TAVI-PREP: A Deep Learning-Based Tool for Automated Measurements Extraction in TAVI Planning
Abstract
:1. Introduction
- Full Automation: We introduce a fully automated pre-TAVI measurement extraction algorithm capable of extracting 22 measurements in approximately 2 min, streamlining clinical workflows.
- Robust Validation: Our algorithm undergoes comparison with two experts in the field, enhancing its reliability and suitability for potential future clinical use. Validation is conducted on the largest cohort to date, involving 200 patients, further ensuring its accuracy and applicability.
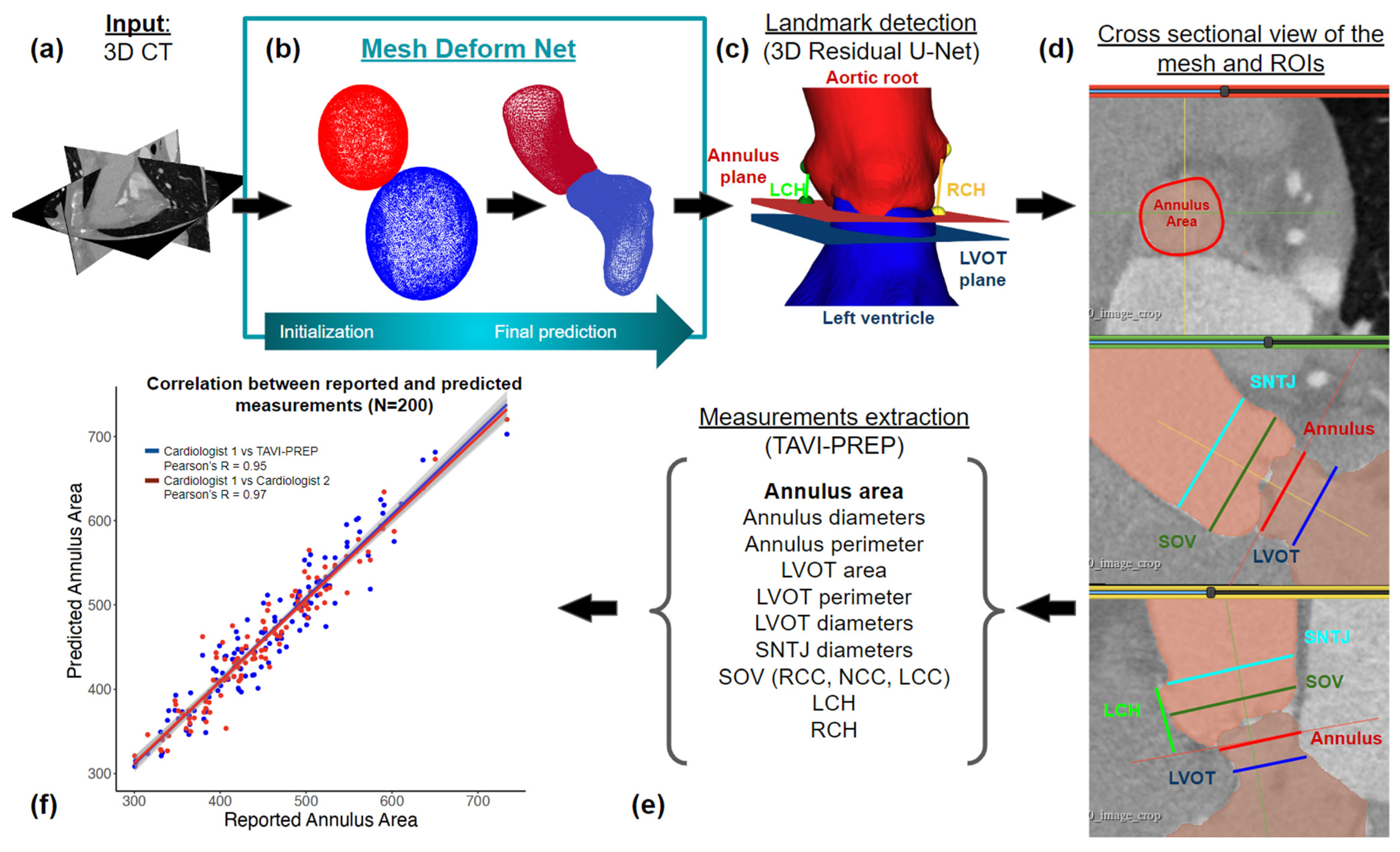
2. Materials and Methods
2.1. Data
2.1.1. Segmentation Dataset
2.1.2. Landmark Detection Dataset
2.1.3. Final Measurements Dataset
2.2. Segmentation
2.3. Landmark Detection
2.4. Derivation of Measurements
2.4.1. Centerline Extraction
2.4.2. Annulus Plane
2.4.3. Left Coronary Height (LCH) and Right Coronary Height (RCH)
2.4.4. Left Ventricular Outflow Track (LVOT)
2.4.5. Sinus of Valsalva (SOV) and Sinotubular Junction (SNTJ)
3. Results
3.1. Segmentation and Landmark Detection Performance
3.2. Manual vs. Automatic Measurements
- Mean relative error, correlation coefficients, and confidence intervals (CIs): Discrepancies were reported as the absolute relative mean of the error and the 95% confidence interval (CI) boundaries, as defined in Equation (1). is the mean, Z is the chosen z-score (1.96 for 95% CI), s is the standard deviation, and n is the number of samples. Pearson correlation coefficients are also reported.
