Development of an Artificial Intelligence-Based Breast Cancer Detection Model by Combining Mammograms and Medical Health Records
Abstract
1. Introduction
1.1. Related Work
1.2. Novelty and Contribution
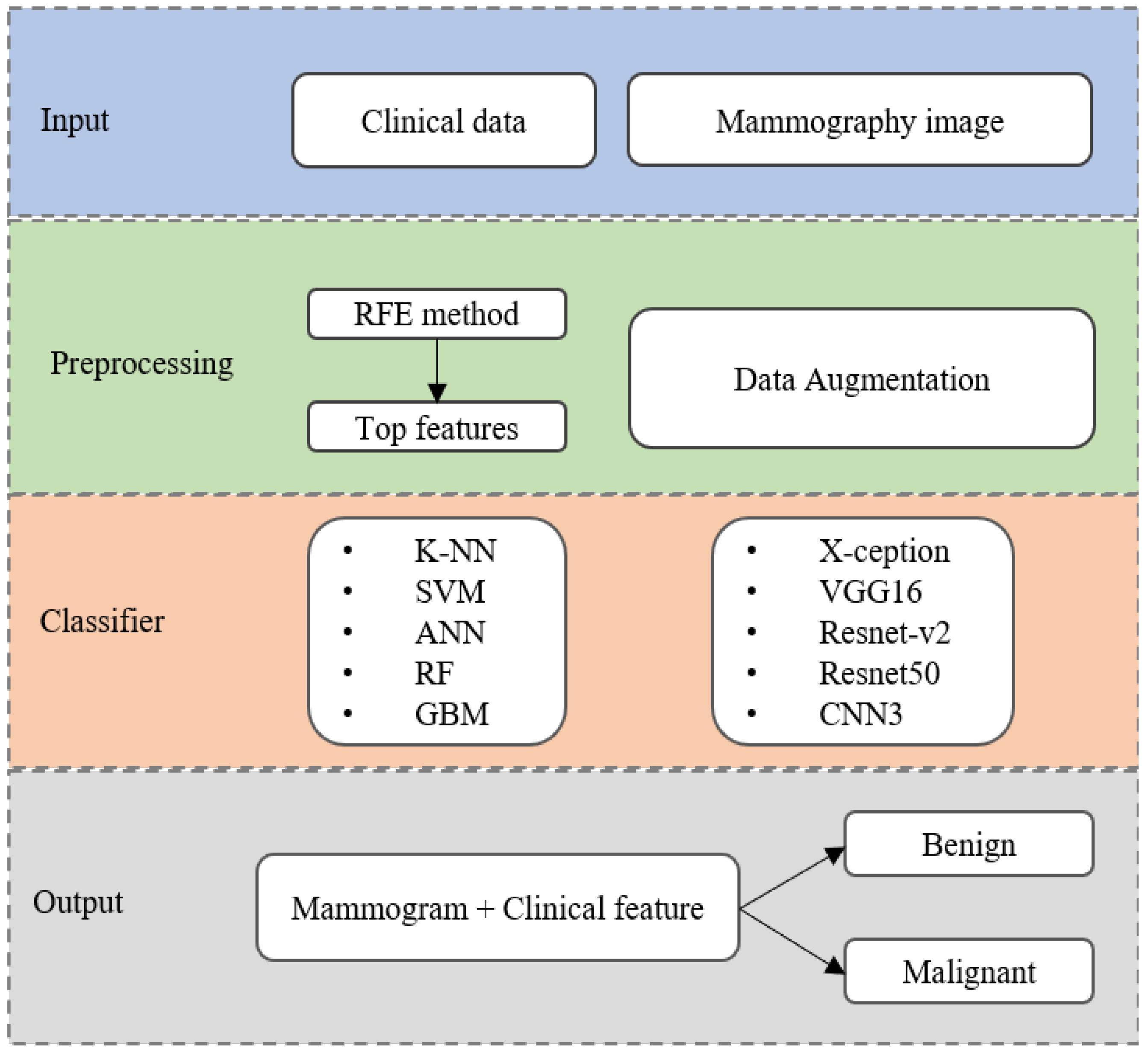
- We have proposed an AI framework based on ML-DL approaches which include various algorithms for each data type. Moreover, this study attempts to use mammography images combined with clinical variables as input for breast cancer detection;
- Multiple deep learning models to detect breast cancer in mammograms, including X-ception, VGG16, ResNet-v2, ResNet50, and CNN3 were employed. An augmentation technique was utilized for creating more training samples to avoid overfitting;
- To determine the most common clinical features related to cancer capability and select an appropriate ML model based on levels of model complexity to achieve high accuracy and expedite the learning process;
- We have developed an effective model combination for breast cancer detection based on the mammogram and clinical features to comprehensively assess at the individual patient level.
