Automated Pneumonia Based Lung Diseases Classification with Robust Technique Based on a Customized Deep Learning Approach
Abstract
1. Introduction
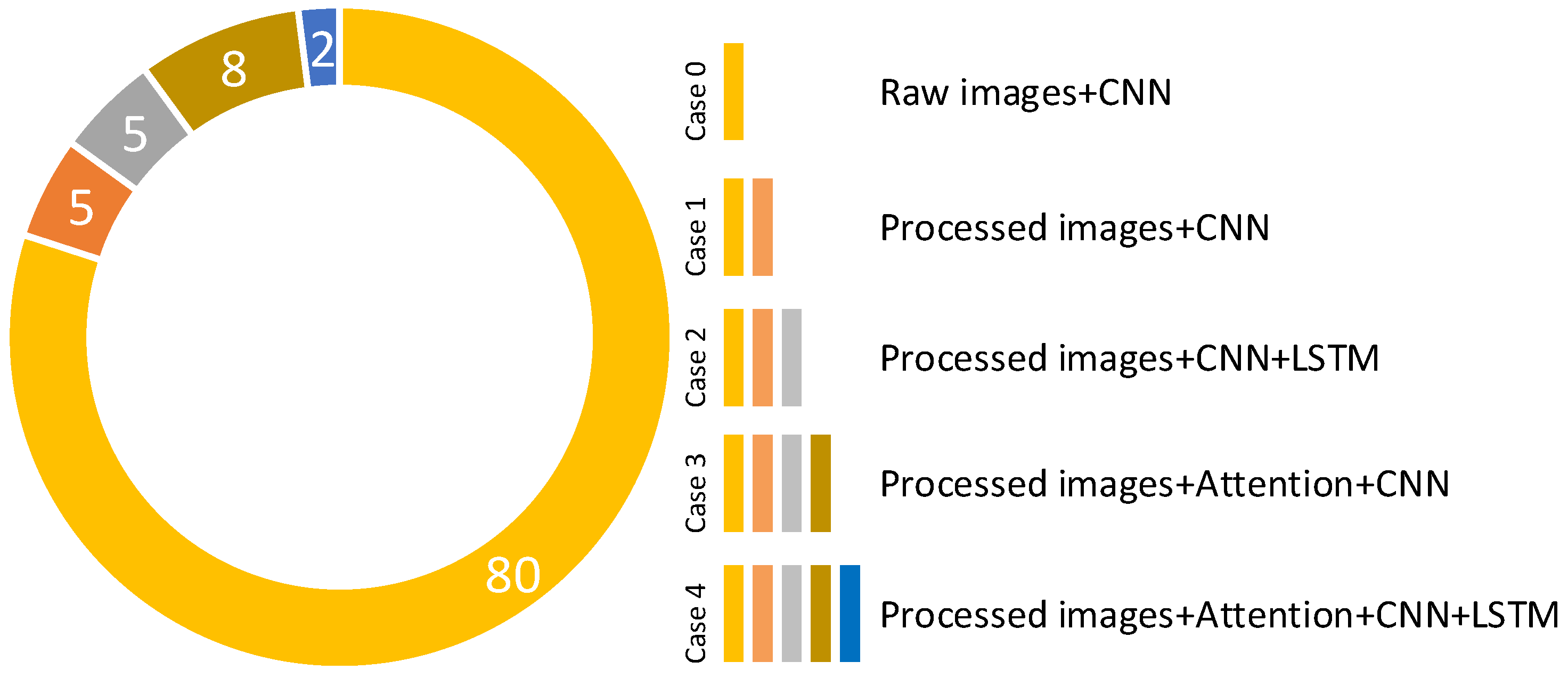
- Different regions on images are marked using the MCW segmentation algorithm. Because of this, it enables the unique information in the data to stand out. The pre-processing operation with the MCW algorithm increased the classification accuracy.
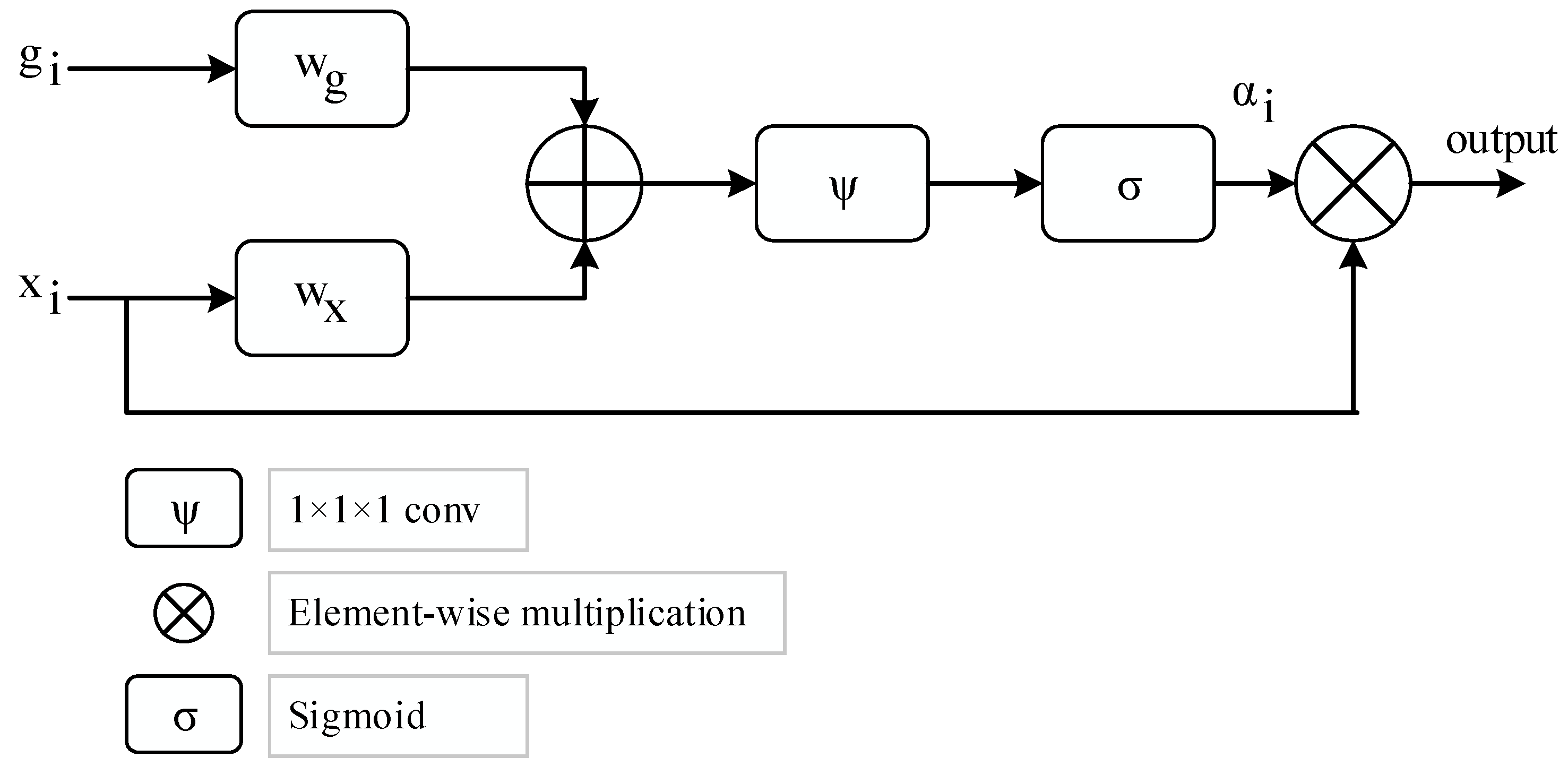
- The attention structure in the CNN models is used to increase the distinctive representation. The LSTM blocks in