Attention TurkerNeXt: Investigations into Bipolar Disorder Detection Using OCT Images
Abstract
:1. Introduction
1.1. Literature Gaps
1.2. Motivation
1.3. Contributions and Novelties
2. Related Works
3. Materials and Methods
3.1. Dataset
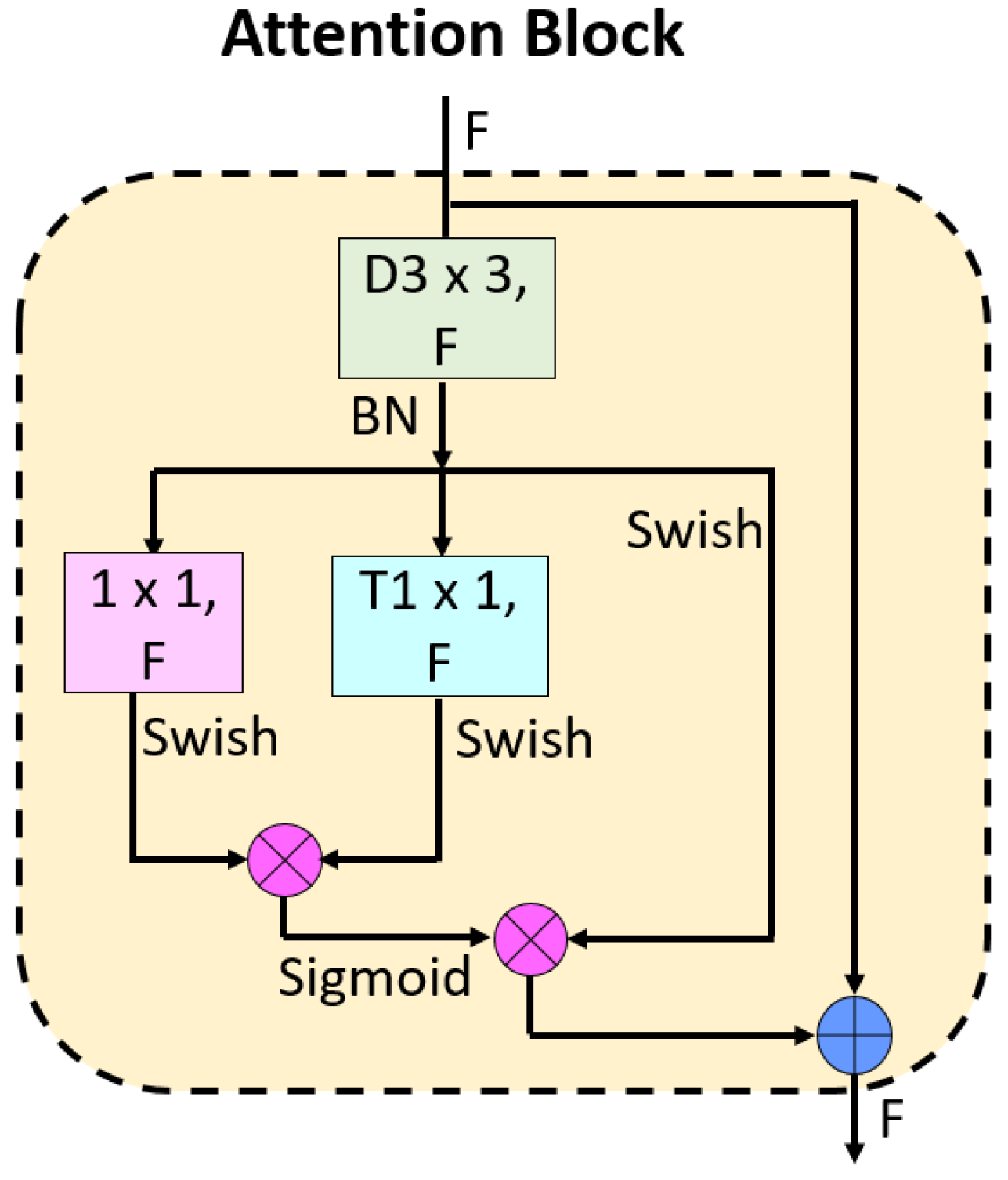
3.2. Attention TurkerNeXt
4. Experimental Results and Discussions
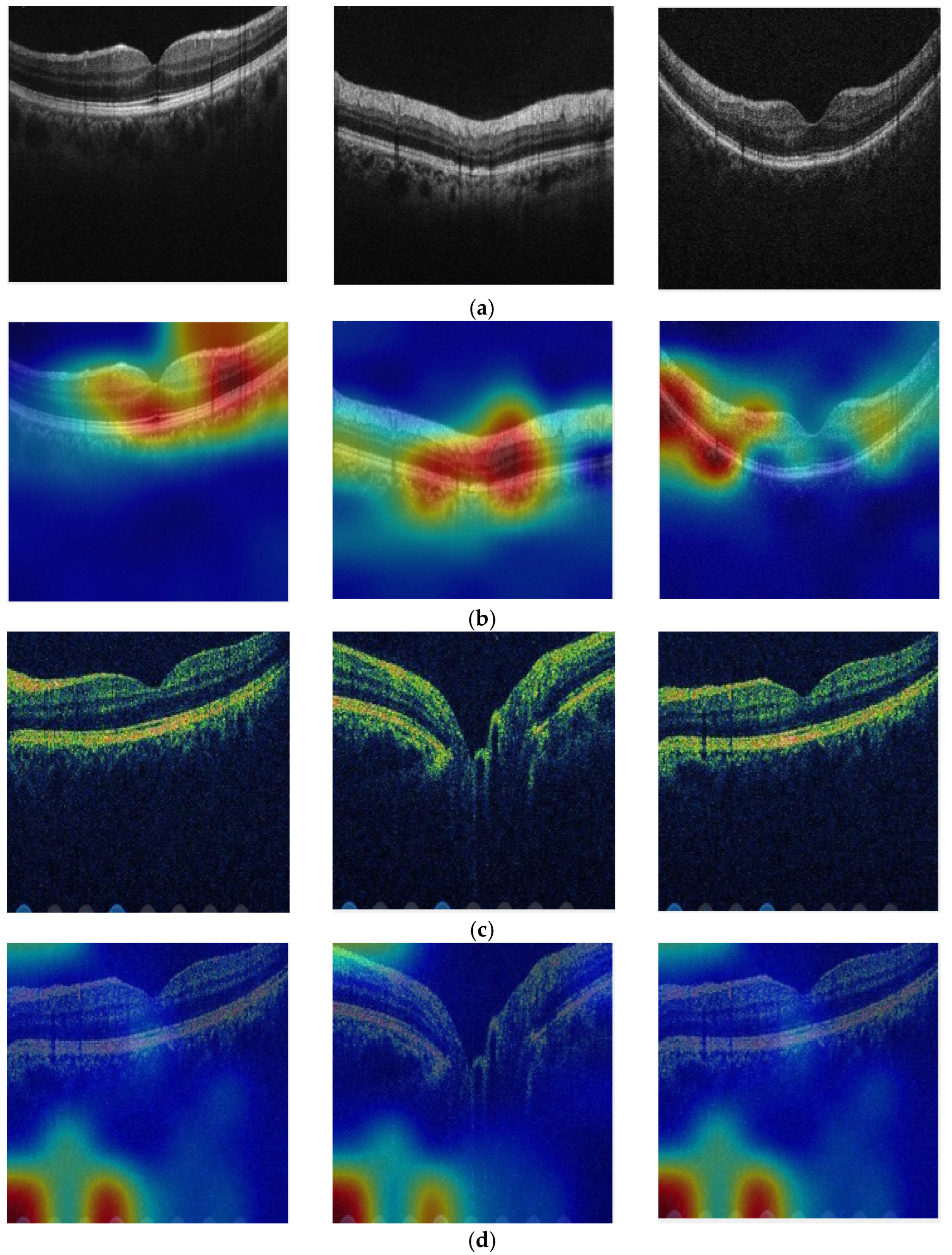
4.1. Explainable Results
4.2. Discussions
- -
- In this research, we have proposed a new deep learning algorithm and this Algorithm is named TurkerNeXt.
- -
- We have collected a new OCT image dataset to detect bipolar disorder.
- -
- We have shown the explainable results.
- -
- The proposed Attention TurkerNeXt model efficiently identified potential biomarkers through an OCT image dataset. We have listed the findings of the proposed Attention TurkerNeXt as below.
- ○
- Attention TurkerNeXt integrates components from Swin Transformers, ConvNeXt, MLP, and ResNet. This amalgamation is unique and tailored for the specific task of identifying biomarkers associated with bipolar disorder in OCT images. The thoughtful integration of these diverse components contributes to the model’s adaptability and effectiveness in capturing complex patterns.
- ○
- The proposed Attention TurkerNeXt block is a novel addition to the architecture. It combines an MLP structure with a shortcut, incorporating depth-wise convolution, point-wise convolution, and transposed convolution simultaneously. The utilization of attention and residual connections within this block enhances the model’s capacity to capture intricate features relevant to bipolar disorder. This block’s architecture is distinct from traditional CNN building blocks, making it a novel contribution to the field.