2. Materials and Methods
2.1. Study Design and Data Preparation
2.2. Breast Cancer Detection Algorithm
2.3. Deep-Learning Classifiers
2.3.1. Data Augmentation
2.3.2. Deep Neural Networks
2.4. Machine-Learning Classifiers
2.4.1. Data Preprocessing
2.4.2. Model Description
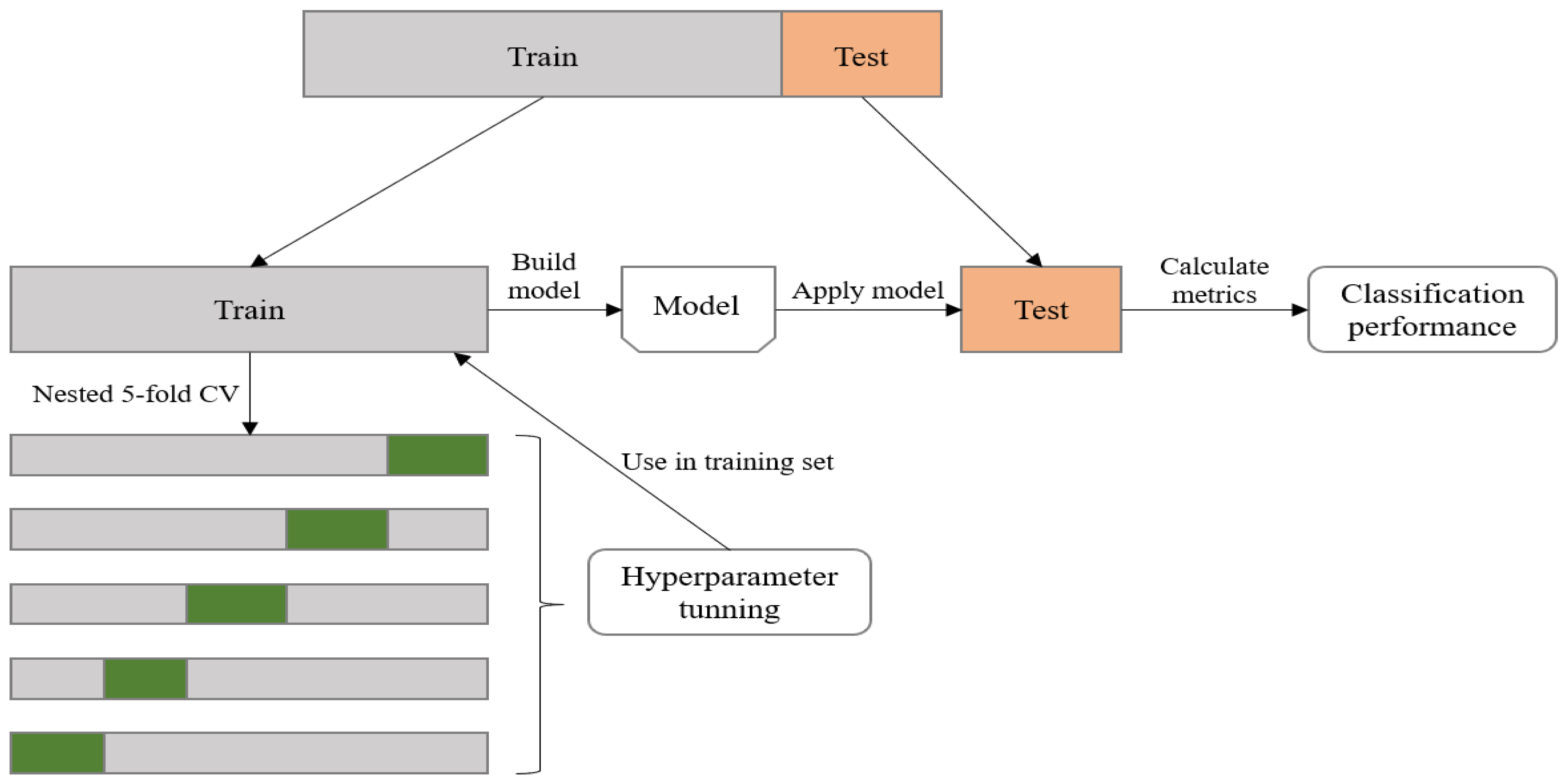
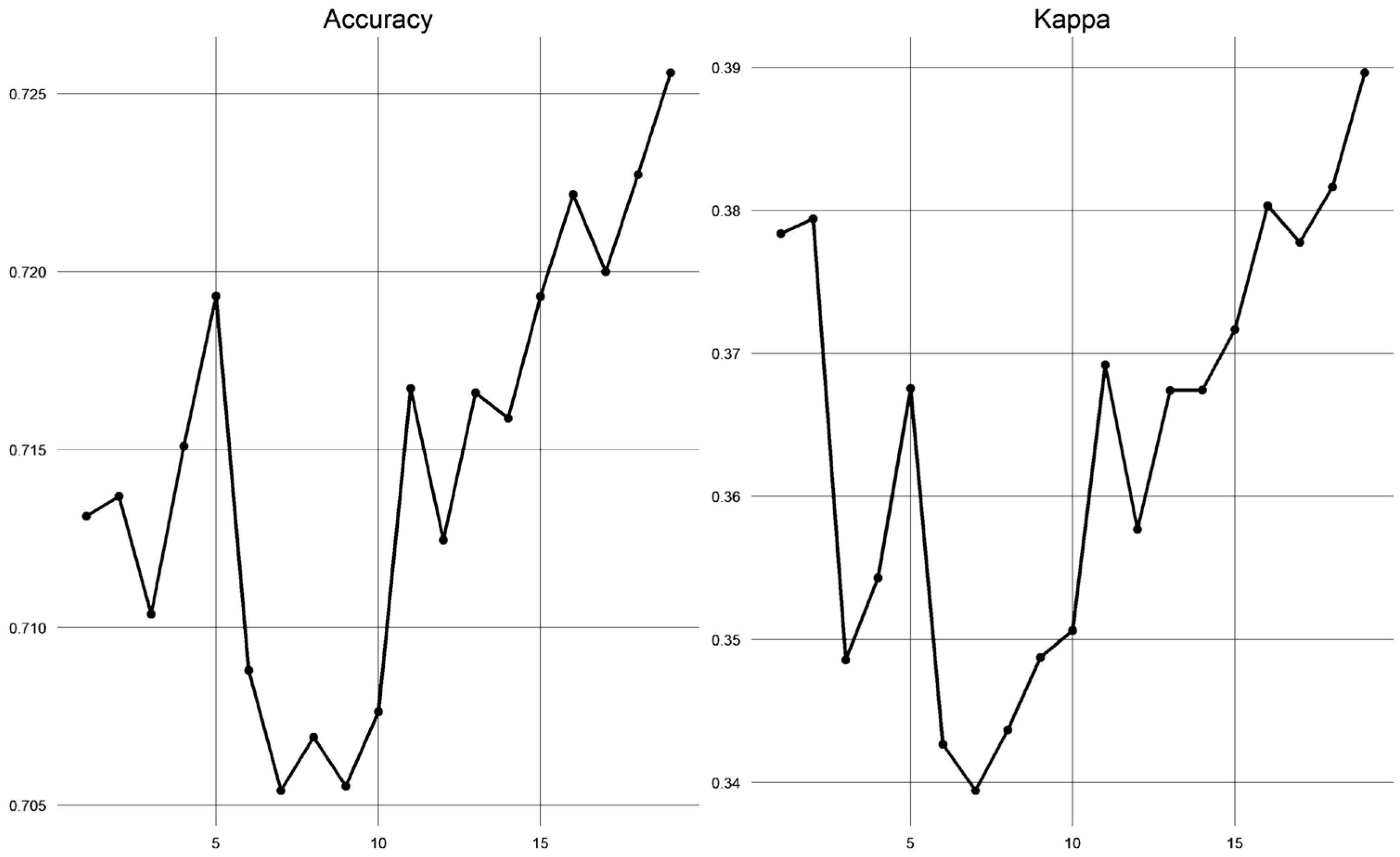
2.4.3. Model Parameter Tuning
2.5. Performance Evaluation Metrics
3. Experimental Results
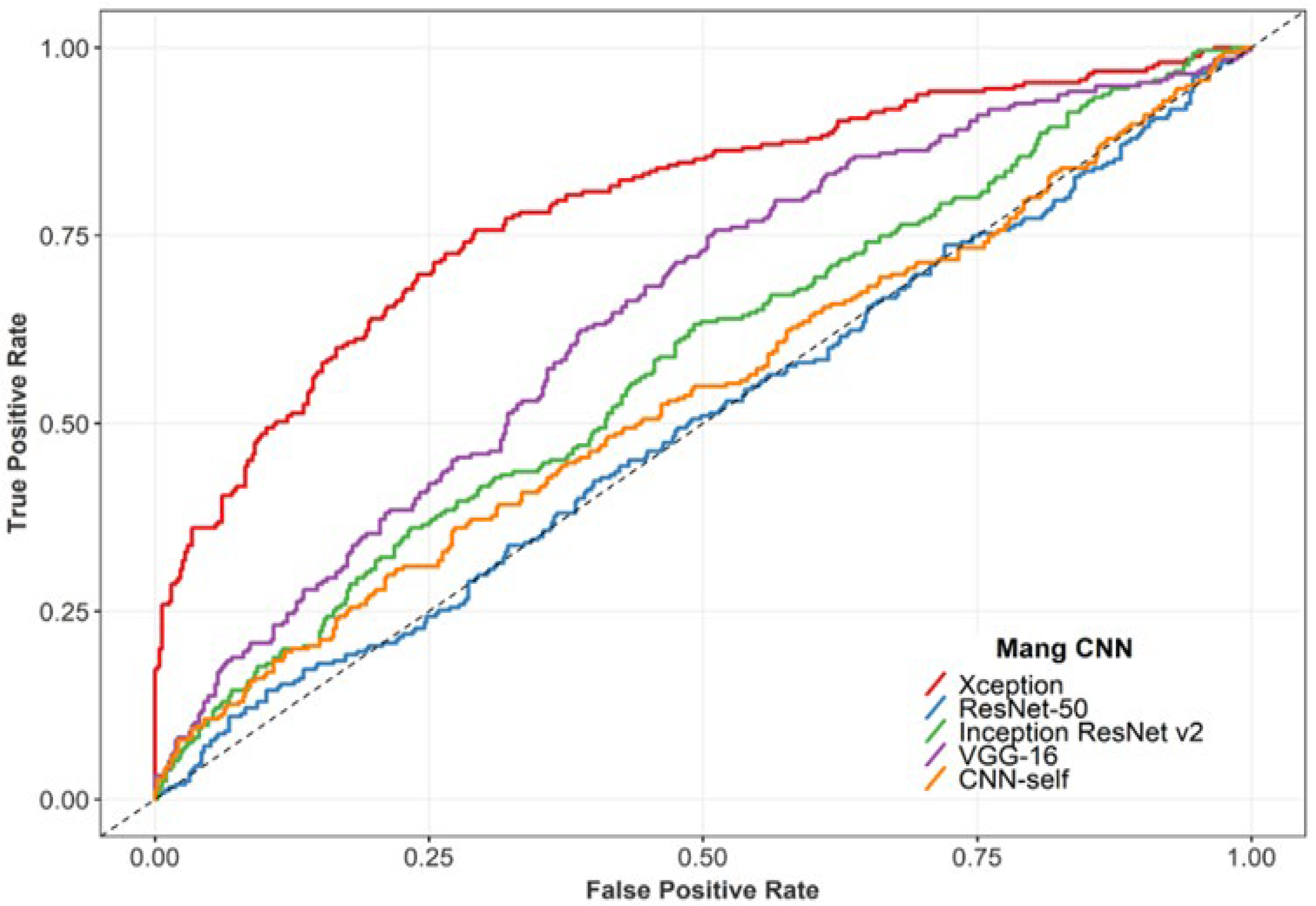
3.1. Performance of the DL Classifiers
3.2. Performance of the ML Classifiers
3.3. Performance of the ML-DL Model
4. Discussion
4.1. Performance Comparison with the Existing Literature
4.2. Limitations and Future Developments
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Variables | Group | Value |
|---|---|---|
| Age at diagnosis (mean ± SD) | 48 ± 11.4 | |
| BMI (mean ± SD) | 23 ± 3.2 | |
| Age at menstruation (mean ± SD) | 15 ± 2.0 | |
| Age at menopause (mean ± SD) | 50 ± 5.2 | |
| No of children (mean-IQR) | 2 (1–3) | |
| Early menstruation | Yes | 14 (3.9%) |
| No | 343 (96.1%) | |
| Late menopause | Yes | 101 (28.3%) |
| No | 256 (71.7%) | |
| Timing of pregnancy | Never had full-term pregnancy | 53 (14.8%) |
| ≤35 age | 267 (74.8%) | |
| >35 age | 37 (10.4%) | |
| Breastfeeding | Yes | 279 (78.2%) |
| No | 78 (21.8%) | |
| First-degree family member with breast cancer | Yes | 30 (8.4%) |
| No | 327 (91.6%) | |
| Any family member with breast cancer | Yes | 4 (1.1%) |
| No | 353 (98.9%) | |
| Past or present use of progesterone | Yes | 50 (14%) |
| No | 307 (86%) | |
| Palpable lump | One breast | 324 (90.8%) |
| Both breasts | 33 (9.2%) | |
| Breast skin flaking or thickened | Yes | 15 (4.2%) |
| No | 342 (95.8%) | |
| Skin dimpling | Yes | 7 (2.0%) |
| No | 350 (98.0%) | |
| Nipple retraction | Yes | 18 (5.0%) |
| No | 339 (95.0%) | |
| Nipple discharge | Yes | 34 (9.5%) |
| No | 323 (90.5%) | |
| Lymph node | Yes | 31 (8.7%) |
| No | 326 (91.3%) | |
| BI-RADS categories | 0 | 9 (2.5%) |
| 1 | 35 (9.9%) | |
| 2 | 34 (9.6%) | |
| 3 | 82 (23.1%) | |
| 4 | 152 (42.8%) | |
| 5 | 43 (12.1%) | |
| Tumor size (mean-IQR) | 20.2 (15–30.3) | |
| Echo pattern | Hypoechoic | 326 (92.9%) |
| Isoechoic | 18 (5.1%) | |
| Hyperechoic | 7 (2.0%) | |
| Calcifications | Yes | 102 (28.8%) |
| No | 252 (71.2%) | |
| Vascular abnormalities | Yes | 108 (30.5%) |
| No | 246 (69.5%) | |
| Architecture distortion | Yes | 22 (6.2%) |
| No | 332 (93.8%) |
References
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2022. CA: Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef] [PubMed]