deep learning models are added to benefit the ability to keep weight information in their memory blocks. Therefore, the attention-CNN LSTM (ACL) model, which was synchronously trained in the attention structure, convolutional layers, and the LSTM model, improved classification performance compared to the CNN model which did not contain attention and LSTM structures.
2. Related Works
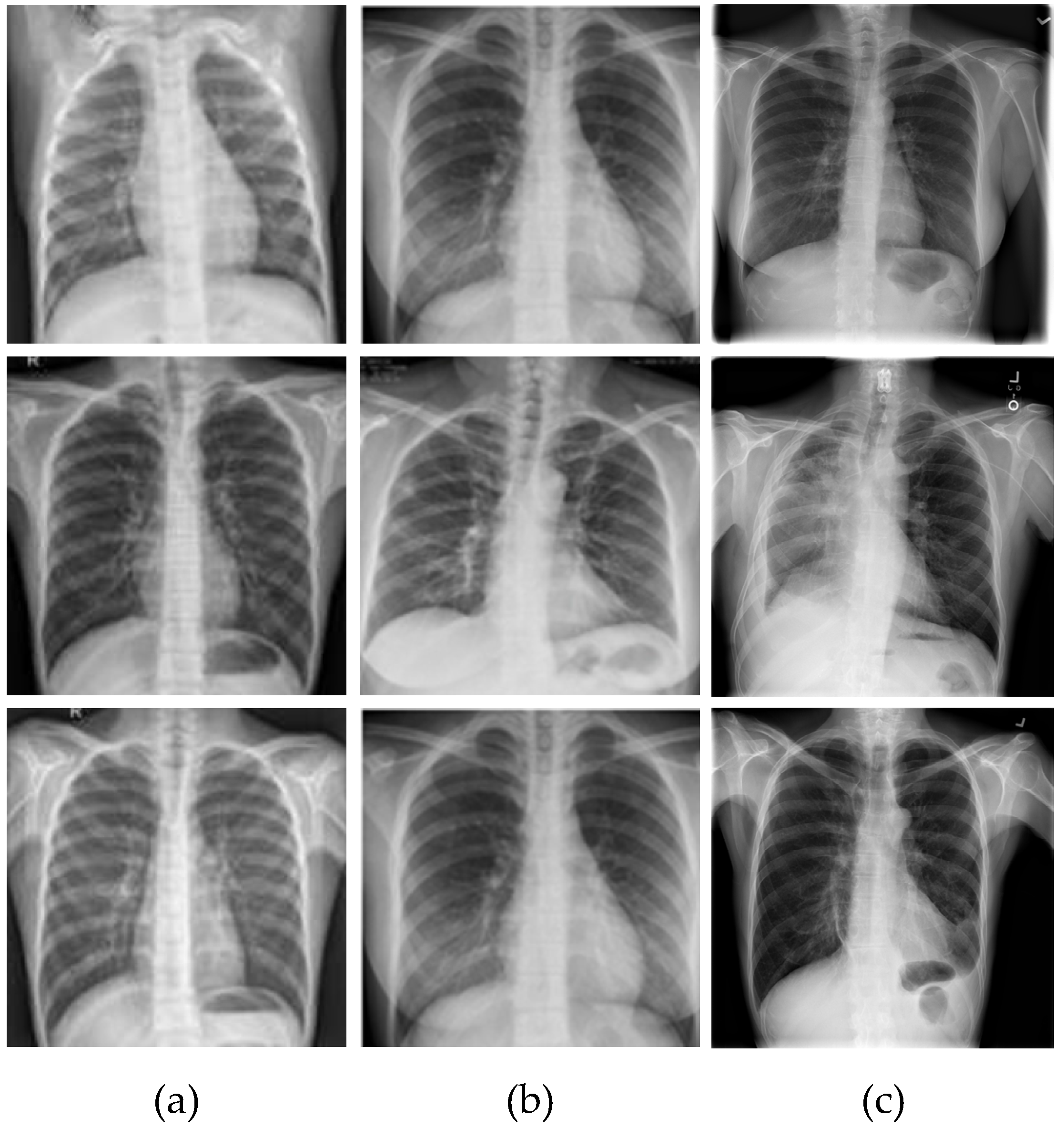
3. Dataset
4. Proposed Methodology
5. Methodology Techniques
5.1. Pre-Processing
- Step-1
- Calculate the segmentation process that divides dark areas into items.