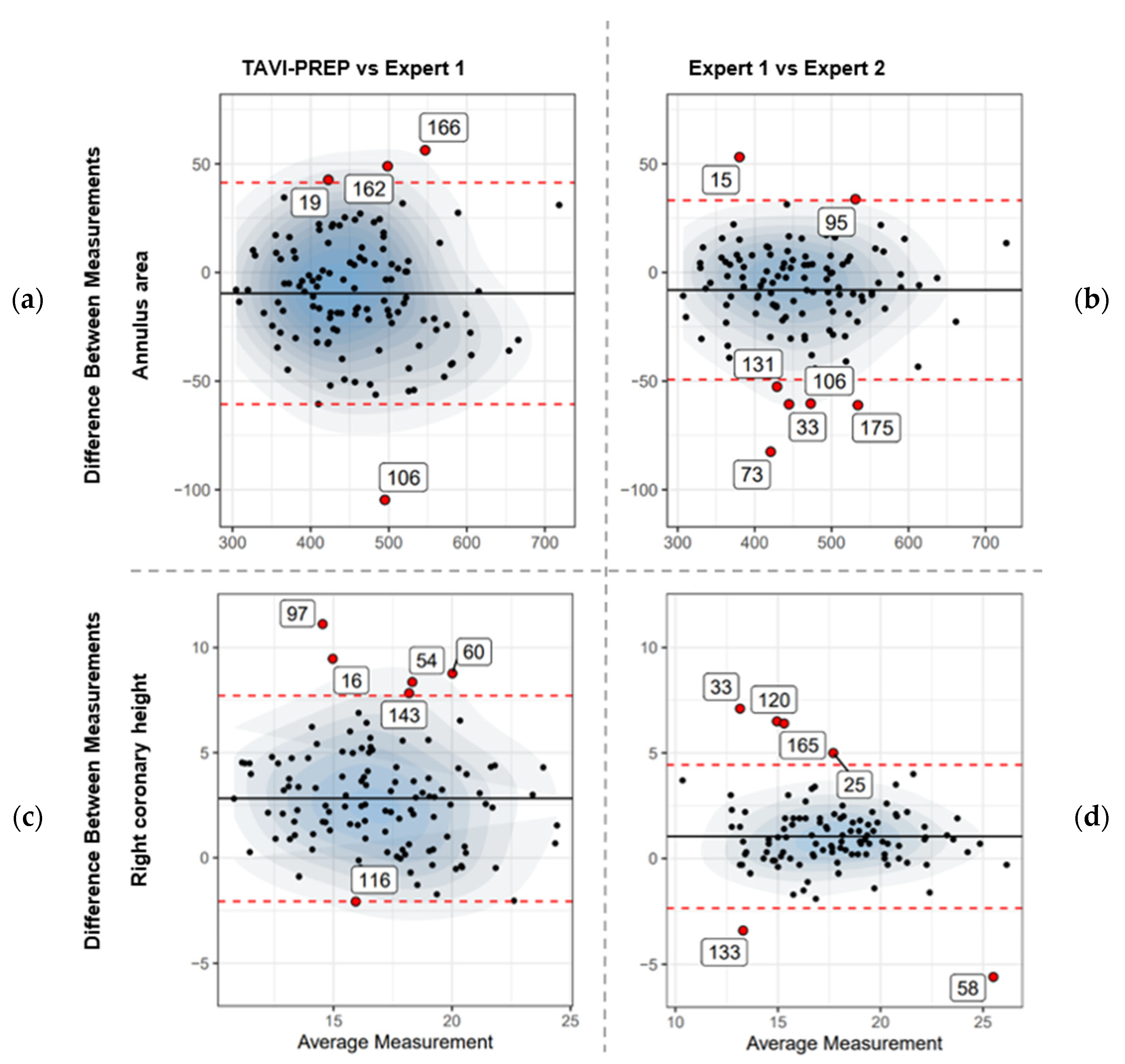
- Bland–Altman plots: A graphical method to analyze the agreement between two quantitative measurements. Plots were created in a pairwise fashion (Expert 1 vs. Expert 2, TAVI-PREP vs. Expert 1, and TAVI-PREP vs. Expert 2). These plots give a comprehensive understanding of how predicted values compare to expected values across the range of measurements.