- Dixon, A.-M. Diagnostic Breast Imaging: Mammography, Sonography, Magnetic Resonance Imaging, and Interventional Procedures. Ultrasound: J. Br. Med. Ultrasound Soc. 2014, 22, 182. [Google Scholar] [CrossRef]
- Sickles, E.; D’Orsi, C.; Bassett, L.; Appleton, C.; Berg, W.; Burnside, E. ACR BI-RADS Atlas, Breast Imaging Reporting and Data System, 5th ed.; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Giger, M.L.; Karssemeijer, N.; Schnabel, J.A. Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer. Annu. Rev. Biomed. Eng. 2013, 15, 327–357. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Pollán, R.; Guevara-López, M.A.; Suárez-Ortega, C.; Díaz-Herrero, G.; Franco-Valiente, J.M.; Rubio-del-Solar, M.; González-de-Posada, N.; Vaz, M.A.P.; Loureiro, J.; Ramos, I. Discovering mammography-based machine learning classifiers for breast cancer diagnosis. J. Med. Syst. 2012, 36, 2259–2269. [Google Scholar] [CrossRef]
- Warren Burhenne, L.J.; Wood, S.A.; D’Orsi, C.J.; Feig, S.A.; Kopans, D.B.; O’Shaughnessy, K.F.; Sickles, E.A.; Tabar, L.; Vyborny, C.J.; Castellino, R.A. Potential contribution of computer-aided detection to the sensitivity of screening mammography. Radiology 2000, 215, 554–562. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Aly, F. Deep learning approaches for data augmentation and classification of breast masses using ultrasound images. Int. J. Adv. Comput. Sci. Appl. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Swain, M.; Kisan, S.; Chatterjee, J.M.; Supramaniam, M.; Mohanty, S.N.; Jhanjhi, N.; Abdullah, A. Hybridized machine learning based fractal analysis techniques for breast cancer classification. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 179–184. [Google Scholar] [CrossRef]
- Saber, A.; Sakr, M.; Abo-Seida, O.M.; Keshk, A.; Chen, H. A novel deep-learning model for automatic detection and classification of breast cancer using the transfer-learning technique. IEEE Access 2021, 9, 71194–71209. [Google Scholar] [CrossRef]
- Çayır, S.; Solmaz, G.; Kusetogullari, H.; Tokat, F.; Bozaba, E.; Karakaya, S.; Iheme, L.O.; Tekin, E.; Özsoy, G.; Ayaltı, S. MITNET: A novel dataset and a two-stage deep learning approach for mitosis recognition in whole slide images of breast cancer tissue. Neural Comput. Appl. 2022, 34, 17837–17851. [Google Scholar] [CrossRef]
- Chakravarthy, S.S.; Rajaguru, H. Automatic detection and classification of mammograms using improved extreme learning machine with deep learning. IRBM 2022, 43, 49–61. [Google Scholar] [CrossRef]
- Altameem, A.; Mahanty, C.; Poonia, R.C.; Saudagar, A.K.J.; Kumar, R. Breast cancer detection in mammography images using deep convolutional neural networks and fuzzy ensemble modeling techniques. Diagnostics 2022, 12, 1812. [Google Scholar] [CrossRef] [PubMed]
