An Italian Multicenter Perspective Harmonization Trial for the Assessment of MET Exon 14 Skipping Mutations in Standard Reference Samples
Abstract
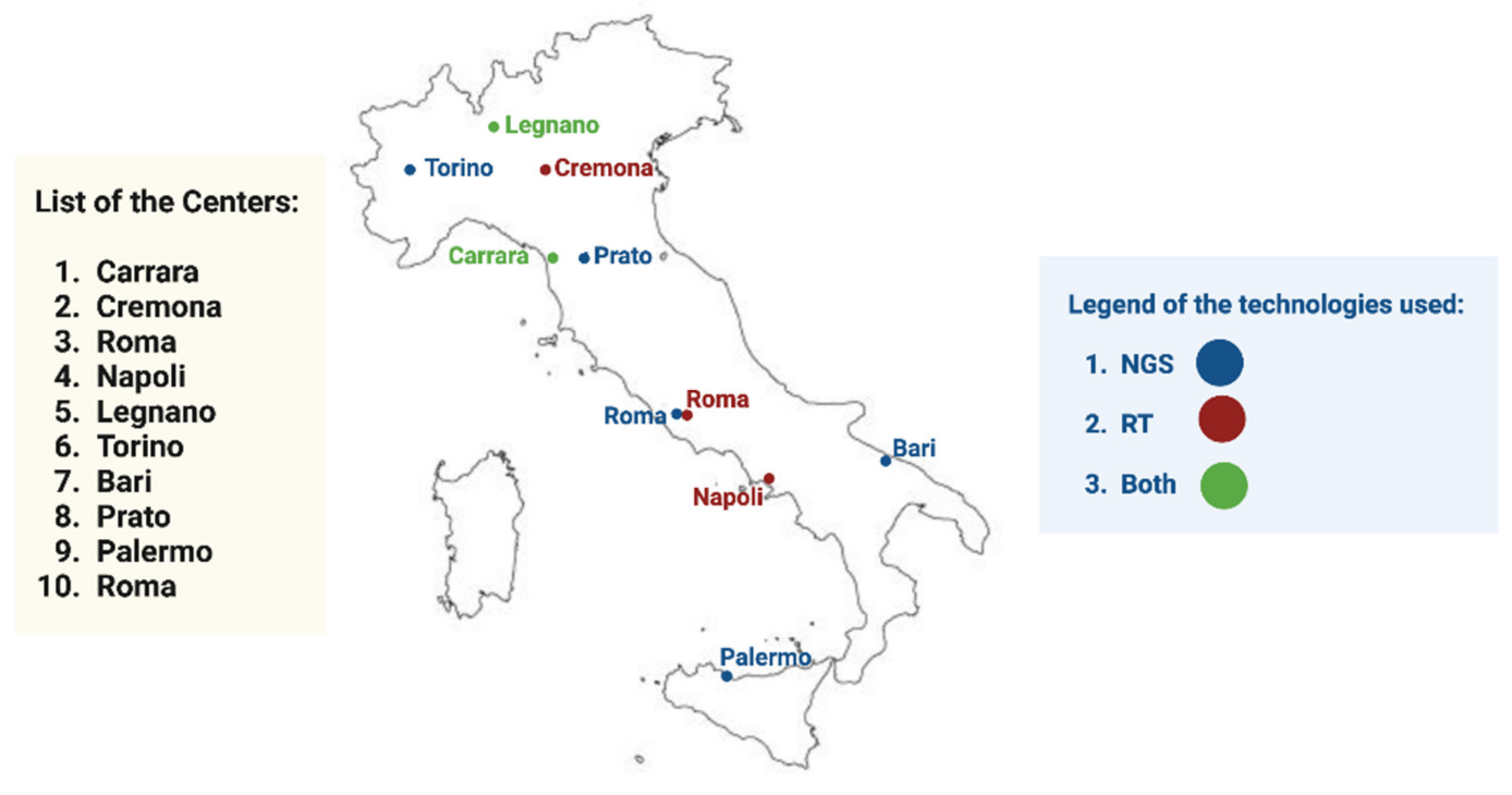
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Validation of Customized Reference Standard Sample
2.3. Technical Approaches
3. Results
3.1. Validation of Customized Reference Standard Sample
3.2. Ring Trial Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar]
- Yuan, M.; Huang, L.-L.; Chen, J.-H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef]
- Remon, J.; Ahn, M.-J.; Girard, N.; Johnson, M.; Kim, D.-W.; Lopes, G.; Pillai, R.N.; Solomon, B.; Villacampa, G.; Zhou, Q. Advanced-Stage Non–Small Cell Lung Cancer: Advances in Thoracic Oncology. J. Thorac. Oncol. 2019, 14, 1134–1155. [Google Scholar] [CrossRef]
- Malapelle, U.; Muscarella, L.A.; Pisapia, P.; Rossi, A. Targeting emerging molecular alterations in the treatment of non-small cell lung cancer: Current challenges and the way forward. Expert Opin. Investig. Drugs 2020, 29, 363–372. [Google Scholar] [CrossRef]
- Russo, A.; Lopes, A.R.; McCusker, M.; Garrigues, S.G.; Ricciardi, G.R.; Arensmeyer, K.E.; Scilla, K.A.; Mehra, R.; Rolfo, C. New Targets in Lung Cancer (Excluding EGFR, ALK, ROS1). Curr. Oncol. Rep. 2020, 22, 48. [Google Scholar] [CrossRef]
- Pan, Y.; Fu, Y.; Zeng, Y.; Liu, X.; Peng, Y.; Hu, C.; Deng, C.; Qiu, Z.; Zou, J.; Liu, Y.; et al. The key to immunotherapy: How to choose better therapeutic biomarkers for patients with non-small cell lung cancer. Biomark. Res. 2022, 10, 9. [Google Scholar] [CrossRef]
- Fujino, T.; Suda, K.; Mitsudomi, T. Lung Cancer with MET exon 14 Skipping Mutation: Genetic Feature, Current Treatments, and Future Challenges. Lung Cancer Targets Ther. 2021, 12, 35–50. [Google Scholar] [CrossRef]
- Cortot, A.B.; Kherrouche, Z.; Descarpentries, C.; Wislez, M.; Baldacci, S.; Furlan, A.; Tulasne, D. Exon 14 Deleted MET Receptor as a New Biomarker and Target in Cancers. Gynecol. Oncol. 2017, 109, djw262. [Google Scholar] [CrossRef]
