Feasibility of Optical Genome Mapping from Placental and Umbilical Cord Sampled after Spontaneous or Therapeutic Pregnancy Termination
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, Transport, and Storage
2.2. Sample Preparation, UHMW DNA Isolation, and Labelling and Chip Loading
2.3. OGM Metrics Data and Data Analysis
2.4. De Novo Assembly, Structural Variant Calling and Data Analysis
2.5. Pulsed-Field Capillary Electrophoresis
3. Results
3.1. High-Molecular-Weight (HMW) DNA Extraction
3.1.1. Umbilical Cord Biopsies
3.1.2. Chorionic/Placental Villi Biopsies
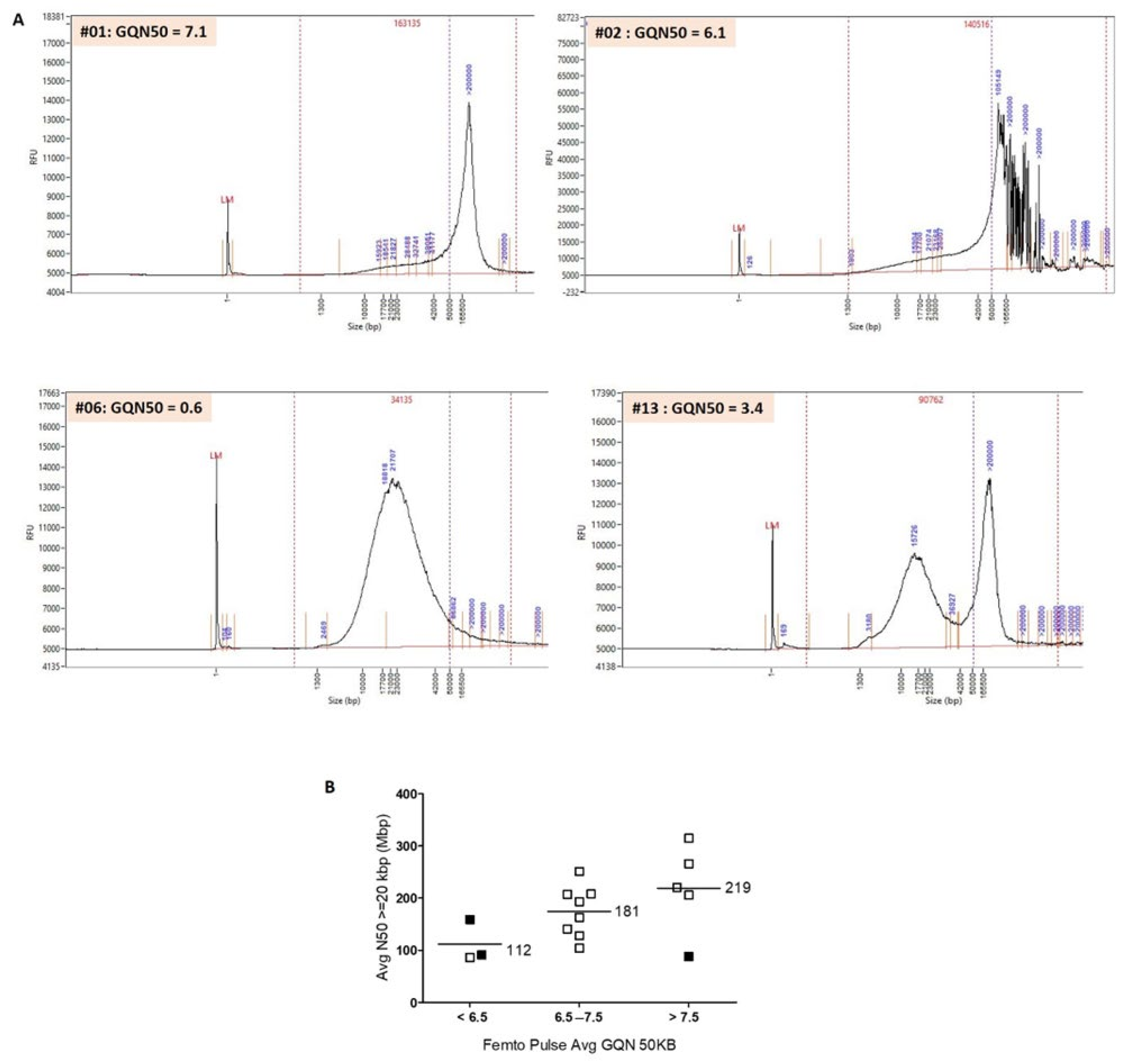
3.2. Evaluation of the Quality of HMW DNA with the Femto Pulse System
3.3. OGM Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dolanc Merc, M.; Peterlin, B.; Lovrecic, L. The Genetic Approach to Stillbirth: A »systematic Review«. Prenat. Diagn. 2023, 43, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Ernst, L.M.; Rand, C.M.; Bao, R.; Andrade, J.; Linn, R.L.; Minturn, L.; Zhang, C.; Kang, W.; Weese-Mayer, D.E. Stillbirth: Genome-Wide Copy Number Variation Profiling in Archived Placental Umbilical Cord Samples with Pathologic and Clinical Correlation. Placenta 2015, 36, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Reddy, U.M.; Page, G.P.; Saade, G.R.; Silver, R.M.; Thorsten, V.R.; Parker, C.B.; Pinar, H.; Willinger, M.; Stoll, B.J.; Heim-Hall, J.; et al. Karyotype versus Microarray Testing for Genetic Abnormalities after Stillbirth. N. Engl. J. Med. 2012, 367, 2185–2193. [Google Scholar] [CrossRef] [PubMed]
- Dalton, S.E.; Workalemahu, T.; Allshouse, A.A.; Page, J.M.; Reddy, U.M.; Saade, G.R.; Pinar, H.; Goldenberg, R.L.; Dudley, D.J.; Silver, R.M. Copy Number Variants and Fetal Growth in Stillbirths. Am. J. Obstet. Gynecol. 2023, 228, 579.e1–579.e11. [Google Scholar] [CrossRef] [PubMed]
- Mantere, T.; Neveling, K.; Pebrel-Richard, C.; Benoist, M.; van der Zande, G.; Kater-Baats, E.; Baatout, I.; van Beek, R.; Yammine, T.; Oorsprong, M.; et al. Optical Genome Mapping Enables Constitutional Chromosomal Aberration Detection. Am. J. Hum. Genet. 2021, 108, 1409–1422. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.A.; Broeckel, U.; Levy, B.; Skinner, S.; Sahajpal, N.S.; Rodriguez, V.; Stence, A.; Awayda, K.; Scharer, G.; Skinner, C.; et al. Multisite Assessment of Optical Genome Mapping for Analysis of Structural Variants in Constitutional Postnatal Cases. J. Mol. Diagn. 2023, 25, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Levy, B.; Baughn, L.B.; Akkari, Y.; Chartrand, S.; LaBarge, B.; Claxton, D.; Lennon, P.A.; Cujar, C.; Kolhe, R.; Kroeger, K.; et al. Optical Genome Mapping in Acute Myeloid Leukemia: A Multicenter Evaluation. Blood Adv. 2023, 7, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Sahajpal, N.S.; Mondal, A.K.; Fee, T.; Hilton, B.; Layman, L.; Hastie, A.R.; Chaubey, A.; DuPont, B.R.; Kolhe, R. Clinical Validation and Diagnostic Utility of Optical Genome Mapping in Prenatal Diagnostic Testing. J. Mol. Diagn. 