- Step-2
- Determine the foreground markers, which contain the linked pixel blots inside of each object.
- Step-3
- Determine background markers or pixels that are not a part of any item.
- Step-4
- Update for decreasing the foreground and background marker locations’ segmentation functions.
- Step-5
- Use the revised parameters to calculate the watershed transform.
- Step-6
- Compute learning parameters.
5.2. Machine Learning Technique
6. Experimental Studies
7. Discussion
8. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| SGDM Options | Values |
|---|---|
| Momentum | 0.9 |
| Initial Learn Rate | 0.001 |
| Learn Rate Schedule | ‘none’ |
| Learn Rate Drop Factor | 0.1 |
| Learn Rate Drop Period | 10 |
| L2 Regularization | 0.0001 |
| Gradient Threshold Method | ‘l2norm’ |
| Gradient Threshold | Inf |
| Max Epochs | 5 |
| Mini-BatchSize | 32 |
| Verbose | 0 |
| Verbose Frequency | 50 |
| Validation Frequency | 30 |
| Validation Patience | Inf |
| Shuffle | ‘every-epoch’ |
| Execution Environment | ‘auto’ |
| Sequence Length | ‘longest’ |
| Sequence Padding Value | 0 |
| Sequence Padding Direction | ‘right’ |
| Dispatch In Background | 0 |
| Reset Input Normalization | 1 |
| Batch Normalization Statistics | ‘population’ |
References
- European Lung Foundation. Acute Lower Respiratory Infections. Available online: https://europeanlung.org/en/information-hub/lung-conditions/acute-lower-respiratory-infections/ (accessed on 2 March 2022).
- American Lung Association. Learn about Pneumonia. Available online: https://www.lung.org/lung-health-diseases/lung-disease-lookup/pneumonia/learn-about-pneumonia (accessed on 2 March 2022).
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 7 September 2022).
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Bernheim, A.; Mei, X.; Huang, M.; Yang, Y.; Fayad, Z.A.; Zhang, N.; Diao, K.; Lin, B.; Zhu, X.; Li, K.; et al. Chest CT findings in coronavirus disease 2019 (COVID-19): Relationship to duration of infection. Radiology 2020, 295, 685–691. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, E115–E117. [Google Scholar] [CrossRef]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021, 24, 1207–1220. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef] [PubMed]
- Tuncer, T.; Dogan, S.; Ozyurt, F. An automated Residual Exemplar Local Binary Pattern and iterative ReliefF based corona detection method using lung X-ray image. Chemom. Intell. Lab. Syst. 2020, 203, 104054. [Google Scholar] [CrossRef]
- Singh, V.; Sharma, N.; Singh, S. A review of imaging techniques for plant disease detection. Artif. Intell. Agric. 2020, 4, 229–242. [Google Scholar] [CrossRef]
- Tu, K.N.; Lie, J.D.; Wan, C.K.V.; Cameron, M.; Austel, A.G.; Nguyen, J.K.; Van, K.; Hyun, D. Osteoporosis: A review of treatment options. P&T 2018, 43, 92–104. [Google Scholar]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA—J. Am. Med. Assoc. 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Dimmeler, S. Cardiovascular disease review series. EMBO Mol. Med. 2011, 3, 697. [Google Scholar] [CrossRef] [PubMed]
- Okinda, C.; Nyalala, I.; Korohou, T.; Okinda, C.; Wang, J.; Achieng, T.; Wamalwa, P.; Mang, T.; Shen, M. A review on computer vision systems in monitoring of poultry: A welfare perspective. Artif. Intell. Agric. 2020, 4, 184–208. [Google Scholar] [CrossRef]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit. Lett. 2020, 138, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Sethy, P.K.; Behera, S.K.; Ratha, P.K.; Biswas, P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int. J. Math. Eng. Manag. Sci. 2020, 5, 643–651. [Google Scholar] [CrossRef]
- Hemdan, E.E.-D.; Shouman, M.A.; Karar, M.E. COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef]
- Horry, M.J.; Chakraborty, S.; Paul, M.; Ulhaq, A.; Pradhan, B.; Saha, M.; Shukla, N. COVID-19 Detection through Transfer Learning Using Multimodal Imaging Data. IEEE Access 2020, 8, 149808–149824. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Rajendra Acharya, U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Altan, A.; Karasu, S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos Solitons Fractals 2020, 140, 110071. [Google Scholar] [CrossRef]
- Tsiknakis, N.; Trivizakis, E.; Vassalou, E.; Papadakis, G.; Spandidos, D.; Tsatsakis, A.; Sánchez-García, J.; López-González, R.; Papanikolaou, N.; Karantanas, A.; et al. Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Exp. Ther. Med. 2020, 20, 727–735. [Google Scholar] [CrossRef]
- Demir, F. DeepCoroNet: A deep LSTM approach for automated detection of COVID-19 cases from chest X-ray images. Appl. Soft Comput. 2021, 103, 107160. [Google Scholar] [CrossRef] [PubMed]
- Ismael, A.M.; Şengür, A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021, 164, 114054. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, N.; Gupta, S.; Prusty, M.R.; Tripathy, R.K. Detection of COVID19 from X-ray images using multiscale Deep Convolutional Neural Network. Appl. Soft Comput. 2022, 119, 108610. [Google Scholar] [CrossRef] [PubMed]
- Demir, F.; Demir, K.; Şengür, A. DeepCov19Net: Automated COVID-19 Disease Detection with a Robust and Effective Technique Deep Learning Approach. New Gener. Comput. 2022, 40, 1053–1075. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Chest X-Ray Images. Available online: https://www.kaggle.com/bachrr/covid-chest-xray (accessed on 1 March 2022).