- Muduli, D.; Dash, R.; Majhi, B. Automated diagnosis of breast cancer using multi-modal datasets: A deep convolution neural network based approach. Biomed. Signal Process. Control 2022, 71, 102825. [Google Scholar] [CrossRef]
- Heenaye-Mamode Khan, M.; Boodoo-Jahangeer, N.; Dullull, W.; Nathire, S.; Gao, X.; Sinha, G.; Nagwanshi, K.K. Multi-class classification of breast cancer abnormalities using Deep Convolutional Neural Network (CNN). PLoS ONE 2021, 16, e0256500. [Google Scholar] [CrossRef]
- Bhowal, P.; Sen, S.; Velasquez, J.D.; Sarkar, R. Fuzzy ensemble of deep learning models using choquet fuzzy integral, coalition game and information theory for breast cancer histology classification. Expert Syst. Appl. 2022, 190, 116167. [Google Scholar] [CrossRef]
- Wakili, M.A.; Shehu, H.A.; Sharif, M.; Sharif, M.; Uddin, H.; Umar, A.; Kusetogullari, H.; Ince, I.F.; Uyaver, S. Classification of Breast Cancer Histopathological Images Using DenseNet and Transfer Learning. Comput. Intell. Neurosci. 2022, 2022, 8904768. [Google Scholar] [CrossRef]
- Alshammari, M.M.; Almuhanna, A.; Alhiyafi, J. Mammography Image-Based Diagnosis of Breast Cancer Using Machine Learning: A Pilot Study. Sensors 2021, 22, 203. [Google Scholar] [CrossRef]
- Moura, D.C.; Guevara López, M.A. An evaluation of image descriptors combined with clinical data for breast cancer diagnosis. Int. J. Comput. Assist. Radiol. Surg. 2013, 8, 561–574. [Google Scholar] [CrossRef]
- Delen, D.; Walker, G.; Kadam, A. Predicting breast cancer survivability: A comparison of three data mining methods. Artif. Intell. Med. 2005, 34, 113–127. [Google Scholar] [CrossRef]
- Burke, H.B.; Rosen, D.B.; Goodman, P.H. Comparing the prediction accuracy of artificial neural networks and other statistical models for breast cancer survival. Adv. Neural Inf. Process. Syst. 1994, 7, 1064–1067. [Google Scholar]
- Xiao, T.; Liu, L.; Li, K.; Qin, W.; Yu, S.; Li, Z. Comparison of transferred deep neural networks in ultrasonic breast masses discrimination. BioMed Res. Int. 2018, 2018, 4605191. [Google Scholar] [CrossRef] [PubMed]
- Meads, C.; Ahmed, I.; Riley, R.D. A systematic review of breast cancer incidence risk prediction models with meta-analysis of their performance. Breast Cancer Res. Treat. 2012, 132, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.H.; Petrick, N.A.; Pezeshk, A.X.; Graff, C.G.; Sharma, D.; Badal, A.; Sahiner, B. Evaluation of data augmentation via synthetic images for improved breast mass detection on mammograms using deep learning. J. Med. Imaging 2019, 7, 012703. [Google Scholar] [CrossRef]