2023, 25, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Goumy, C.; Guy Ouedraogo, Z.; Soler, G.; Eymard-Pierre, E.; Laurichesse, H.; Delabaere, A.; Gallot, D.; Bouchet, P.; Perthus, I.; Pebrel-Richard, C.; et al. Optical Genome Mapping for Prenatal Diagnosis: A Prospective Study. Clin. Chim. Acta 2023, 551, 117594. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.; Vestergaard, E.M.; Graakjaer, J.; Petkov, Y.; Bache, I.; Fagerberg, C.; Kibæk, M.; Svaneby, D.; Petersen, O.B.; Brasch-Andersen, C.; et al. 17q12 Deletion and Duplication Syndrome in Denmark—A Clinical Cohort of 38 Patients and Review of the Literature. Am. J. Med. Genet. Part A 2016, 170, 2934–2942. [Google Scholar] [CrossRef] [PubMed]
- Sahajpal, N.S.; Mondal, A.K.; Ananth, S.; Pundkar, C.; Jones, K.; Williams, C.; Fee, T.; Weissman, A.; Tripodi, G.; Oza, E.; et al. Optical Genome Mapping and Single Nucleotide Polymorphism Microarray: An Integrated Approach for Investigating Products of Conception. Genes 2022, 13, 643. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Chai, H.; Zhou, Q.; Wen, J.; Reddy, U.M.; Kastury, R.; Jiang, Y.; Mak, W.; Bale, A.E.; Zhang, H.; et al. Exome Sequencing Analysis on Products of Conception: A Cohort Study to Evaluate Clinical Utility and Genetic Etiology for Pregnancy Loss. Genet. Med. 2021, 23, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, M.; Smits, G.; Ceysens, G.; Costa, E.; Coulon, R.; Daelemans, C.; De Coninck, C.; Derisbourg, S.; Gajewska, K.; Garofalo, G.; et al. Implementation of Fetal Clinical Exome Sequencing: Comparing Prospective and Retrospective Cohorts. Genet. Med. 2022, 24, 344–363. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, S.E.; Rodenburg, K.; Corominas, J.; Ben-Yosef, T.; Reurink, J.; Kremer, H.; Whelan, L.; Plomp, A.S.; Berger, W.; Farrar, G.J.; et al. Optical Genome Mapping and Revisiting Short-Read Genome Sequencing Data Reveal Previously Overlooked Structural Variants Disrupting Retinal Disease-Associated Genes. Genet. Med. 2022, 25, 100345. [Google Scholar] [CrossRef] [PubMed]

| Samples Characteristics | Saphyr Quality Control Metrics and Results | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Type | Optimal Size/Weight | Pregnancy Outcome | Storage Time and Temperature before Freezing | HMW DNA Concentration Mean | Coefficient of Variation % | Avg N50 ≥ 150 kbp (Mbp) | Avg N50 ≥ 20 kbp (Mbp) | Avg Label Density (per 100 kbp) | Avg Map Rate % | Effective Coverage | Result |