- COVID-19 Chest X-Ray Images (Pneumonia). Available online: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia (accessed on 1 March 2022).
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray: Hospital-Scale Chest X-ray Database and Benchmarks on Weakly Supervised Classification and Localization of Common Thorax Diseases. In Deep Learning and Convolutional Neural Networks for Medical Imaging and Clinical Informatics; Advances in Computer Vision and Pattern Recognition; Springer: Cham, Switzerland, 2019; pp. 369–392. [Google Scholar]
- Wang, L.; Gong, P.; Biging, G.S. Individual tree-crown delineation and treetop detection in high-spatial-resolution aerial imagery. Photogramm. Eng. Remote Sensing 2004, 70, 351–357. [Google Scholar] [CrossRef]
- Huang, H.; Li, X.; Chen, C. Individual tree crown detection and delineation from very-high-resolution UAV images based on bias field and marker-controlled watershed segmentation algorithms. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 2253–2262. [Google Scholar] [CrossRef]
- Alakwaa, W.; Nassef, M.; Badr, A. Lung cancer detection and classification with 3D convolutional neural network (3D-CNN). Int. J. Biol. Biomed. Eng. 2017, 11, 66–73. [Google Scholar] [CrossRef]
- Demir, F.; Siddique, K.; Alswaitti, M.; Demir, K.; Sengur, A. A Simple and Effective Approach Based on a Multi-Level Feature Selection for Automated Parkinson’s Disease Detection. J. Pers. Med. 2022, 12, 55. [Google Scholar] [CrossRef]
- Demir, F.; Akbulut, Y. A new deep technique using R-CNN model and L1NSR feature selection for brain MRI classification. Biomed. Signal Process. Control 2022, 75, 103625. [Google Scholar] [CrossRef]
- Petmezas, G.; Haris, K.; Stefanopoulos, L.; Kilintzis, V.; Tzavelis, A.; Rogers, J.A.; Katsaggelos, A.K.; Maglaveras, N. Automated Atrial Fibrillation Detection using a Hybrid CNN-LSTM Network on Imbalanced ECG Datasets. Biomed. Signal Process. Control 2021, 63, 102194. [Google Scholar] [CrossRef]
