAF protein | Negative | 39 (60.9) |
| Positive | 25 (39.1) | |
| Score 0 | 39 (60.9) | |
| Score 1 | 11 (17.2) | |
| Score 2 | 9 (14.1) | |
| Score 3 | 5 (7.8) | |
| PD1-IIC | Negative | 23 (35.9) |
| Positive | 41 (64.1) | |
| PDL1-NC | Negative | 30 (46.9) |
| Positive | 34 (53.1) | |
| PDL1-IIC | Negative | 31 (48.4) |
| Positive | 33 (51.6) |
| Variable | Total 64 (100) | BRAF Protein Expression | p-Value | ||
|---|---|---|---|---|---|
| Negative N = 39 | Positive N = 25 | ||||
| Age (y) | Mean | 57.9 ± 13.4 | 58 ± 11 | 0.9 | |
| Age Group | ≤55 | 29 | 17 | 12 | 0.7 |
| >55 | 35 | 22 | 13 | ||
| Sex | F | 29 | 18 | 11 | 0.8 |
| M | 35 | 21 | 14 | ||
| Laterality | Rt | 52 | 30 | 22 | 0.5 |
| Lt | 12 | 9 | 3 | ||
| Grade | G1 | 9 | 7 | 2 | 0.04 |
| G2 | 38 | 25 | 13 | ||
| G3 | 17 | 7 | 10 | ||
| T stage | T1 | 5 | 2 | 3 | 0.5 |
| T2 | 15 | 8 | 7 | ||
| T3 | 40 | 28 | 12 | ||
| T4 | 4 | 1 | 3 | ||
| N stage | N0 | 37 | 31 | 6 | 0.00 |
| N1 | 24 | 7 | 17 | ||
| N2 | 3 | 1 | 2 | ||
| M stage | M0 | 59 | 36 | 23 | 0.9 |
| M1 | 5 | 3 | 2 | ||
| LVI | No | 35 | 25 | 10 | 0.06 |
| Yes | 29 | 14 | 15 | ||
| Dukes | A-B | 35 | 28 | 7 | 0.001 |
| C-D | 29 | 11 | 18 | ||
| Relapse | No | 44 | 31 | 13 | 0.02 |
| Yes | 20 | 8 | 12 | ||
| OS | Dead | 35 | 14 | 21 | 0.00 |
| Survived | 29 | 25 | 4 | ||
| Variable | Total | PD1-IIC | p | PDL1-NC | p | PDL1-IIC | p | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Negative | Positive | ||||||
| N | 64 | 23 | 41 | 28 | 36 | 35 | 29 | ||||
| Age | M ± SD | 58.7 ± 13.6 | 57.5 ± 12.1 | 0.7 | 58.3 ± 13 | 57.6 ± 12.4 | 0.8 | 59.3 ± 12.1 | 56.3 ± 13.2 | 0.4 | |
| ≤55 | 29 | 10 | 19 | 0.8 | 12 | 17 | 0.7 | 14 | 15 | 0.3 | |
| >55 | 35 | 13 | 22 | 16 | 19 | 21 | 14 | ||||
| Sex | F | 29 | 10 | 19 | 0.8 | 14 | 15 | 0.5 | 16 | 13 | 0.9 |
| M | 35 | 13 | 22 | 14 | 21 | 19 | 16 | ||||
| Location | Rt | 47 | 19 | 33 | 0.8 | 21 | 31 | 0.2 | 28 | 24 | 0.7 |
| Lt | 12 | 4 | 8 | 7 | 5 | 7 | 5 | ||||
| Grade | G1 | 9 | 2 | 7 | 0.3 | 5 | 4 | 0.1 | 6 | 3 | |
| G2 | 38 | 18 | 20 | 18 | 20 | 21 | 17 | 0.3 | |||
| G3 | 17 | 3 | 14 | 5 | 12 | 8 | 9 | ||||
| T stage | T1 | 5 | 0 | 5 | 0.02 | 2 | 3 | 0.6 | 1 | 4 | 0.5 |
| T2 | 15 | 5 | 10 | 8 | 7 | 9 | 6 | ||||