- ○
- Attention TurkerNeXt stands out by providing explainable results, a critical feature in medical applications. The attention mechanisms integrated into the model contribute to its interpretability, allowing clinicians and researchers to understand the basis for the model’s predictions. This emphasis on explainability is a novel and crucial aspect, especially in the context of medical image analysis.
- ○
- The use of the patchify approach in the Stem block to generate the first feature map is a novel strategy. This method, employing a 4 × 4-sized convolution, batch normalization, and swish activation functions, contributes to the model’s initial feature extraction and sets the stage for subsequent processing.
- ○
- The graphical output of Attention TurkerNeXt, along with the presented transition table, provides a comprehensive view of the model’s architecture. This transparency is crucial in understanding the flow of information through different layers, contributing to the novelty of the model’s design.
- ○
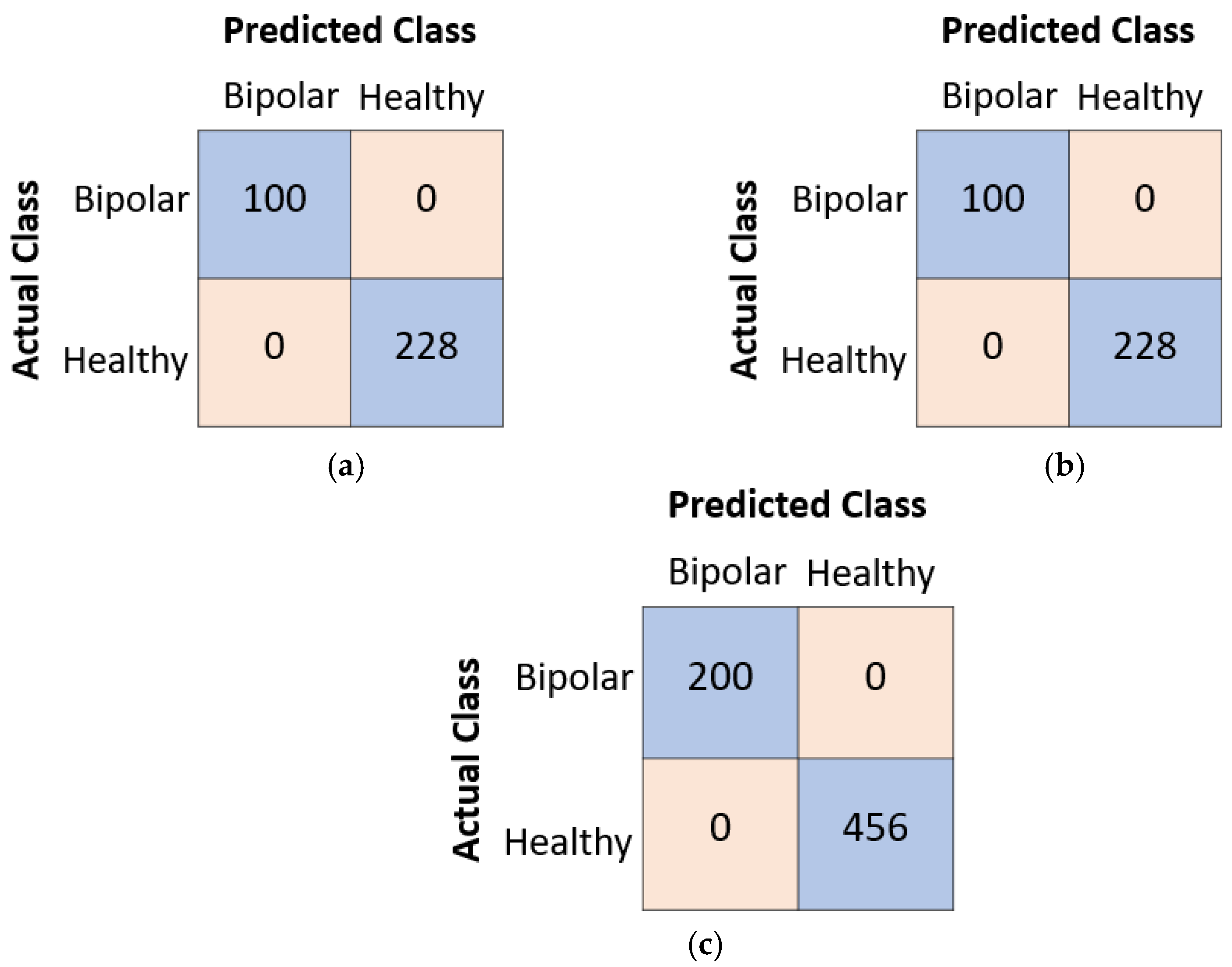
- Achieving a 100% classification accuracy on both the validation and test sets for bipolar disorder detection in OCT images is a remarkable and novel accomplishment. This level of accuracy is indicative of the model’s ability to discern subtle patterns and features associated with bipolar disorder, setting it apart from existing models.