- Frampton, G.M.; Ali, S.M.; Rosenzweig, M.; Chmielecki, J.; Lu, X.; Bauer, T.M.; Akimov, M.; Bufill, J.A.; Lee, C.; Jentz, D.; et al. Activation of MET via Diverse Exon 14 Splicing Alterations Occurs in Multiple Tumor Types and Confers Clinical Sensitivity to MET Inhibitors. Cancer Discov. 2015, 5, 850–859. [Google Scholar] [CrossRef]
- Awad, M.M.; Oxnard, G.R.; Jackman, D.M.; Savukoski, D.O.; Hall, D.; Shivdasani, P.; Heng, J.C.; Dahlberg, S.E.; Jänne, P.A.; Verma, S.; et al. MET Exon 14 Mutations in Non–Small-Cell Lung Cancer Are Associated with Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J. Clin. Oncol. 2016, 34, 721–730. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Paik, P.K.; Drilon, A.; Fan, P.-D.; Yu, H.; Rekhtman, N.; Ginsberg, M.S.; Borsu, L.; Schultz, N.; Berger, M.F.; Rudin, C.M.; et al. Response to MET Inhibitors in Patients with Stage IV Lung Adenocarcinomas Harboring MET Mutations Causing Exon 14 Skipping. Cancer Discov. 2015, 5, 842–849. [Google Scholar] [CrossRef]
- Wolf, J.; Seto, T.; Han, J.Y.; Reguart, N.; Garon, E.B.; Groen, H.J.M.; Tan, D.S.W.; Hida, T.; de Jonge, M.; Orlov, S.V.; et al. GEOMETRY mono-1 Investigators. Capmatinib in MET Exon 14-Mutated or MET-Amplified Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 944–957. [Google Scholar] [CrossRef]
- Socinski, M.A.; Pennell, N.A.; Davies, K.D. MET Exon 14 Skipping Mutations in Non-Small-Cell Lung Cancer: An Overview of Biology, Clinical Outcomes, and Testing Considerations. JCO Precis Oncol. 2021, 5, PO.20.00516. [Google Scholar]
- Davies, K.D.; Lomboy, A.; Lawrence, C.A.; Yourshaw, M.; Bocsi, G.T.; Camidge, D.R.; Aisner, D.L. DNA-Based versus RNA-Based De-tection of MET Exon 14 Skipping Events in Lung Cancer. J Thorac. Oncol. 2019, 14, 737–741. [Google Scholar] [CrossRef]
- Kim, E.K.; Kim, K.A.; Lee, C.Y.; Kim, S.; Chang, S.; Cho, B.C.; Shim, H.S. Molecular Diagnostic Assays and Clinicopathologic Implications of MET Exon 14 Skipping Mutation in Non–small-cell Lung Cancer. Clin. Lung Cancer 2018, 20, e123–e132. [Google Scholar] [CrossRef]