| T3 | 40 | 14 | 26 | 16 | 24 | 23 | 17 | ||||
| T4 | 4 | 4 | 0 | 2 | 2 | 2 | 2 | ||||
| T1+T2 | 17 | 4 | 13 | 0.2 | 10 | 7 | 0.1 | 10 | 7 | 0.6 | |
| T3+T4 | 47 | 19 | 28 | 18 | 29 | 25 | 22 | ||||
| N stage | N0 | 37 | 8 | 29 | 0.00 | 22 | 15 | 0.00 | 35 | 12 | 0.00 |
| N1 | 24 | 13 | 11 | 6 | 18 | 10 | 14 | ||||
| N2 | 3 | 2 | 1 | 0 | 3 | 0 | 3 | ||||
| M stage | M0 | 59 | 18 | 41 | 0.00 | 27 | 32 | 0.2 | 35 | 24 | 0.01 |
| M1 | 5 | 5 | 0 | 1 | 4 | 0 | 5 | ||||
| LVI | No | 35 | 13 | 22 | 0.8 | 18 | 17 | 0.1 | 22 | 13 | 0.1 |
| Yes | 29 | 10 | 19 | 10 | 19 | 13 | 16 | ||||
| Dukes | A | 10 | 3 | 7 | 0.00 | 8 | 2 | 0.00 | 6 | 4 | 0.00 |
| B | 26 | 3 | 23 | 14 | 12 | 20 | 6 | ||||
| C | 23 | 12 | 11 | 5 | 18 | 9 | 14 | ||||
| D | 5 | 5 | 0 | 1 | 4 | 0 | 5 | ||||
| A-B | 35 | 6 | 29 | 0.00 | 22 | 13 | 0.00 | 25 | 10 | 0.00 | |
| C-D | 29 | 17 | 12 | 6 | 23 | 10 | 19 | ||||
| Relapse | No | 44 | 12 | 32 | 0.03 | 22 | 22 | 0.1 | 25 | 19 | 0.6 |
| Yes | 20 | 11 | 9 | 6 | 14 | 10 | 10 | ||||
| Alive | Dead | 35 | 14 | 21 | 0.4 | 13 | 22 | 0.2 | 14 | 21 | 0.01 |
| Survived | 29 | 9 | 20 | 15 | 14 | 21 | 8 | ||||
| Variable | PDL1 | p-Value | |||||
|---|---|---|---|---|---|---|---|
| NC−/IIC− | NC+/IIC− | NC−/IIC+ | NC+/IIC+ | ||||
| Number | 18 | 17 | 10 | 19 | |||
| Age (y) | ≤55 | 29 | 7 | 7 | 5 | 10 | 0.8 |
| >55 | 35 | 11 | 10 | 5 | 9 | ||
| Sex | F | 29 | 8 | 8 | 6 | 7 | 0.7 |
| M | 35 | 10 | 9 | 4 | 12 | ||
| Laterality | RT | 52 | 13 | 15 | 8 | 16 | 0.6 |
| LT | 12 | 5 | 2 | 2 | 3 | ||
| Grade | G1 | 9 | 5 | 1 | 0 | 3 | 0.3 |
| G2 | 38 | 10 | 11 | 8 | 9 | ||
| G3 | 17 | 3 | 5 | 2 | 7 | ||
| T stage | T1 | 5 | 1 | 0 | 1 | 3 | 0.5 |
| T2 | 15 | 6 | 3 | 2 | 4 | ||
| T3 | 40 | 10 | 13 | 6 | 11 | ||
| T4 | 4 | 1 | 1 | 1 | 1 | ||
| T1-2 | 17 | 7 | 3 | 3 | 4 | 0.4 | |
| T3-4 | 47 | 11 | 714 | 7 | 15 | ||
| N stage | No | 37 | 15 | 10 | 7 | 5 | 0.002 |
| N1 | 24 | 3 | 7 | 3 | 11 | ||
| N2 | 3 | 0 | 0 | 0 | 3 | ||
| M stage | No | 59 | 18 | 17 | 9 | 15 | 0.05 |
| Yes | 5 | 0 | 0 | 1 | 4 | ||
| LVI | No | 35 | 12 | 10 | 6 | 7 | 0.3 |
| Yes | 29 | 6 | 7 | 4 | 12 | ||
| Dukes | A | 10 | 5 | 1 | 3 | 1 | 0.001 |
| B | 26 | 11 | 9 | 3 | 3 | ||