- -
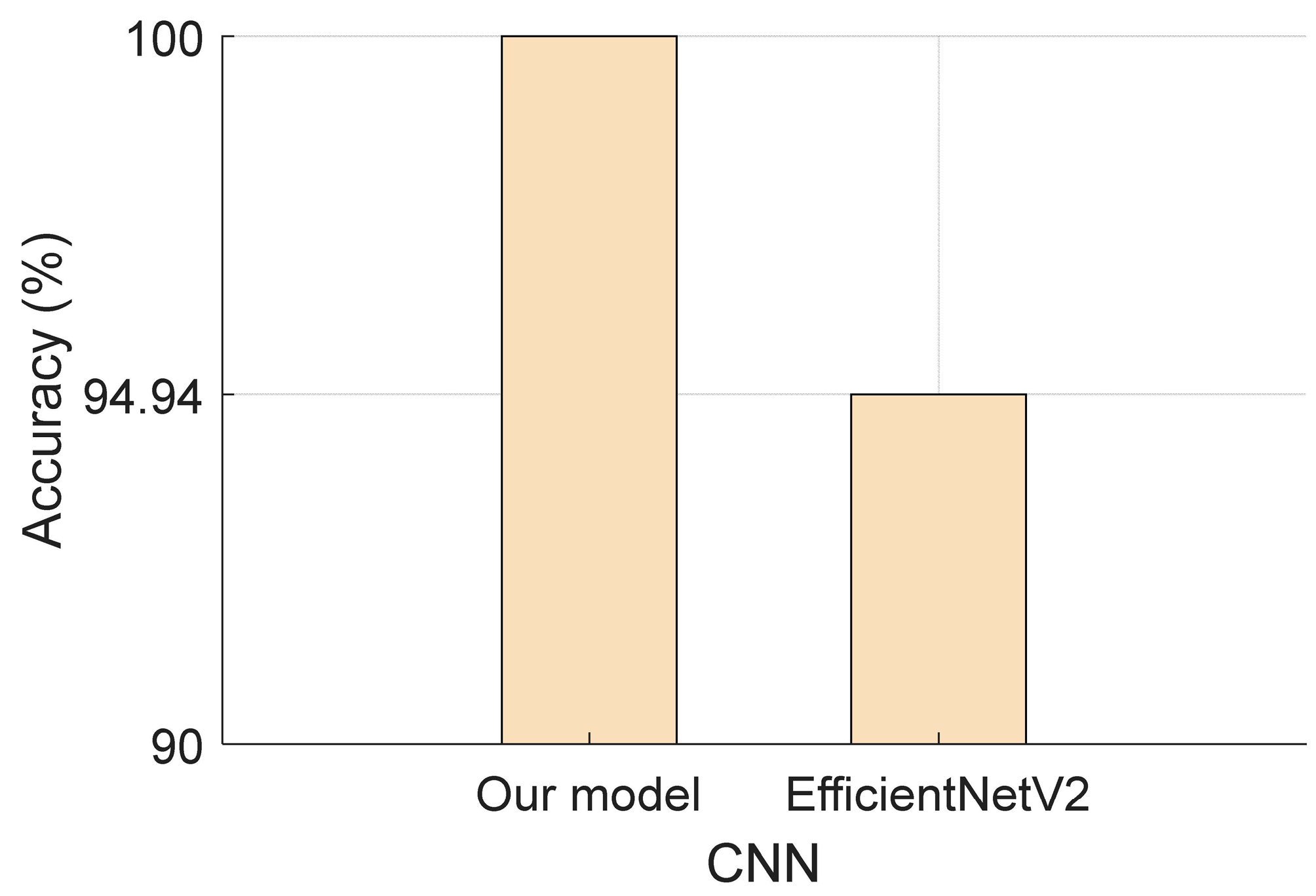
- A comparative analysis revealed that the Attention TurkerNeXt outperformed EfficientNetV2, achieving a validation accuracy of 94.94%.
- -
- The proposed model achieves perfect classification performance, indicating its reliability and robustness.
- -
- The model does not just provide outcomes; it gives explainable results, enabling better understanding and trustworthiness.
- -
- Compared to existing models like EfficientNetV2, Attention TurkerNeXt showcases superior classification capability, especially in the context of the collected dataset.