- Oyelade, O.N.; Ezugwu, A.E. A deep learning model using data augmentation for detection of architectural distortion in whole and patches of images. Biomed. Signal Process. Control 2021, 65, 102366. [Google Scholar] [CrossRef]
- Costa, A.C.; Oliveira, H.C.; Vieira, M.A. Data augmentation: Effect in deep convolutional neural network for the detection of architectural distortion in digital mammography. Assoc. Bras. Fis. Médica (ABFM) 2019, 51, 51041896. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst. 2014, 27, 3320–3328. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Albashish, D.; Al-Sayyed, R.; Abdullah, A.; Ryalat, M.H.; Almansour, N.A. Deep CNN model based on VGG16 for breast cancer classification. In Proceedings of the 2021 International Conference on Information Technology (ICIT), Amman, Jordan, 14–15 July 2021. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Yue, H.; Lin, Y.; Wu, Y.; Wang, Y.; Li, Y.; Guo, X.; Huang, Y.; Wen, W.; Zhao, G.; Pang, X. Deep learning for diagnosis and classification of obstructive sleep apnea: A nasal airflow-based multi-resolution residual network. Nat. Sci. Sleep 2021, 13, 361. [Google Scholar] [CrossRef]
- Kooi, T.; Litjens, G.; Van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef]
- Sun, W.; Tseng, T.-L.B.; Zhang, J.; Qian, W. Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput. Med. Imaging Graph. 2017, 57, 4–9. [Google Scholar] [CrossRef]
- Dasarathy, B.V. Nearest Neighbor (NN) Norms: NN Pattern Classification Techniques; IEEE Computer Society: Los Alamitos, CA, USA, 1991; ISBN 978-0818689307. [Google Scholar]
- Liu, B. Web Data Mining: Exploring Hyperlinks, Contents, and Usage Data; Springer: Berlin/Heidelberg, Germany, 2011; Volume 1. [Google Scholar]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef] [PubMed]
- Sepandi, M.; Taghdir, M.; Rezaianzadeh, A.; Rahimikazerooni, S. Assessing breast cancer risk with an artificial neural network. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 1017. [Google Scholar] [PubMed]
- Biau, G.; Devroye, L. On the layered nearest neighbour estimate, the bagged nearest neighbour estimate and the random forest method in regression and classification. J. Multivar. Anal. 2010, 101, 2499–2518. [Google Scholar] [CrossRef]
- Ezhilraman, S.V.; Srinivasan, S.; Suseendran, G. Breast Cancer Detection using Gradient Boost Ensemble Decision Tree Classifier. Int. J. Eng. Adv. Technol. 2019, 9, 2169–2173. [Google Scholar] [CrossRef]
- Kuhn, M. Caret: Classification and Regression Training; ascl:1505.003; Astrophysics Source Code Library: Houghton, MI, USA, 2015. [Google Scholar]
- Huynh, B.Q.; Li, H.; Giger, M.L. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J. Med. Imaging 2016, 3, 034501. [Google Scholar] [CrossRef]