- Demir, F. DeepBreastNet: A novel and robust approach for automated breast cancer detection from histopathological images. Biocybern. Biomed. Eng. 2021, 41, 1123–1139. [Google Scholar] [CrossRef]
- Demir, F.; Tașcı, B. An Effective and Robust Approach Based on R-CNN+LSTM Model and NCAR Feature Selection for Ophthalmological Disease Detection from Fundus Images. J. Pers. Med. 2021, 11, 1276. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Atila, O.; Şengür, A. Attention guided 3D CNN-LSTM model for accurate speech based emotion recognition. Appl. Acoust. 2021, 182, 108260. [Google Scholar] [CrossRef]
- Ucar, F.; Korkmaz, D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses 2020, 140, 109761. [Google Scholar] [CrossRef]
- Nour, M.; Cömert, Z.; Polat, K. A Novel Medical Diagnosis model for COVID-19 infection detection based on Deep Features and Bayesian Optimization. Appl. Soft Comput. 2020, 97, 106580. [Google Scholar] [CrossRef] [PubMed]
- Turkoglu, M. COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl. Intell. 2021, 51, 1213–1226. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020, 121, 103805. [Google Scholar] [CrossRef]

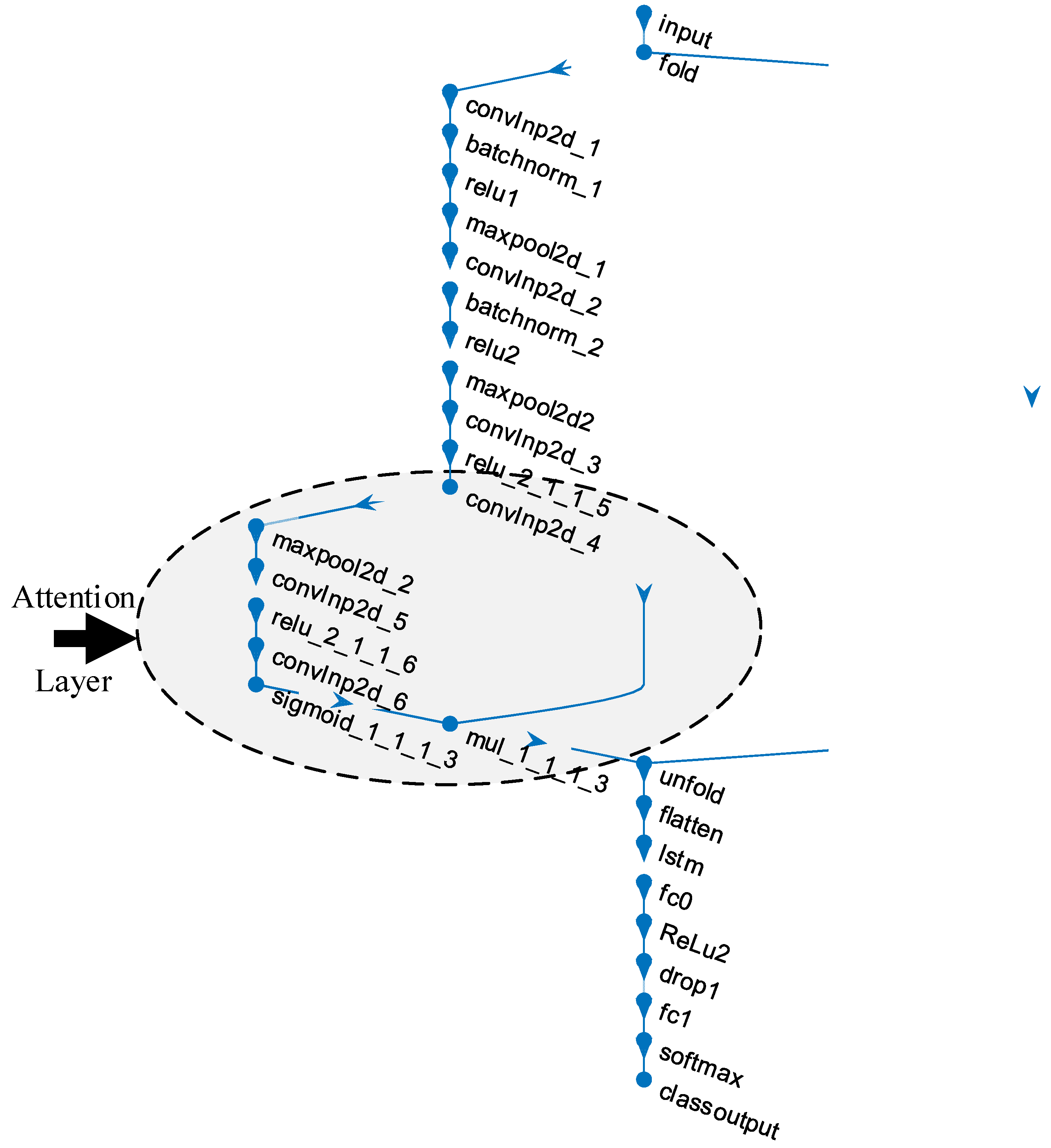

| Layer # | Layer Name | Layer | Layer Info |
|---|---|---|---|
| 1 | input | Sequence Input | Sequence input with 100 × 100 × 3 dimensions |
| 2 | fold | Sequence Folding | Sequence folding |
| 3 | convInp2d_1 | Convolution | 16 3 × 3 × 3 convolutions with stride: 1 and padding: 0 |
| 4 | batchnorm_1 | BN | BN with 16 channels |
| 5 | relu1 | ReLU | ReLU |
| 6 | maxpool2d_1 | Max Pooling | 3 × 3 max pooling with stride: 1 and padding: 0 |
| 7 | convInp2d_2 | Convolution | 16 3 × 3 × 16 convolutions with stride: 1 and padding: 0 |
| 8 | batchnorm_2 | BN | BN with 16 channels |
| 9 | relu2 | ReLU | ReLU |
| 10 | maxpool2d2 | Max Pooling | 3 × 3 max pooling with stride: 1 and padding: 0 |