- -
- The model’s ability to identify new potential biomarkers can greatly enhance diagnostic methods in medical research.
- -
- The proposed CNN has only 1.6 million parameters. Therefore, the proposed Attention TurkerNeXt is a lightweight model.
- -
- Larger and more diverse OCT datasets can be gathered. OCT images from other macular degenerative disorders can be employed to identify patterns indicative of bipolar disorder.
- -
- Attention TurkerNeXt can be tested for other computer vision problems.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cotrena, C.; Branco, L.D.; Kochhann, R.; Shansis, F.M.; Fonseca, R.P. Quality of life, functioning and cognition in bipolar disorder and major depression: A latent profile analysis. Psychiatry Res. 2016, 241, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Uher, R. Gene–environment interactions in severe mental illness. Front. Psychiatry 2014, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Berk, M.; Berk, L.; Dodd, S.; Cotton, S.; Macneil, C.; Daglas, R.; Conus, P.; Bechdolf, A.; Moylan, S.; Malhi, G.S. Stage managing bipolar disorder. Bipolar Disord. 2014, 16, 471–477. [Google Scholar] [CrossRef]
- Berk, M.; Kapczinski, F.; Andreazza, A.C.; Dean, O.M.; Giorlando, F.; Maes, M.; Yücel, M.; Gama, C.S.; Dodd, S.; Dean, B. Pathways underlying neuroprogression in bipolar disorder: Focus on inflammation, oxidative stress and neurotrophic factors. Neurosci. Biobehav. Rev. 2011, 35, 804–817. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.; Maes, M. Bipolar disorder: Role of immune-inflammatory cytokines, oxidative and nitrosative stress and tryptophan catabolites. Curr. Psychiatry Rep. 2015, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Andreazza, A.C.; Young, L.T. The neurobiology of bipolar disorder: Identifying targets for specific agents and synergies for combination treatment. Int. J. Neuropsychopharmacol. 2014, 17, 1039–1052. [Google Scholar] [CrossRef]
- Duong, A.; Syed, B.; Scola, G. Biomarkers for bipolar disorder: Current insights. Curr. Biomark. Find. 2015, 2015, 79–92. [Google Scholar]
- Scola, G.; Andreazza, A.C. Current state of biomarkers in bipolar disorder. Curr. Psychiatry Rep. 2014, 16, 514. [Google Scholar] [CrossRef]
- Singh, I.; Rose, N. Biomarkers in psychiatry. Nature 2009, 460, 202–207. [Google Scholar] [CrossRef]
- Kempton, M.J.; Geddes, J.R.; Ettinger, U.; Williams, S.C.; Grasby, P.M. Meta-analysis, database, and meta-regression of 98 structural imaging studies in bipolar disorder. Arch. Gen. Psychiatry 2008, 65, 1017–1032. [Google Scholar] [CrossRef]
- Vita, A.; De Peri, L.; Sacchetti, E. Gray matter, white matter, brain, and intracranial volumes in first-episode bipolar disorder: A meta-analysis of magnetic resonance imaging studies. Bipolar Disord. 2009, 11, 807–814. [Google Scholar] [CrossRef]
- Moorhead, T.W.J.; McKirdy, J.; Sussmann, J.E.; Hall, J.; Lawrie, S.M.; Johnstone, E.C.; McIntosh, A.M. Progressive gray matter loss in patients with bipolar disorder. Biol. Psychiatry 2007, 62, 894–900. [Google Scholar] [CrossRef]
- Frey, B.N.; Zunta-Soares, G.B.; Caetano, S.C.; Nicoletti, M.A.; Hatch, J.P.; Brambilla, P.; Mallinger, A.G.; Soares, J.C. Illness duration and total brain gray matter in bipolar disorder: Evidence for neurodegeneration? Eur. Neuropsychopharmacol. 2008, 18, 717–722. [Google Scholar] [CrossRef]
- Papiol, S.; Molina, V.; Desco, M.; Rosa, A.; Reig, S.; Sanz, J.; Palomo, T.; Fananas, L. Gray matter deficits in bipolar disorder are associated with genetic variability at interleukin-1 beta gene (2q13). Genes Brain Behav. 2008, 7, 796–801. [Google Scholar] [CrossRef]