| #01 | UC | ~500 mg L = 1 cm ø = 5 mm | TA | 18 h; RT | 114.9 | 44 | 339 | 251 | 18.62 | 68.2 | 134X | Pass |
| #02 | TA | 20 h; RT | 69.8 | 7 | 239 | 159 | 12.35 | 53.6 | 90X | Fail | ||
| #03 | TA | 13 h; RT | 36.4 | 29 | 366 | 213 | 17.75 | 68.2 | 115X | Pass | ||
| #04 | IUFD | 120 h; RT | 91.3 | 52 | 247 | 76 | 12.85 | 9.6 | 6X | Fail | ||
| #05 | TA | 20 h; RT | 78.8 | 15 | 198 | 100 | 10 | 17.9 | 36X | Fail | ||
| #06 | TA | 50 h; RT | 61.2 | 18 | 210 | 91 | 7.54 | 0.4 | 0X | Fail | ||
| #07 | TA | 16 h; RT | 50.6 | 23 | 196 | 88 | 13.64 | 41.4 | 23X | Fail | ||
| #08 | IUFD | 4 h; RT | 58.2 | 4 | 278 | 243 | 15.04 | 93.5 | 156X | Pass | ||
| #09 | SAB | 13 h; RT | 80.8 | 3 | 271 | 206 | 14.46 | 90.6 | 149X | Pass | ||
| #10 | CV | 10–20 mg | TA | 2 h 30; RT | 70.1 | 8 | 207 | 104 | 14.77 | 76.6 | 130X | Pass |
| #11 | TA | 72 h; 4 °C | 172 | 5 | 243 | 128 | 15.08 | 89.4 | 147X | Pass | ||
| #12 | IUFD | 1 h; RT | 145 | 6 | 289 | 220 | 13.96 | 86.7 | 170X | Pass | ||
| #13 | TA | 22 h; RT | 89.7 | 25 | 227 | 86 | 14.6 | 71.1 | 139X | Pass | ||
| #14 | TA | 4 h; RT | 59 | 3 | 299 | 207 | 12.47 | 74.6 | 121X | Pass | ||
| #15 | TA | 1 h; RT | 115.3 | 9 | 358 | 315 | 14.35 | 89 | 150X | Pass | ||
| Sample Description | Saphyr Results | Femto Pulse Results | |||||
|---|---|---|---|---|---|---|---|
| ID | Type | Avg N50 ≥ 150 kbp (Mbp) | Avg N50 ≥ 20 kbp (Mbp) | Avg Label Density (per 100 kbp) | Issue | Avg GQN50 kb | Smear Size (kb) |
| #01 | UC | 339 | 251 | 18.62 | Pass | 7.1 | 163 |
| #02 | UC | 239 | 159 | 12.35 | Fail | 6.1 | 140 |
| #05 | UC | 198 | 100 | 10 | Fail | 8.2 | 201 |
| #06 | UC | 210 | 91 | 7.54 | Fail | 0.6 | 34 |
| #07 | UC | 196 | 88 | 13.64 | Fail | 8.3 | 162 |
| #09 | UC | 271 | 206 | 14.46 | Pass | 7.9 | 147 |
| #10 | CV | 207 | 104 | 14.77 | Pass | 7.1 | 146 |
| #11 | CV | 243 | 128 | 15.08 | Pass | 6.5 | 182 |
| #12 | CV | 289 | 220 | 13.96 | Pass | 7.9 | 175 |
| #13 | CV | 227 | 86 | 14.6 | Pass | 3.4 | 91 |
| #14 | CV | 299 | 207 | 12.47 | Pass | 6.9 | 156 |
| #15 | CV | 358 | 315 | 14.35 | Pass | 8.7 | 165 |
| Cell culture control | 301 | 208 | 13.32 | Pass | 7.2 | 162 | |
| Cell culture control | 240 | 141 | 14.78 | Pass | 7.5 | 157 | |
| Cell culture control | 228 | 163 | 15.4 | Pass | 7.2 | 168 | |
| Cell culture control | 255 | 193 | 13.21 | Pass | 7.5 | 223 | |
| Cell culture control | 305 | 266 | 15.27 | Pass | 7.9 | 180 | |
| Sample Description | OGM Results | Results of Standard Cytogenetic Techniques | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Fetal Phenotype | Gestational Age (WG) | OMG Aneuploidy and Polyploidy | OGM CNVs | Number of SV | Number of OMIM Genes | Number of OMIM Morbid Genes | Karyotype | CMA |