3.3. Confidence Intervals and Pearson Correlation Coefficients
3.3.1. Annulus and LVOT
3.3.2. SNTJ
3.3.3. Sinus
3.3.4. Coronary Heights (LCH and RCH)
3.4. Bland–Altman
3.5. Edge Cases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reardon, M.J.; Van Mieghem, N.M.; Popma, J.J.; Kleiman, N.S.; Søndergaard, L.; Mumtaz, M.; Adams, D.H.; Deeb, G.M.; Maini, B.; Gada, H.; et al. Surgical or transcatheter aortic-valve replacement in intermediate-risk patients. N. Engl. J. Med. 2017, 376, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Leon, M.B.; Smith, C.R.; Mack, M.J.; Makkar, R.R.; Svensson, L.G.; Kodali, S.K.; Thourani, V.H.; Tuzcu, E.M.; Miller, D.C.; Herrmann, H.C.; et al. Transcatheter or Surgical Aortic-Valve Replacement in Intermediate-Risk Patients. N. Engl. J. Med. 2016, 374, 1609–1620. [Google Scholar] [CrossRef] [PubMed]
- Makkar, R.R.; Thourani, V.H.; Mack, M.J.; Kodali, S.K.; Kapadia, S.; Webb, J.G.; Yoon, S.H.; Trento, A.; Svensson, L.G.; Herrmann, H.C.; et al. Five-year outcomes of transcatheter or surgical aortic-valve replacement. N. Engl. J. Med. 2020, 382, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Mack, M.J.; Leon, M.B.; Thourani, V.H.; Makkar, R.; Kodali, S.K.; Russo, M.; Kapadia, S.R.; Malaisrie, S.C.; Cohen, D.J.; Pibarot, P.; et al. Transcatheter Aortic-Valve Replacement with a Balloon-Expandable Valve in Low-Risk Patients. N. Engl. J. Med. 2019, 380, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Popma, J.J.; Deeb, G.M.; Yakubov, S.J.; Mumtaz, M.; Gada, H.; O’Hair, D.; Bajwa, T.; Heiser, J.C.; Merhi, W.; Kleiman, N.S.; et al. Transcatheter Aortic-Valve Replacement with a Self-Expanding Valve in Low-Risk Patients. N. Engl. J. Med. 2019, 380, 1706–1715. [Google Scholar] [CrossRef] [PubMed]
- Vahanian, A.; Beyersdorf, F.; Praz, F.; Milojevic, M.; Baldus, S.; Bauersachs, J.; Capodanno, D.; Conradi, L.; De Bonis, M.; De Paulis, R.; et al. 2021 ESC/EACTS Guidelines for the management of valvular heart disease. Eur. Heart J. 2021, 43, 561–632. [Google Scholar] [CrossRef] [PubMed]
- Sundt, T.M.; Jneid, H. Guideline Update on Indications for Transcatheter Aortic Valve Implantation Based on the 2020 American College of Cardiology/American Heart Association Guidelines for Management of Valvular Heart Disease. JAMA Cardiol. 2021, 6, 1088. [Google Scholar] [CrossRef]
- Horehledova, B.; Mihl, C.; Hendriks, B.M.; Eijsvoogel, N.G.; Vainer, J.; Veenstra, L.F.; Wildberger, J.E.; Das, M. Do CTA measurements of annular diameter, perimeter and area result in different TAVI prosthesis sizes? Int. J. Cardiovasc. Imaging 2018, 34, 1819–1829. [Google Scholar] [CrossRef]
- Bleakley, C.; Monaghan, M.J. The Pivotal Role of Imaging in TAVR Procedures. Curr. Cardiol. Rep. 2018, 20, 9. [Google Scholar] [CrossRef]
- Akinseye, O.A.; Jha, S.K.; Ibebuogu, U.N. Clinical outcomes of coronary occlusion following transcatheter aortic valve replacement: A systematic review. Cardiovasc. Revascularization Med. 2018, 19, 229–236. [Google Scholar] [CrossRef]