- Ladouceur, C.D.; Almeida, J.R.; Birmaher, B.; Axelson, D.A.; Nau, S.; Kalas, C.; Monk, K.; Kupfer, D.J.; Phillips, M.L. Subcortical gray matter volume abnormalities in healthy bipolar offspring: Potential neuroanatomical risk marker for bipolar disorder? J. Am. Acad. Child Adolesc. Psychiatry 2008, 47, 532–539. [Google Scholar] [CrossRef]
- Chu, E.M.-Y.; Kolappan, M.; Barnes, T.R.; Joyce, E.M.; Ron, M.A. A window into the brain: An in vivo study of the retina in schizophrenia using optical coherence tomography. Psychiatry Res. Neuroimaging 2012, 203, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Yeap, S.; Kelly, S.P.; Sehatpour, P.; Magno, E.; Garavan, H.; Thakore, J.H.; Foxe, J.J. Visual sensory processing deficits in Schizophrenia and their relationship to disease state. Eur. Arch. Psychiatry Clin. Neurosci. 2008, 258, 305–316. [Google Scholar] [CrossRef]
- Tasci, G.; Gun, M.V.; Keles, T.; Tasci, B.; Barua, P.D.; Tasci, I.; Dogan, S.; Baygin, M.; Palmer, E.E.; Tuncer, T. QLBP: Dynamic patterns-based feature extraction functions for automatic detection of mental health and cognitive conditions using EEG signals. Chaos Solitons Fractals 2023, 172, 113472. [Google Scholar] [CrossRef]
- Tasci, G.; Loh, H.W.; Barua, P.D.; Baygin, M.; Tasci, B.; Dogan, S.; Tuncer, T.; Palmer, E.E.; Tan, R.-S.; Acharya, U.R. Automated accurate detection of depression using twin Pascal’s triangles lattice pattern with EEG Signals. Knowl.-Based Syst. 2023, 260, 110190. [Google Scholar] [CrossRef]
- Şuheda, K.; TASCİ, B. Electroencephalogram-Based Major Depressive Disorder Classification Using Convolutional Neural Network and Transfer Learning. Turk. J. Sci. Technol. 2023, 18, 207–214. [Google Scholar]
- Tasci, B.; Tasci, G.; Dogan, S.; Tuncer, T. A novel ternary pattern-based automatic psychiatric disorders classification using ECG signals. Cogn. Neurodyn. 2022, 1–14. [Google Scholar] [CrossRef]
- Tatli, S.; Macin, G.; Tasci, I.; Tasci, B.; Barua, P.D.; Baygin, M.; Tuncer, T.; Dogan, S.; Ciaccio, E.J.; Acharya, U.R. Transfer-transfer model with MSNet: An automated accurate multiple sclerosis and myelitis detection system. Expert Syst. Appl. 2024, 236, 121314. [Google Scholar] [CrossRef]
- Tasci, B.; Tasci, G.; Ayyildiz, H.; Kamath, A.P.; Barua, P.D.; Tuncer, T.; Dogan, S.; Ciaccio, E.J.; Chakraborty, S.; Acharya, U.R. Automated schizophrenia detection model using blood sample scattergram images and local binary pattern. Multimed. Tools Appl. 2023, 1–29. [Google Scholar] [CrossRef]
- Tasci, I.; Tasci, B.; Barua, P.D.; Dogan, S.; Tuncer, T.; Palmer, E.E.; Fujita, H.; Acharya, U.R. Epilepsy detection in 121 patient populations using hypercube pattern from EEG signals. Inf. Fusion 2023, 96, 252–268. [Google Scholar] [CrossRef]
- Thomas, A.; Sunija, A.P.; Manoj, R.; Ramachandran, R.; Ramachandran, S.; Varun, P.G.; Palanisamy, P. RPE layer detection and baseline estimation using statistical methods and randomization for classification of AMD from retinal OCT. Comput. Methods Programs Biomed. 2021, 200, 105822. [Google Scholar] [CrossRef]
- Perdomo, O.; Rios, H.; Rodríguez, F.J.; Otálora, S.; Meriaudeau, F.; Müller, H.; González, F.A. Classification of diabetes-related retinal diseases using a deep learning approach in optical coherence tomography. Comput. Methods Programs Biomed. 2019, 178, 181–189. [Google Scholar] [CrossRef]
- Lee, C.S.; Baughman, D.M.; Lee, A.Y. Deep learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmol. Retin. 2017, 1, 322–327. [Google Scholar] [CrossRef]
- Perdomo, O.; Otálora, S.; González, F.A.; Meriaudeau, F.; Müller, H. Oct-net: A convolutional network for automatic classification of normal and diabetic macular edema using sd-oct volumes. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 1423–1426. [Google Scholar]