- Samala, R.K.; Chan, H.-P.; Hadjiiski, L.M.; Helvie, M.A.; Cha, K.H.; Richter, C.D. Multi-task transfer learning deep convolutional neural network: Application to computer-aided diagnosis of breast cancer on mammograms. Phys. Med. Biol. 2017, 62, 8894. [Google Scholar] [CrossRef]
- Ahn, C.K.; Heo, C.; Jin, H.; Kim, J.H. A novel deep learning-based approach to high accuracy breast density estimation in digital mammography. In Medical Imaging 2017: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2017. [Google Scholar]
- Jiao, Z.; Gao, X.; Wang, Y.; Li, J. A deep feature based framework for breast masses classification. Neurocomputing 2016, 197, 221–231. [Google Scholar] [CrossRef]
- Qiu, Y.; Wang, Y.; Yan, S.; Tan, M.; Cheng, S.; Liu, H.; Zheng, B. An initial investigation on developing a new method to predict short-term breast cancer risk based on deep learning technology. In Medical Imaging 2016: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2016. [Google Scholar]
- Chougrad, H.; Zouaki, H.; Alheyane, O. Deep convolutional neural networks for breast cancer screening. Comput. Methods Programs Biomed. 2018, 157, 19–30. [Google Scholar] [CrossRef]
- Mohapatra, S.; Muduly, S.; Mohanty, S.; Ravindra, J.; Mohanty, S.N. Evaluation of deep learning models for detecting breast cancer using histopathological mammograms Images. Sustain. Oper. Comput. 2022, 3, 296–302. [Google Scholar] [CrossRef]
- Li, C.; Xu, J.; Liu, Q.; Zhou, Y.; Mou, L.; Pu, Z.; Xia, Y.; Zheng, H.; Wang, S. Multi-view mammographic density classification by dilated and attention-guided residual learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 1003–1013. [Google Scholar] [CrossRef]
- Ting, F.F.; Tan, Y.J.; Sim, K.S. Convolutional neural network improvement for breast cancer classification. Expert Syst. Appl. 2019, 120, 103–115. [Google Scholar] [CrossRef]
- Geras, K.J.; Wolfson, S.; Shen, Y.; Wu, N.; Kim, S.; Kim, E.; Heacock, L.; Parikh, U.; Moy, L.; Cho, K. High-resolution breast cancer screening with multi-view deep convolutional neural networks. arXiv 2017, arXiv:1703.07047. [Google Scholar]
- Boughorbel, S.; Al-Ali, R.; Elkum, N. Model comparison for breast cancer prognosis based on clinical data. PLoS ONE 2016, 11, e0146413. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Lee, J.C.; Park, B.; Bae, J.; Lim, M.H.; Kang, D.; Yoo, K.-Y.; Park, S.K.; Kim, Y.; Kim, S. Computational discrimination of breast cancer for Korean women based on epidemiologic data only. J. Korean Med. Sci. 2015, 30, 1025–1034. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, G.; Kang, Y.; Zhao, Y.; Qu, Q. Breast tumor detection in digital mammography based on extreme learning machine. Neurocomputing 2014, 128, 175–184. [Google Scholar] [CrossRef]