| 11 | convInp2d_3 | Convolution | 16 3 × 3 × 16 convolutions with stride: 1 and padding: 0 |
| 12 | relu_2_1_1_5 | ReLU | ReLU |
| 13 | convInp2d_4 | Convolution | 16 3 × 3 × 16 convolutions with stride: 1 and padding: 0 |
| 14 | maxpool2d_2 | Max Pooling | 3 × 3 max pooling with stride: 1 and padding: 0 |
| 15 | convInp2d_5 | Convolution | 16 3 × 3 × 16 convolutions with stride: 1 and padding: 0 |
| 16 | relu_2_1_1_6 | ReLU | ReLU |
| 17 | convInp2d_6 | Convolution | 16 3 × 3 × 16 convolutions with stride: 1 and padding: 0 |
| 18 | sigmoid_1_1_1_3 | sigmoidLayer | sigmoidLayer |
| 19 | mul_1_1_1_3 | ElementWiseMultiplication | Element Wise Multiplication of 2 inputs |
| 20 | unfold | Sequence Unfolding | Sequence unfolding |
| 21 | flatten | Flatten | Flatten |
| 22 | lstm | LSTM | LSTM with 100 hidden units |
| 23 | fc0 | FC | 100 FC layer |
| 24 | ReLu2 | ReLU | ReLU |
| 25 | drop1 | Dropout | 40% dropout |
| 26 | fc1 | FC | 3 FC layer |
| 27 | softmax | Softmax | softmax |
| 28 | class output | Classification Output | crossentropyex |
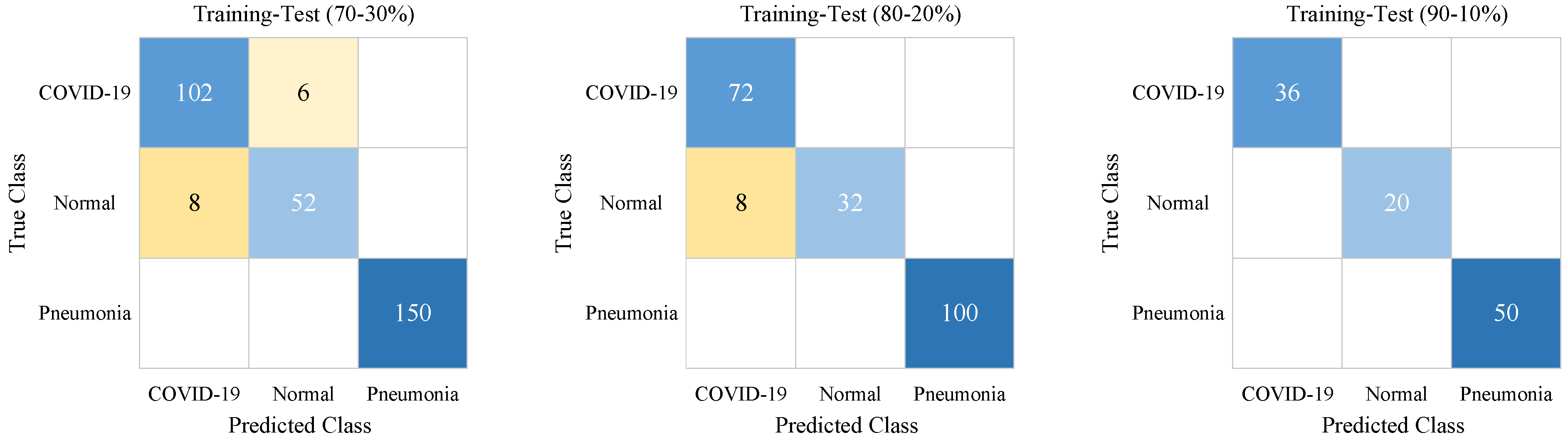
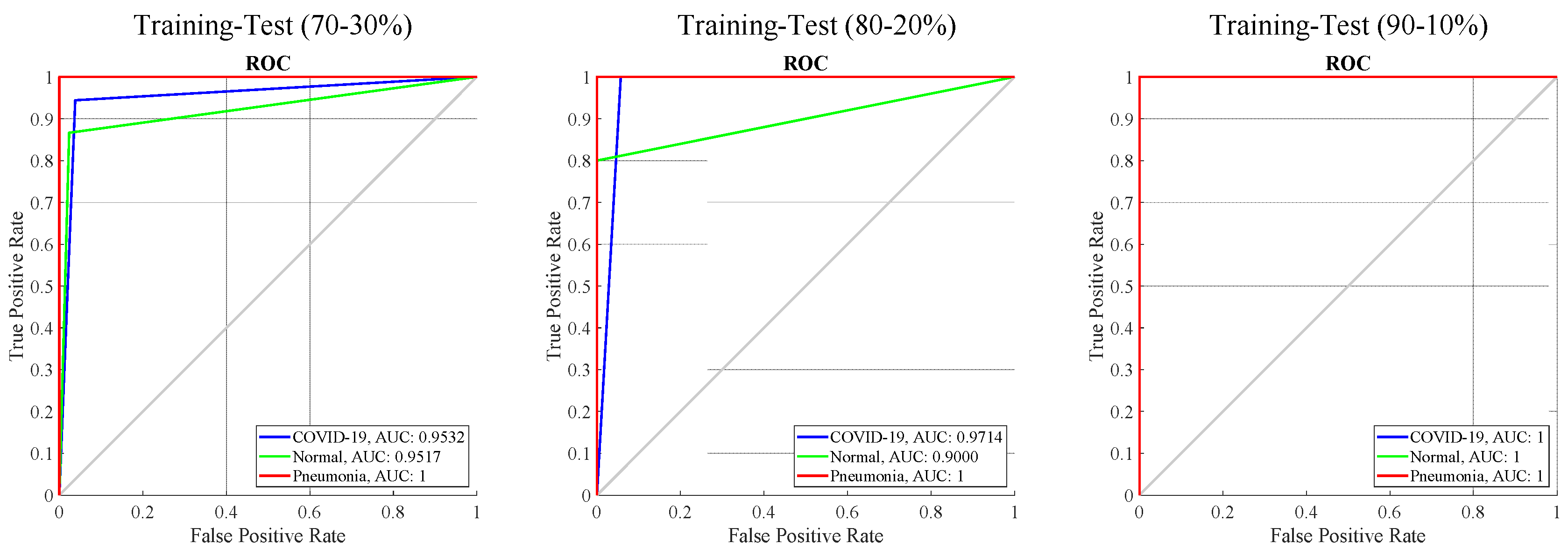
| Training-Test Ratios | Classes | Se | Sp | Pr | F-score |
|---|---|---|---|---|---|
| 70–30% | COVID-19 | 0.94 | 0.96 | 0.93 | 0.94 |
| Normal | 0.87 | 0.98 | 0.90 | 0.88 | |
| Pneumonia | 1.0 | 1.0 | 1.0 | 1.0 | |
| 80–20% | COVID-19 | 1.0 | 0.94 | 0.90 | 0.95 |
| Normal | 0.80 | 1.0 | 1.0 | 0.89 | |
| Pneumonia | 1.0 | 1.0 | 1.0 | 1.0 | |
| 90–10% | COVID-19 | 1.0 | 1.0 | 1.0 | 1.0 |
| Normal | 1.0 | 1.0 | 1.0 | 1.0 | |
| Pneumonia | 1.0 | 1.0 | 1.0 | 1.0 |
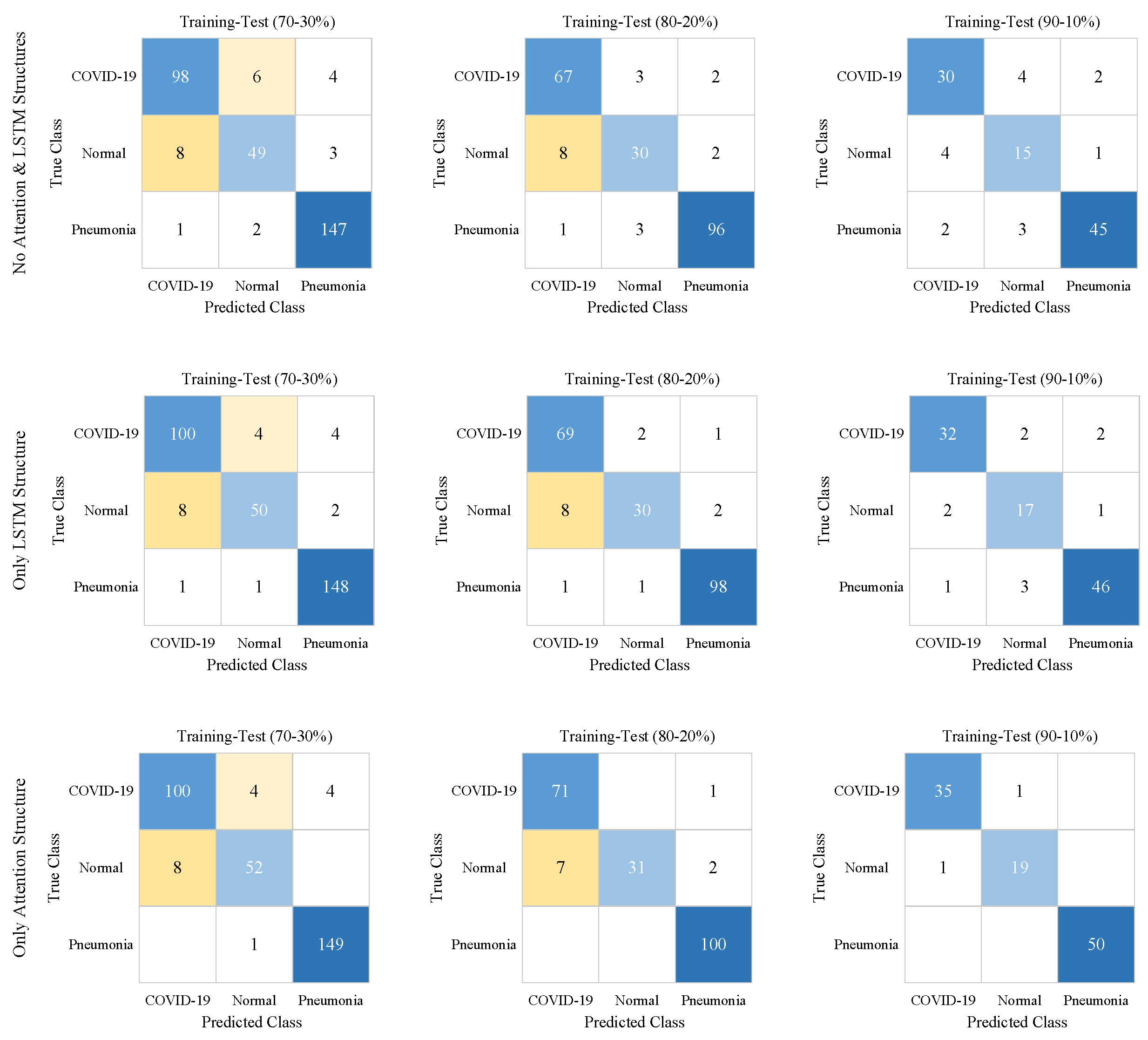
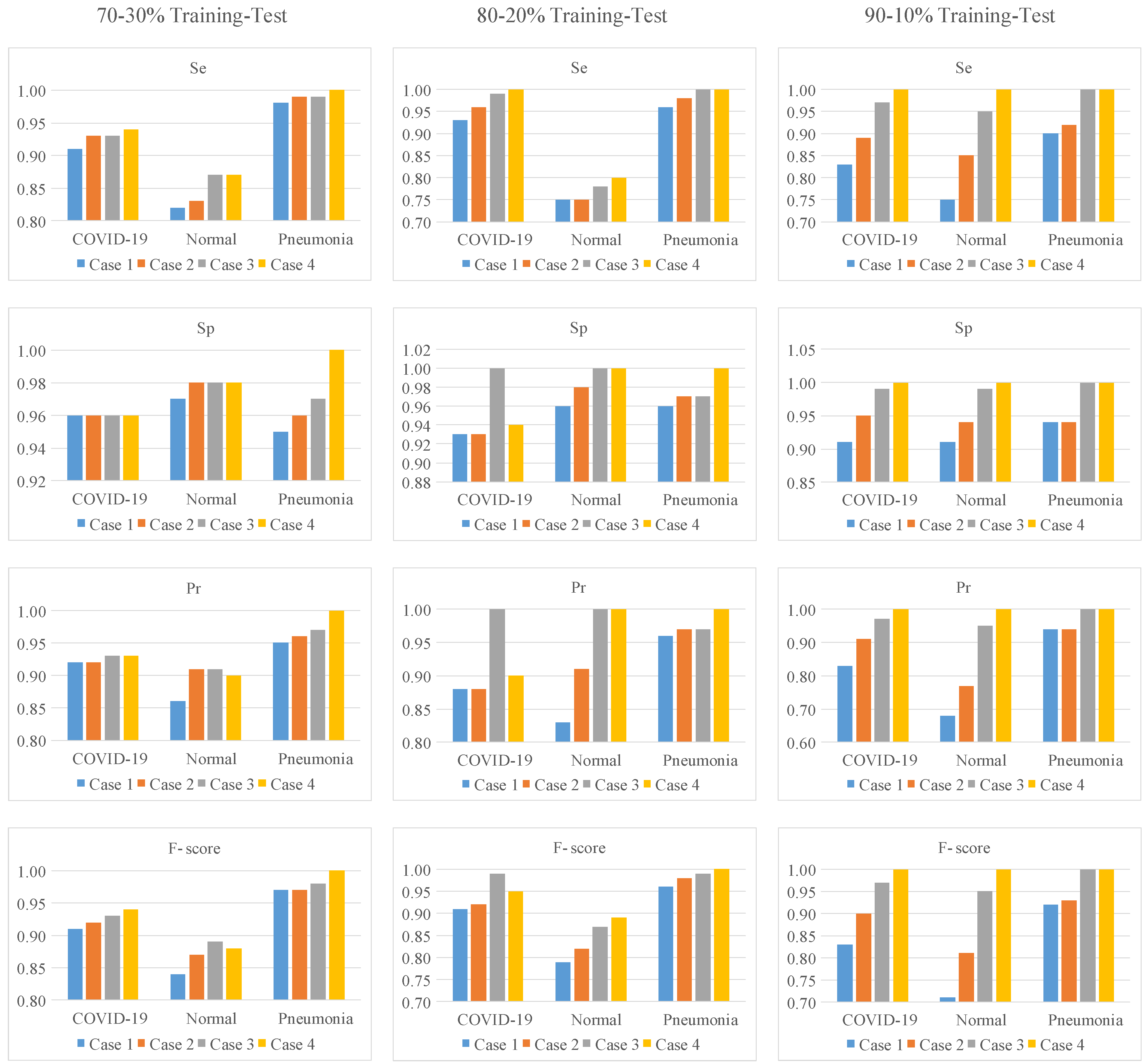
| Case # | Models | Training-Test Ratios | Classes | Se | Sp | Pr | F-score | Acc |
|---|---|---|---|---|---|---|---|---|
| 1 | No Attention and LSTM Structures | 70–30% | COVID-19 | 0.91 | 0.96 | 0.92 | 0.91 | 0.92 |
| Normal | 0.82 | 0.97 | 0.86 | 0.84 | ||||