- Zhang, Q.; Liu, Z.; Li, J.; Liu, G. Identifying diabetic macular edema and other retinal diseases by optical coherence tomography image and multiscale deep learning. Diabetes Metab. Syndr. Obes. 2020, 13, 4787–4800. [Google Scholar] [CrossRef]
- Abdullahi, M.M.; Chakraborty, S.; Kaushik, P.; Sami, B.S. Detection of dry and wet age-related macular degeneration using deep learning. In Proceedings of the 2nd International Conference on Industry 4.0 and Artificial Intelligence (ICIAI 2021), Virtual, 5–8 March 2022; pp. 211–214. [Google Scholar]
- Saleh, N.; Abdel Wahed, M.; Salaheldin, A.M. Transfer learning-based platform for detecting multi-classification retinal disorders using optical coherence tomography images. Int. J. Imaging Syst. Technol. 2022, 32, 740–752. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Zheng, Q.; Liu, J.; Ji, Y.; Zhang, Y.; Chen, X.; Liu, B. Elevated levels of monocyte-lymphocyte ratio and platelet-lymphocyte ratio in adolescents with non-suicidal self-injury. BMC Psychiatry 2022, 22, 618. [Google Scholar] [CrossRef]
- Tolstikhin, I.; Houlsby, N.; Kolesnikov, A.; Beyer, L.; Zhai, X.; Unterthiner, T.; Yung, J.; Keysers, D.; Uszkoreit, J.; Lucic, M. MLP-Mixer: An all-MLP Architecture for Vision. arXiv 2021, arXiv:2105.01601. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Kamran, S.A.; Tavakkoli, A.; Zuckerbrod, S.L. Improving robustness using joint attention network for detecting retinal degeneration from optical coherence tomography images. In Proceedings of the 2020 IEEE International Conference On Image Processing (ICIP), Abu Dhabi, United Arab Emirates, 25–28 October 2020; pp. 2476–2480. [Google Scholar]
- Saraiva, A.A.; Santos, D.B.S.; Pimentel, P.M.C.; Sousa, J.V.M.; Ferreira, N.; Batista Neto, J.E.S.; Soares, S.; Valente, A. Classification of optical coherence tomography using convolutional neural networks. In Proceedings of the BIOSTEC 2020: 13th International Joint Conference on Biomedical Engineering Systems and Technologies, Valletta, Malta, 24–26 February 2020. [Google Scholar]
- Khan, A.; Pin, K.; Aziz, A.; Han, J.W.; Nam, Y. Optical coherence tomography image classification using hybrid deep learning and ant colony optimization. Sensors 2023, 23, 6706. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Wang, J.; Han, Z.; Ma, J.; Wang, C.; Qi, M. An interpretable transformer network for the retinal disease classification using optical coherence tomography. Sci. Rep. 2023, 13, 3637. [Google Scholar] [CrossRef]
- Fang, L.; Wang, C.; Li, S.; Rabbani, H.; Chen, X.; Liu, Z. Attention to lesion: Lesion-aware convolutional neural network for retinal optical coherence tomography image classification. IEEE Trans. Med. Imaging 2019, 38, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Xie, Q.; Xie, P.; Fan, F.; Gao, X.; Zhu, J. HCTNet: A Hybrid ConvNet-Transformer Network for Retinal Optical Coherence Tomography Image Classification. Biosensors 2022, 12, 542. [Google Scholar] [CrossRef]
- Barua, P.D.; Chan, W.Y.; Dogan, S.; Baygin, M.; Tuncer, T.; Ciaccio, E.J.; Islam, N.; Cheong, K.H.; Shahid, Z.S.; Acharya, U.R. Multilevel deep feature generation framework for automated detection of retinal abnormalities using OCT images. Entropy 2021, 23, 1651. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Le, Q. Efficientnetv2: Smaller models and faster training. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 10096–10106. [Google Scholar]



| Diagnosis | Bipolar Disorder | Healthy Control | ||
|---|---|---|---|---|
| Sex | 10 female | 10 male | 15 female | 15 male |
| Mean age, years | 36.5 ± 4.25 | 40.4 ± 8.8 | 28.7 ± 5.36 | 30.4 ± 3.55 |
| Age range, years | 25–59 | 18–62 | 23–48 | 27–56 |
| Beck Depression Inventory (BDI) | 4.37 ± 3.15 | 4.66 ± 2.87 | - | - |