- Saadi, R.P.; Tagliari, A.P.; Saadi, E.K.; Miglioranza, M.H.; Polanczyck, C.A. Preoperative TAVR Planning: How to Do It. J. Clin. Med. 2022, 11, 2582. [Google Scholar] [CrossRef]
- Blanke, P.; Weir-McCall, J.R.; Achenbach, S.; Delgado, V.; Hausleiter, J.; Jilaihawi, H.; Marwan, M.; Nørgaard, B.L.; Piazza, N.; Schoenhagen, P.; et al. Computed Tomography Imaging in the Context of Transcatheter Aortic Valve Implantation (TAVI)/Transcatheter Aortic Valve Replacement (TAVR): An Expert Consensus Document of the Society of Cardiovascular Computed Tomography. JACC Cardiovasc. Imaging 2019, 12, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Schmidkonz, C.; Marwan, M.; Klinghammer, L.; Mitschke, M.; Schuhbaeck, A.; Arnold, M.; Lell, M.; Achenbach, S.; Pflederer, T. Interobserver variability of CT angiography for evaluation of aortic annulus dimensions prior to transcatheter aortic valve implantation (TAVI). Eur. J. Radiol. 2014, 83, 1672–1678. [Google Scholar] [CrossRef] [PubMed]
- Schuhbaeck, A.; Achenbach, S.; Pflederer, T.; Marwan, M.; Schmid, J.; Nef, H.; Rixe, J.; Hecker, F.; Schneider, C.; Lell, M.; et al. Reproducibility of aortic annulus measurements by computed tomography. Eur. Radiol. 2014, 24, 1878–1888. [Google Scholar] [CrossRef]
- Sardar, P.; Abbott, J.D.; Kundu, A.; Aronow, H.D.; Granada, J.F.; Giri, J. Impact of Artificial Intelligence on Interventional Cardiology: From Decision-Making Aid to Advanced Interventional Procedure Assistance. JACC Cardiovasc. Interv. 2019, 12, 1293–1303. [Google Scholar] [CrossRef] [PubMed]
- Itchhaporia, D. Artificial intelligence in cardiology. Trends Cardiovasc. Med. 2022, 32, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA J. Am. Med. Assoc. 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Tahir, A.M.; Mutlu, O.; Bensaali, F.; Ward, R.; Ghareeb, A.N.; Helmy, S.M.; Othman, K.T.; Al-Hashemi, M.A.; Abujalala, S.; Chowdhury, M.E.; et al. Latest Developments in Adapting Deep Learning for Assessing TAVR Procedures and Outcomes. J. Clin. Med. 2023, 12, 4774. [Google Scholar] [CrossRef] [PubMed]
- Krüger, N.; Meyer, A.; Tautz, L.; Hüllebrand, M.; Wamala, I.; Pullig, M.; Kofler, M.; Kempfert, J.; Sündermann, S.; Falk, V.; et al. Cascaded neural network-based CT image processing for aortic root analysis. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 507–519. [Google Scholar] [CrossRef]
- Ma, Q.; Lemarchand, L.; Chan-Sock-Line, D.; Rigal, L.; Simon, A.; Haigron, P. Fully-automatic aortic valve landmarks detection with two-stage-based convolutional neural networks. In Medical Imaging 2023: Image Processing; Išgum, I., Colliot, O., Eds.; SPIE: Paris, France, 2023; p. 13. [Google Scholar] [CrossRef]
- Saitta, S.; Sturla, F.; Gorla, R.; Oliva, O.A.; Votta, E.; Bedogni, F.; Redaelli, A. A CT-based deep learning system for automatic assessment of aortic root morphology for TAVI planning. Comput. Biol. Med. 2023, 163, 107147. [Google Scholar] [CrossRef]
- Astudillo, P.; Mortier, P.; Bosmans, J.; De Backer, O.; de Jaegere, P.; Iannaccone, F.; De Beule, M.; Dambre, J. Automatic Detection of the Aortic Annular Plane and Coronary Ostia from Multidetector Computed Tomography. J. Interv. Cardiol. 2020, 2020, 9843275. [Google Scholar] [CrossRef]