| C | 23 | 2 | 7 | 3 | 11 | ||
| D | 5 | 0 | 0 | 1 | 4 | ||
| A-B | 35 | 16 | 9 | 6 | 4 | 0.001 | |
| C-D | 29 | 2 | 8 | 4 | 15 | ||
| Relapse | No | 44 | 15 | 10 | 7 | 12 | 0.4 |
| Yes | 20 | 3 | 7 | 3 | 7 | ||
| Alive | Dead | 35 | 7 | 7 | 6 | 15 | 0.05 |
| Survived | 29 | 11 | 10 | 4 | 4 | ||
| BRAF Protein Score | BRAF Protein Positivity | IIC PD1 | NC PDL1 | IIC PDL1 | PDL1gp | |||
|---|---|---|---|---|---|---|---|---|
| BRAF protein Score | Correlation Coefficient | 1.000 | 0.966 ** | −0.124 | 0.181 | 0.139 | 0.191 | |
| Sig. (2-tailed) | . | 0.000 | 0.329 | 0.152 | 0.274 | 0.132 | ||
| N | 64 | 64 | 64 | 64 | 64 | 64 | ||
| BRAF protein Positive | Correlation Coefficient | 0.966 ** | 1.000 | −0.135 | 0.190 | 0.172 | 0.222 | |
| Sig. (2-tailed) | 0.000 | 0.289 | 0.133 | 0.174 | 0.078 | |||
| N | 64 | 64 | 64 | 64 | 64 | 64 | ||
| IIC PD1 | Correlation Coefficient | −0.124 | −0.135 | 1.000 | 0.062 | −0.234 | −0.172 | |
| Sig. (2-tailed) | 0.329 | 0.289 | . | 0.629 | 0.063 | 0.174 | ||
| N | 64 | 64 | 64 | 64 | 64 | 64 | ||
| NC PDL1 | Correlation Coefficient | 0.181 | 0.190 | 0.062 | 1.000 | 0.170 | 0.591 ** | |
| Sig. (2-tailed) | 0.152 | 0.133 | 0.629 | . | 0.179 | 0.000 | ||
| N | 64 | 64 | 64 | 64 | 64 | 64 | ||
| IIC PDL1 | Correlation Coefficient | 0.139 | 0.172 | −0.234 | 0.170 | 1.000 | 0.895 ** | |
| Sig. (2-tailed) | 0.274 | 0.174 | 0.063 | 0.179 | . | 0.000 | ||
| N | 64 | 64 | 64 | 64 | 64 | 64 | ||
| PDL1gp | Correlation Coefficient | 0.191 | 0.222 | −0.172 | 0.591 ** | 0.895 ** | 1.000 | |
| Sig. (2-tailed) | 0.132 | 0.078 | 0.174 | 0.000 | 0.000 | . | ||
| N | 64 | 64 | 64 | 64 | 64 | 64 | ||
| OS | DFS | |||||||
|---|---|---|---|---|---|---|---|---|
| Exp(B) | 95.0% CI for Exp(B) | Sig. | Exp(B) | 95.0% CI for Exp(B) | Sig. | |||
| Lower | Upper | Lower | Upper | |||||
| Age (>55 vs ≤55) | 2.343 | 0.660 | 8.320 | 0.188 | 1.240 | 0.509 | 3.022 | 0.636 |
| Sex | 1.887 | 0.609 | 5.842 | 0.271 | 1.931 | 0.879 | 4.244 | 0.101 |
| Location (left vs. right) | 433.2 | 1.437 | 130573.113 | 0.037 | 11.139 | 0.340 | 364.5 | 0.176 |
| Grade (G3 vs G1/2) | 0.512 | 0.180 | 1.461 | 0.211 | 0.589 | 0.313 | 1.108 | 0.100 |