| Pneumonia | 0.98 | 0.95 | 0.95 | 0.97 | ||||
| 80–20% | COVID-19 | 0.93 | 0.93 | 0.88 | 0.91 | 0.91 | ||
| Normal | 0.75 | 0.96 | 0.83 | 0.79 | ||||
| Pneumonia | 0.96 | 0.96 | 0.96 | 0.96 | ||||
| 90–10% | COVID-19 | 0.83 | 0.91 | 0.83 | 0.83 | 0.85 | ||
| Normal | 0.75 | 0.91 | 0.68 | 0.71 | ||||
| Pneumonia | 0.9 | 0.94 | 0.94 | 0.92 | ||||
| 2 | Only LSTM Structure | 70–30% | COVID-19 | 0.93 | 0.96 | 0.92 | 0.92 | 0.94 |
| Normal | 0.83 | 0.98 | 0.91 | 0.87 | ||||
| Pneumonia | 0.99 | 0.96 | 0.96 | 0.97 | ||||
| 80–20% | COVID-19 | 0.96 | 0.93 | 0.88 | 0.92 | 0.93 | ||
| Normal | 0.75 | 0.98 | 0.91 | 0.82 | ||||
| Pneumonia | 0.98 | 0.97 | 0.97 | 0.98 | ||||
| 90–10% | COVID-19 | 0.89 | 0.95 | 0.91 | 0.90 | 0.90 | ||
| Normal | 0.85 | 0.94 | 0.77 | 0.81 | ||||
| Pneumonia | 0.92 | 0.94 | 0.94 | 0.93 | ||||
| 3 | Only Attention Structure | 70–30% | COVID-19 | 0.93 | 0.96 | 0.93 | 0.93 | 0.95 |
| Normal | 0.87 | 0.98 | 0.91 | 0.89 | ||||
| Pneumonia | 0.99 | 0.97 | 0.97 | 0.98 | ||||
| 80–20% | COVID-19 | 0.99 | 1.0 | 1.0 | 0.99 | 0.95 | ||
| Normal | 0.78 | 1.0 | 1.0 | 0.87 | ||||
| Pneumonia | 1.0 | 0.97 | 0.97 | 0.99 | ||||
| 90–10% | COVID-19 | 0.97 | 0.99 | 0.97 | 0.97 | 0.98 | ||
| Normal | 0.95 | 0.99 | 0.95 | 0.95 | ||||
| Pneumonia | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| 4 | Proposed Approach | 70–30% | COVID-19 | 0.94 | 0.96 | 0.93 | 0.94 | 0.96 |
| Normal | 0.87 | 0.98 | 0.90 | 0.88 | ||||
| Pneumonia | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| 80–20% | COVID-19 | 1.0 | 0.94 | 0.90 | 0.95 | 0.96 | ||
| Normal | 0.80 | 1.0 | 1.0 | 0.89 | ||||
| Pneumonia | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| 90–10% | COVID-19 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| Normal | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| Pneumonia | 1.0 | 1.0 | 1.0 | 1.0 |
| Authors | Methods | Dataset | Classes # | Acc (%) | Se (%) | Sp (%) |