- Kong, F.; Wilson, N.; Shadden, S.C. A Deep-Learning Approach for Direct Whole-Heart Mesh Reconstruction. Med. Image Anal. 2021, 74, 102222. [Google Scholar] [CrossRef]
- Pizer, S.M.; Amburn, E.P.; Austin, J.D.; Cromartie, R.; Geselowitz, A.; Greer, T.; ter Haar Romeny, B.; Zimmerman, J.B.; Zuiderveld, K. Adaptive histogram equalization and its variations. Comput. Vis. Graph. Image Process. 1987, 39, 355–368. [Google Scholar] [CrossRef]
- Asgar, A.W.; Ouzounian, M.; Adams, C.; Afilalo, J.; Fremes, S.; Lauck, S.; Leipsic, J.; Piazza, N.; Rodes-Cabau, J.; Welsh, R.; et al. CCS Tavi Quality Working Group Tavi Quality Report Team. 2019. Available online: https://ccs.ca/app/uploads/2022/04/CCS_2019_TAVI_Report_ENG.pdf (accessed on 30 August 2023).
- Delgado, V.; Ng, A.C.; Schuijf, J.D.; Van Der Kley, F.; Shanks, M.; Tops, L.F.; van de Veire, N.R.; De Roos, A.; Kroft, L.J.; Schalij, M.J.; et al. Automated assessment of the aortic root dimensions with multidetector row computed tomography. Annals of Thoracic Surgery 2011, 91, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Toubal, I.E.; Duan, Y.; Yang, D. Deep learning semantic segmentation for high-resolution medical volumes. In Proceedings of the 2020 IEEE Applied Imagery Pattern Recognition Workshop (AIPR), Washington, DC, USA, 13–15 October 2020; pp. 1–9. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Cao, J.; Tagliasacchi, A.; Olson, M.; Zhang, H.; Su, Z. Point cloud skeletons via laplacian based contraction. In Proceedings of the 2010 Shape Modeling International Conference, Aix en Provence, France, 21–23 June 2010; pp. 187–197. [Google Scholar] [CrossRef]
- Meyer, L.; Gilson, A.; Scholz, O.; Stamminger, M. CherryPicker: Semantic skeletonization and topological reconstruction of cherry trees. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 18–22 June 2023. [Google Scholar] [CrossRef]
- Zeng, A.; Wu, C.; Lin, G.; Xie, W.; Hong, J.; Huang, M.; Zhuang, J.; Bi, S.; Pan, D.; Ullah, N.; et al. ImageCAS: A Large-Scale Dataset and Benchmark for Coronary Artery Segmentation based on Computed Tomography Angiography Images. Comput. Med. Imaging Graph. 2023, 109, 102287. [Google Scholar] [CrossRef]
- Elattar, M.; Wiegerinck, E.; van Kesteren, F.; Dubois, L.; Planken, N.; Vanbavel, E.; Baan, J.; Marquering, H. Automatic aortic root landmark detection in CTA images for preprocedural planning of transcatheter aortic valve implantation. Int. J. Cardiovasc. Imaging 2016, 32, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Le Couteulx, S.; Caudron, J.; Dubourg, B.; Cauchois, G.; Dupré, M.; Michelin, P.; Durand, E.; Eltchaninoff, H.; Dacher, J.N. Multidetector computed tomography sizing of aortic annulus prior to transcatheter aortic valve replacement (TAVR): Variability and impact of observer experience. Diagn. Interv. Imaging 2018, 99, 279–289. [Google Scholar] [CrossRef]