| #12 | IUFD, severe intrauterine growth retardation | 22 + 4 | Trisomy 16 | Gain chr17:34764405-36276597 | 19 | 7 | 2 | 47,XX,+16 | arr[GRCh37] (16)x3,17q12(34817422_36168104)x3 mat |
| #13 | Polymalformative syndrome | 17 + 4 | Triploidy | No | 34 | 11 | 2 | 69,XXX | ND |
| #14 | Cystic hygroma | 13 + 5 | Monosomy X | No | 29 | 15 | 6 | 45,X | ND |
| #01 | Clastic cerebral vascular lesion with voluminous parenchymal destruction, low hair implantation | 30 | No | No | 64 | 27 | 8 | ND | arr(X,Y)x1,(1-22)x2 |
| #03 | Aortic atresia with major hypoplasia of the left heart, hypertelorism, sacral dimple, short corpus callosum | 24 + 6 | No | No | 29 | 8 | 3 | ND | arr(X,1-22)x2 |
| #08 | IUFD, cleft lip | 25 + 5 | No | No | 34 | 11 | 3 | ND | arr(X,1-22)x2 |
| #09 | SAB (recurrence), cystic hygroma, facial dysmorphism | 18 + 5 | No | No | 53 | 23 | 7 | ND | arr(X,1-22)x2 |
| #10 | Facial meningocele | 14 + 4 | No | No | 33 | 17 | 5 | ND | arr(X,1-22)x2 |
| #11 | Acrania, history of spina bifida | 13 + 5 | No | No | 20 | 7 | 2 | ND | arr(X,1-22)x2 |
| #15 | Cardiac chamber imbalance, omphalocele, holoprosencephaly, radial agenesis | 34 + 5 | No | No | 36 | 18 | 4 | ND | arr(X,Y)x1,(1-22)x2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goumy, C.; Ouedraogo, Z.G.; Bellemonte, E.; Eymard-Pierre, E.; Soler, G.; Perthus, I.; Pebrel-Richard, C.; Gouas, L.; Salaun, G.; Véronèse, L.; et al. Feasibility of Optical Genome Mapping from Placental and Umbilical Cord Sampled after Spontaneous or Therapeutic Pregnancy Termination. Diagnostics 2023, 13, 3576. https://doi.org/10.3390/diagnostics13233576
Goumy C, Ouedraogo ZG, Bellemonte E, Eymard-Pierre E, Soler G, Perthus I, Pebrel-Richard C, Gouas L, Salaun G, Véronèse L, et al. Feasibility of Optical Genome Mapping from Placental and Umbilical Cord Sampled after Spontaneous or Therapeutic Pregnancy Termination. Diagnostics. 2023; 13(23):3576. https://doi.org/10.3390/diagnostics13233576
Chicago/Turabian StyleGoumy, Carole, Zangbéwendé Guy Ouedraogo, Elodie Bellemonte, Eleonore Eymard-Pierre, Gwendoline Soler, Isabelle Perthus, Céline Pebrel-Richard, Laetitia Gouas, Gaëlle Salaun, Lauren Véronèse, and et al. 2023. "Feasibility of Optical Genome Mapping from Placental and Umbilical Cord Sampled after Spontaneous or Therapeutic Pregnancy Termination" Diagnostics 13, no. 23: 3576. https://doi.org/10.3390/diagnostics13233576
APA StyleGoumy, C., Ouedraogo, Z. G., Bellemonte, E., Eymard-Pierre, E., Soler, G., Perthus, I., Pebrel-Richard, C., Gouas, L., Salaun, G., Véronèse, L., Laurichesse, H., Darcha, C., & Tchirkov, A. (2023). Feasibility of Optical Genome Mapping from Placental and Umbilical Cord Sampled after Spontaneous or Therapeutic Pregnancy Termination. Diagnostics, 13(23), 3576. https://doi.org/10.3390/diagnostics13233576