| Classifier | Caret Package | Fine-Tuned Hyperparameter |
|---|---|---|
| k-NN | knn | k (neighbors) |
| SVM | svmRadial | σ (Gaussian kernel), C (Cost) |
| ANN | nnet | size (hidden unit), decay (weight decay) |
| RF | rf | mtry (randomly selected variables) |
| GBM | gbm | interaction.depth, n.trees, shrinkage |
| Classifiers | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | F1S | MCC | Kappa |
|---|---|---|---|---|---|---|---|
| Xception | 72.5 # | 75.7 | 70.8 | 0.79 | 0.66 | 0.45 | 0.44 |
| VGG16 | 58.3 | 75.7 | 48.9 | 0.49 | 0.56 | 0.24 | 0.21 |
| Resnet-v2 | 55.2 | 63.1 | 50.9 | 0.58 | 0.50 | 0.13 | 0.12 |
| Resnet50 | 50.5 | 51.0 | 50.2 | 0.50 | 0.42 | 0.01 | 0.01 |
| CNN3 | 53.8 | 50.6 | 55.5 | 0.54 | 0.43 | 0.06 | 0.06 |
| Variables | Benign | Malignant | p-Value | |
|---|---|---|---|---|
| Early menstruation | Yes | 4 (1.8%) | 10 (7.4%) | 0.015 |
| No | 217 (98.2%) | 126 (92.6%) | ||
| Late menopause | Yes | 43 (19.5%) | 58 (42.6%) | <0.001 |
| No | 178 (80.5%) | 78 (57.4%) | ||
| Breast skin flaking or thickened | Yes | 5 (2.3%) | 10 (7.4%) | 0.028 |
| No | 216 (97.7%) | 126 (92.6%) | ||
| Nipple retraction | Yes | 3 (1.4%) | 15 (11.0%) | 0.001 |
| No | 218 (98.6%) | 121 (89.0%) | ||
| Lymph node | Yes | 4 (1.8%) | 27 (19.9%) | <0.001 |
| No | 217 (98.2%) | 109 (80.1%) | ||
| Calcification | Yes | 28 (12.7%) | 74 (54.4%) | <0.001 |
| No | 193 (87.3%) | 62 (45.6%) | ||
| Enhancedvascularity | Yes | 31 (14.0%) | 77 (56.6%) | <0.001 |
| No | 190 (86.0%) | 59 (43.4%) | ||
| Architecturedistortion | Yes | 6 (2.7%) | 16 (11.8%) | 0.001 |
| No | 215 (97.3%) | 120 (88.2%) | ||
| Lymph node | Yes | 44 (19.9%) | 76 (55.9%) | <0.001 |
| No | 177 (80.1%) | 60 (44.1%) | ||
| Size lump # | 20 (11.1–25.3) | 25 (16.6–35.2) | 0.002 | |
| BI-RADS # | 2.86 (2.0–4.0) | 4 (4.0–5.0) | <0.001 | |
| Classifiers | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | F1S | MCC | Kappa |
|---|---|---|---|---|---|---|---|
| k-NN | 66.2 | 66.7 | 64.7 | 0.76 | 0.60 | 0.31 | 0.30 |
| SVM | 73.2 | 74.5 | 70.8 | 0.81 | 0.67 | 0.44 | 0.43 |
| ANN | 78.9 | 81.4 | 75.0 | 0.82 | 0.73 | 0.55 | 0.54 |
| RF | 69.0 | 67.9 | 73.3 | 0.79 | 0.64 | 0.40 | 0.40 |
| GBM | 81.7 # | 83.7 | 78.6 | 0.84 | 0.77 | 0.61 | 0.60 |
| Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | |
|---|---|---|---|---|
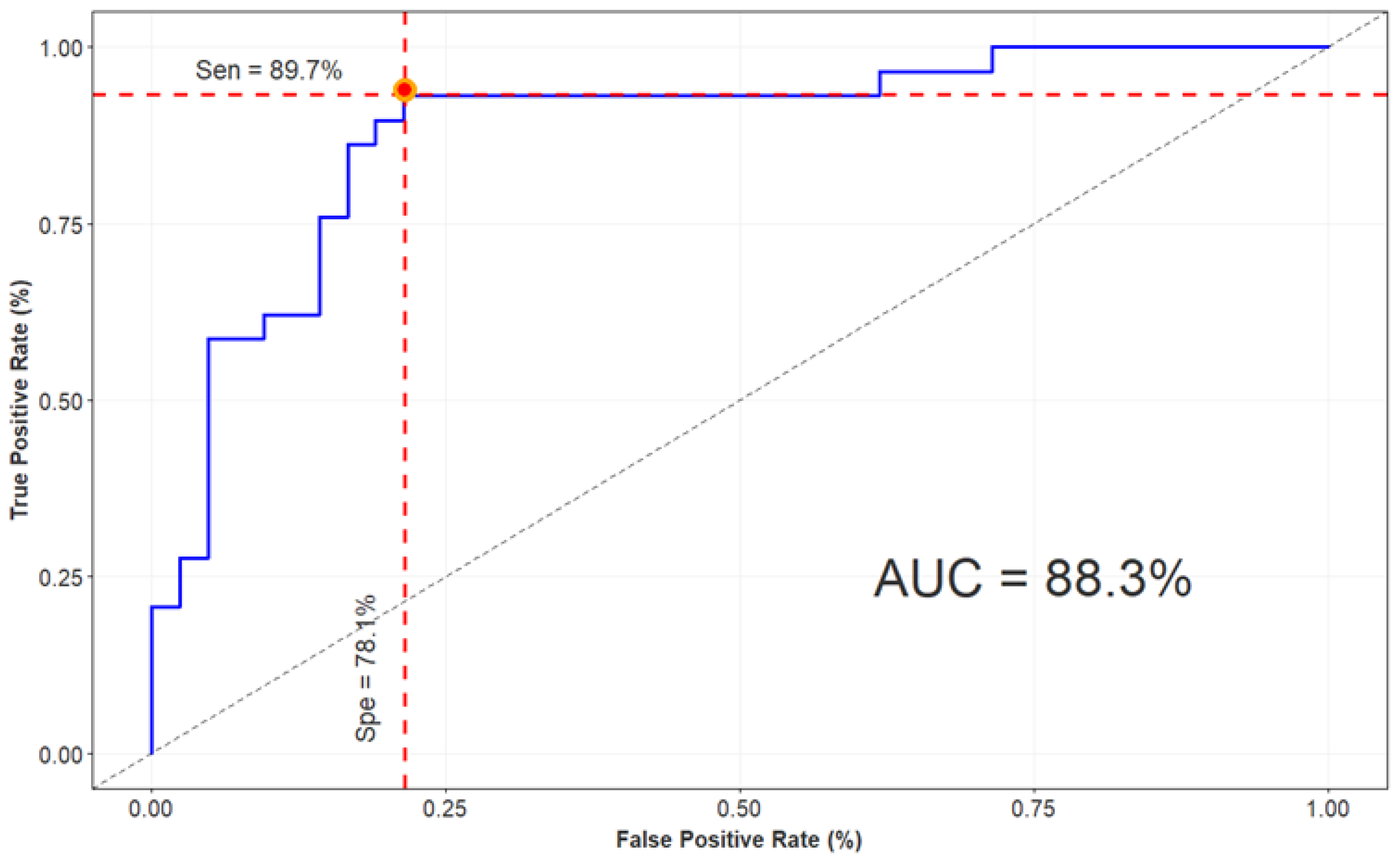
| X-ception + GBM | 84.5 | 89.7 | 78.1 | 0.88 |
| Author (Year) | Database (Population) | Category | Classifiers | Classes | Performance |
|---|---|---|---|---|---|
| This paper | Privacy dataset (731) | Mammography + clinical data | Xception + GBM | Benign, malignant | Acc: 84.5% AUC: 0.88 |