| T stage | 11.482 | 1.491 | 88.393 | 0.019 | 0.947 | 0.369 | 2.431 | 0.909 |
| LN stage | 0.815 | 0.068 | 9.822 | 0.872 | 0.547 | 0.096 | 3.131 | 0.498 |
| Metastasis | 0.184 | 0.004 | 8.693 | 0.390 | 0.940 | 0.072 | 12.263 | 0.962 |
| LVI | 2.503 | 0.703 | 8.905 | 0.157 | 1.272 | 0.446 | 3.627 | 0.653 |
| Dukes Staging | 0.810 | 0.100 | 6.536 | 0.843 | 0.831 | 0.205 | 3.364 | 0.795 |
| BRAF Score | 0.074 | 0.003 | 1.643 | 0.100 | 1.326 | 0.489 | 3.591 | 0.579 |
| BRAF Positive | 11.405 | 0.076 | 1711.9 | 0.341 | 0.512 | 0.060 | 4.381 | 0.541 |
| PD1 stroma | 0.233 | 0.060 | 0.903 | 0.035 | 1.056 | 0.333 | 3.352 | 0.926 |
| PDL1 tissue | 3.250 | 1.088 | 9.713 | 0.035 | 2.241 | 1.003 | 5.006 | 0.049 |
| PDL1stroma | 0.380 | 0.102 | 1.411 | 0.148 | 1.131 | 0.480 | 2.668 | 0.778 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahiem, A.T.; Eladl, E.; Toraih, E.A.; Fawzy, M.S.; Abdelwahab, K.; Elnaghi, K.; Emarah, Z.; Shaalan, A.A.M.; Ehab, Z.; Soliman, N.A. Prognostic Value of BRAF, Programmed Cell Death 1 (PD1), and PD Ligand 1 (PDL1) Protein Expression in Colon Adenocarcinoma. Diagnostics 2023, 13, 237. https://doi.org/10.3390/diagnostics13020237
Ibrahiem AT, Eladl E, Toraih EA, Fawzy MS, Abdelwahab K, Elnaghi K, Emarah Z, Shaalan AAM, Ehab Z, Soliman NA. Prognostic Value of BRAF, Programmed Cell Death 1 (PD1), and PD Ligand 1 (PDL1) Protein Expression in Colon Adenocarcinoma. Diagnostics. 2023; 13(2):237. https://doi.org/10.3390/diagnostics13020237
Chicago/Turabian StyleIbrahiem, Afaf T., Entsar Eladl, Eman A. Toraih, Manal S. Fawzy, Khaled Abdelwahab, Khaled Elnaghi, Ziad Emarah, Aly A. M. Shaalan, Ziad Ehab, and Nahed A. Soliman. 2023. "Prognostic Value of BRAF, Programmed Cell Death 1 (PD1), and PD Ligand 1 (PDL1) Protein Expression in Colon Adenocarcinoma" Diagnostics 13, no. 2: 237. https://doi.org/10.3390/diagnostics13020237
APA StyleIbrahiem, A. T., Eladl, E., Toraih, E. A., Fawzy, M. S., Abdelwahab, K., Elnaghi, K., Emarah, Z., Shaalan, A. A. M., Ehab, Z., & Soliman, N. A. (2023). Prognostic Value of BRAF, Programmed Cell Death 1 (PD1), and PD Ligand 1 (PDL1) Protein Expression in Colon Adenocarcinoma. Diagnostics, 13(2), 237. https://doi.org/10.3390/diagnostics13020237