| Young Mania Rating Scale (YMRS) | 2 ± 1.51 | 2.88 ± 4.62 | - | - |
| From Left to Right | From Top to Bottom | |||||
|---|---|---|---|---|---|---|
| Train Images | Test Images | Total | Train Images | Test Images | Total | |
| Bipolar Disorder | 303 | 100 | 403 | 303 | 100 | 403 |
| Healthy control | 684 | 228 | 912 | 684 | 228 | 912 |
| Layer | Input Size | Operation | Output Size |
|---|---|---|---|
| Stem | 224 × 224 | 4 × 4, 96, stride: 4 | 56 × 56 |
| Layer 1 | 56 × 56 | 28 × 28 | |
| Layer 2 | 28 × 28 | 14 × 14 | |
| Layer 3 | 14 × 14 | 7 × 7 | |
| Layer 4 | 7 × 7 | 7 × 7 | |
| Output size | 7 × 7 | Global average pooling, fully connected layer, softmax | Number of classes |
| Total learnable parameters | ~1.6 million | ||
| Performance Evaluation Metrics | Case | ||
|---|---|---|---|
| Bottom to Top | Left to Right | Merged | |
| Accuracy | 100% | 100% | 100% |
| Sensitivity | 100% | 100% | 100% |
| Specificity | 100% | 100% | 100% |
| Precision | 100% | 100% | 100% |
| F1-score | 100% | 100% | 100% |
| Geometric mean | 100% | 100% | 100% |
| Study | Model | Dataset | Results (%) |
|---|---|---|---|
| [36] | Joint-Attention Network MobileNet-v2 | OCT2017 500 training images 500 testing images | Accuracy: 95.60 Specificity: 97.10 Sensitivity: 95.60 |
| Joint-Attention Network ResNet50-v1 | Srinivasan2014 2916 training images 315 testing images | Accuracy: 100.0 Specificity: 100.0 Sensitivity: 100.0 | |
| [37] | CNN | 16,896 images 100:1 | Accuracy: 94.35 |
| [38] | Transfer learning, Ant colony optimization | 2397 training images 601 testing images | Accuracy: 99.10 |
| [39] | Swin-Poly Transformer network | OCT-C8 25,600 training images 2800 validation images 2800 testing images | Accuracy: 97.12 Precision: 97.13 Recall: 97.13 F1-Score: 97.10 |
| [40] | Lesion-aware convolution neural network | 2000 images 10-fold CV | Accuracy: 90.10 Sensitivity: 86.80 Precision: 86.20 |
| [41] | Hybrid ConvNet–Transformer Network | Srinivasan2014 3231 images 60:20:20 | Accuracy: 86.18 Sensitivity:85.40 Precision: 88.53 |
| OCT2017 84.484 images 60:20:20 | Accuracy: 91.56 Sensitivity:88.57 Precision: 88.11 | ||
| [42] | CNN, iterative ReliefF | Srinivasan2014 3194 images 10-fold CV | Accuracy: 100.0 Precision: 100.0 F1-score: 100.0 |
| OCT image dataset 11,000 images 10:1 | Accuracy: 97.30 Precision: 97.32 F1-score: 97.30 | ||
| Proposed Model | Attention TurkerNeXt | Collected Dataset 2630 images 60:15:25 | From left to right Accuracy: 100.0 Sensitivity: 100.0 Specificity: 100.0 From top to bottom Accuracy: 100.0 Sensitivity: 100.0 Specificity: 100.0 Merged Accuracy: 100.0 Sensitivity: 100.0 Specificity: 100.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arslan, S.; Kaya, M.K.; Tasci, B.; Kaya, S.; Tasci, G.; Ozsoy, F.; Dogan, S.; Tuncer, T. Attention TurkerNeXt: Investigations into Bipolar Disorder Detection Using OCT Images. Diagnostics 2023, 13, 3422. https://doi.org/10.3390/diagnostics13223422
Arslan S, Kaya MK, Tasci B, Kaya S, Tasci G, Ozsoy F, Dogan S, Tuncer T. Attention TurkerNeXt: Investigations into Bipolar Disorder Detection Using OCT Images. Diagnostics. 2023; 13(22):3422. https://doi.org/10.3390/diagnostics13223422
Chicago/Turabian StyleArslan, Sermal, Mehmet Kaan Kaya, Burak Tasci, Suheda Kaya, Gulay Tasci, Filiz Ozsoy, Sengul Dogan, and Turker Tuncer. 2023. "Attention TurkerNeXt: Investigations into Bipolar Disorder Detection Using OCT Images" Diagnostics 13, no. 22: 3422. https://doi.org/10.3390/diagnostics13223422
APA StyleArslan, S., Kaya, M. K., Tasci, B., Kaya, S., Tasci, G., Ozsoy, F., Dogan, S., & Tuncer, T. (2023). Attention TurkerNeXt: Investigations into Bipolar Disorder Detection Using OCT Images. Diagnostics, 13(22), 3422. https://doi.org/10.3390/diagnostics13223422