| Chougrad et al. [48] (2018) | DDSM (5316) | Mammography, mass-lesion classification | VGG16 + Resnet50 | Benign, malignant | Acc: 97.3% AUC: 0.98 |
| Mohapatra et al. [49] (2017) | Mini-DDSM (1952) | Mammography | AlexNet + VGG16 | Benign, cancer, normal | Acc: 65.0% AUC: 0.72 |
| Li et al. [50] (2020) | Privacy dataset + publicly INbreast (1985) | Mammographic density | Resnet-v2 + CNN | Four BI-RADS categories | Acc: 70% AUC: 0.84 |
| Ting et al. [51] (2019) | MIAS (221) | Mammography | CNN | Benign, malignant, normal | Acc: 74.9% AUC: 0.86 |
| Sun et al. [35] (2017) | FFDM (1874) | Mammogram images with ROIs containing mass extracted | CNN | Benign, malignant | Acc: 82.4% AUC: 0.88 |
| Boughorbel et al. [53] (2016) | METABRIC breast cancer dataset (1981 patients and 11 variables) | Clinical variables and histological | KNN + SVM + Boosted trees | Survived, not survived | AUC: 0.72 |
| Sepandi et al. [39] (2018) | Privacy dataset (655 women and 23 variables) | Demographic and clinical variables | ANN | Benign, malignant | AUC: 0.95 |
| Lee at el. [54] (2015) | Hospital in Korea (4574 cases) | Epidemiological data | SVM + ANN + NB | Case-control | AUC: 0.64 |
| Wang et al. [55] (2014) | Privacy dataset (482 images) | Digital mammography, feature extraction: geometrical, textural | ELM | Image with/without tumor | AUC: 0.85 |
| Moura et al. [19] (2013) | DDSM + BCDR (1762 and 362 instances) | Clinical data + image description | Several ML classifiers | Benign, malignant | AUC: 0.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trang, N.T.H.; Long, K.Q.; An, P.L.; Dang, T.N. Development of an Artificial Intelligence-Based Breast Cancer Detection Model by Combining Mammograms and Medical Health Records. Diagnostics 2023, 13, 346. https://doi.org/10.3390/diagnostics13030346
Trang NTH, Long KQ, An PL, Dang TN. Development of an Artificial Intelligence-Based Breast Cancer Detection Model by Combining Mammograms and Medical Health Records. Diagnostics. 2023; 13(3):346. https://doi.org/10.3390/diagnostics13030346
Chicago/Turabian StyleTrang, Nguyen Thi Hoang, Khuong Quynh Long, Pham Le An, and Tran Ngoc Dang. 2023. "Development of an Artificial Intelligence-Based Breast Cancer Detection Model by Combining Mammograms and Medical Health Records" Diagnostics 13, no. 3: 346. https://doi.org/10.3390/diagnostics13030346
APA StyleTrang, N. T. H., Long, K. Q., An, P. L., & Dang, T. N. (2023). Development of an Artificial Intelligence-Based Breast Cancer Detection Model by Combining Mammograms and Medical Health Records. Diagnostics, 13(3), 346. https://doi.org/10.3390/diagnostics13030346