- Paolisso, P.; Gallinoro, E.; Andreini, D.; Mileva, N.; Esposito, G.; Bermpeis, K.; Bertolone, D.T.; Munhoz, D.; Belmonte, M.; Fabbricatore, D.; et al. Prospective evaluation of the learning curve and diagnostic accuracy for Pre-TAVI cardiac computed tomography analysis by cardiologists in training: The LEARN-CT study. J. Cardiovasc. Comput. Tomogr. 2022, 16, 404–411. [Google Scholar] [CrossRef]




| Measurements | Expert 1 vs. Expert 2 n = 115 | TAVI-PREP vs. Expert 1 n = 200 | Saitta et al. [21] n = 178 | Astudillo et al. [22] n = 100 | Elattar et al. [32] n = 40 |
|---|---|---|---|---|---|
| Annulus area [mm2] | −8.08 [−49.30, 33.14] | −9.65 [−60.65, 41.36] | NA | NA | NA |
| Annulus perimeter [mm] | −0.83 [−4.56, 2.89] | −0.72 [−5.35, 3.90] | −1.8 [−8.06, 11.74] | NA | NA |
| Annulus area-derived diameter [mm] | −0.21 [−1.33, 0.91] | −0.24 [−1.56, 1.09] | 0.07 [−0.24, 0.38] * | NA | NA |
| Annulus perimeter-derived diameter [mm] | −0.27 [−1.44, 0.91] | −0.23 [−1.71, 1.25] | NA | NA | NA |
| Annulus diameter minimum [mm] | −0.17 [−1.70, 1.35] | −0.10 [−1.80, 1.59] | 0.89 [−2.8, 4.62] | NA | NA |
| Annulus diameter maximum [mm] | −0.24 [−2.06, 1.59] | 0.04 [−2.11, 2.20] | 0.51 [−2.79, 3.81] | NA | NA |
| Annulus diameter average [mm] | −0.20 [−1.54, 1.14] | −0.03 [−1.58, 1.52] | 0.52 [−2.96, 4.00] | NA | 0.48 [−2.26, 3.24] |
| SNTJ diameter average [mm] | 0.79 [−1.52, 3.09] | −0.33 [−1.98, 1.31] | 0.05 [−1.98, 2.07] | NA | NA |
| Left coronary height (LCH) [mm] | 0.45 [−2.28, 3.17] | −0.05 [−4.00, 3.89] | NA | 0.54 [−2.46, 3.54] | NA |
| Right coronary height (RCH) [mm] | 0.45 [−2.35, 4.40] | 2.82 [−2.06, 7.71] | NA | −0.16 [−4.09, 3.78] | NA |
| Measurements | Expert 1 vs. Expert 2 n = 115 | TAVI-PREP vs. Expert 1 n = 200 | Saitta et al. [21] n = 178 | Astudillo et al. [22] n = 100 | Elattar et al. [32] n = 40 |
|---|---|---|---|---|---|
| Annulus diameter average | 0.95 [0.93, 0.96] | 0.93 [0.91, 0.95] | NA | NA | 0.84 |
| Left coronary height (LCH) | 0.92 [0.89, 0.94] | 0.80 [0.74, 0.85] | NA | 0.80 | NA |
| Right coronary height (RCH) | 0.86 [0.82, 0.90] | 0.72 [0.64, 0.78] | NA | 0.80 | NA |
| Average LCH-RCH | 0.89 [0.85, 0.92] | 0.76 [0.69, 0.82] | NA | 0.80 | 0.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santaló-Corcoy, M.; Corbin, D.; Tastet, O.; Lesage, F.; Modine, T.; Asgar, A.; Ben Ali, W. TAVI-PREP: A Deep Learning-Based Tool for Automated Measurements Extraction in TAVI Planning. Diagnostics 2023, 13, 3181. https://doi.org/10.3390/diagnostics13203181
Santaló-Corcoy M, Corbin D, Tastet O, Lesage F, Modine T, Asgar A, Ben Ali W. TAVI-PREP: A Deep Learning-Based Tool for Automated Measurements Extraction in TAVI Planning. Diagnostics. 2023; 13(20):3181. https://doi.org/10.3390/diagnostics13203181
Chicago/Turabian StyleSantaló-Corcoy, Marcel, Denis Corbin, Olivier Tastet, Frédéric Lesage, Thomas Modine, Anita Asgar, and Walid Ben Ali. 2023. "TAVI-PREP: A Deep Learning-Based Tool for Automated Measurements Extraction in TAVI Planning" Diagnostics 13, no. 20: 3181. https://doi.org/10.3390/diagnostics13203181