|---|---|---|---|---|---|---|
| Ozturk et al. [21] | DarkCovidNet | Public | 3 | 87.02 | 92.18 | 89.96 |
| Wang et al. [9] | COVID-Net | Public | 3 | 92.64 | 91.37 | 95.76 |
| Apostolopoulos et al. [19] | The pre-trained CNNs | Public | 3 | 96.78 | 98.66 | 96.46 |
| Ucar and Korkmaz [41] | COVIDiagnosis-Net | Public | 3 | 98.26 | 98.33 | 99.10 |
| Nour et al. [42] | Deep CNN, SVM | Public | 3 | 98.97 | 89.39 | 99.75 |
| Turkoglu [43] | AlexNet, Feature Selection, SVM | Public | 3 | 99.18 | 99.13 | 99.21 |
| Togacar et al. [44] | Deep features, SqueezeNet, SVM | Public | 3 | 99.27 | 98.33 | 99.69 |
| Demir et al. [27] | DeepCov19Net | Public | 3 | 99.75 | 99.33 | 99.79 |
| Demir [24] | DeepCoroNet | Public | 3 | 100.00 | 100.00 | 100.00 |
| Ismael and Sengur [25] | ResNet50 Features + SVM | Public | 2 | 94.74 | 91.00 | 98.89 |
| Muralidharan et al. [26] | FB2DEWT + CNN | Public | 3 | 96.00 | 96.00 | 96.00 |
| Proposed Method | Processed images, ACL model | Public | 3 | 100.00 | 100.00 | 100.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbulut, Y. Automated Pneumonia Based Lung Diseases Classification with Robust Technique Based on a Customized Deep Learning Approach. Diagnostics 2023, 13, 260. https://doi.org/10.3390/diagnostics13020260
Akbulut Y. Automated Pneumonia Based Lung Diseases Classification with Robust Technique Based on a Customized Deep Learning Approach. Diagnostics. 2023; 13(2):260. https://doi.org/10.3390/diagnostics13020260
Chicago/Turabian StyleAkbulut, Yaman. 2023. "Automated Pneumonia Based Lung Diseases Classification with Robust Technique Based on a Customized Deep Learning Approach" Diagnostics 13, no. 2: 260. https://doi.org/10.3390/diagnostics13020260
APA StyleAkbulut, Y. (2023). Automated Pneumonia Based Lung Diseases Classification with Robust Technique Based on a Customized Deep Learning Approach. Diagnostics, 13(2), 260. https://doi.org/10.3390/diagnostics13020260