- Pisapia, P.; Malapelle, U.; Roma, G.; Saddar, S.; Zheng, Q.; Pepe, F.; Bruzzese, D.; Vigliar, E.; Bellevicine, C.; Luthra, R.; et al. Consistency and reproducibility of next-generation sequencing in cytopathology: A second worldwide ring trial study on improved cytological molecular reference specimens. Cancer Cytopathol. 2019, 127, 285–296. [Google Scholar] [CrossRef]
- Malapelle, U.; Mayo-de-Las-Casas, C.; Molina-Vila, M.A.; Rosell, R.; Savic, S.; Bihl, M.; Bubendorf, L.; Salto-Tellez, M.; de Biase, D.; Tallini, G.; et al. Molecular Cytopathology Meeting Group. Consistency and reproducibility of next-generation sequencing and other multigene muta-tional assays: A worldwide ring trial study on quantitative cytological molecular reference specimens. Cancer Cytopathol. 2017, 125, 615–626. [Google Scholar]
- Malapelle, U.; Pepe, F.; Pisapia, P.; Altimari, A.; Bellevicine, C.; Brunnström, H.; Bruno, R.; Büttner, R.; Cirnes, L.; de Andrea, C.E.; et al. Reference standards for gene fusion molecular assays on cytological samples: An international validation study. J. Clin. Pathol. 2021, 76, 47–52. [Google Scholar] [CrossRef]
- Lu, S.; Fang, J.; Li, X.; Cao, L.; Zhou, J.; Guo, Q.; Liang, Z.; Cheng, Y.; Jiang, L.; Yang, N.; et al. Once-daily savolitinib in Chinese patients with pulmonary sarcomatoid carcinomas and other non-small-cell lung cancers harbouring MET exon 14 skipping alterations: A multicentre, single-arm, open-label, phase 2 study. Lancet Respir. Med. 2021, 9, 1154–1164. [Google Scholar] [CrossRef]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non–Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef]
- Reungwetwattana, T.; Liang, Y.; Zhu, V.; Ou, S.-H.I. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: The Why, the How, the Who, the Unknown, and the Inevitable. Lung Cancer 2016, 103, 27–37. [Google Scholar] [CrossRef]
- Poirot, B.; Doucet, L.; Benhenda, S.; Champ, J.; Meignin, V.; Lehmann-Che, J. MET Exon 14 Alterations and New Resistance Mutations to Tyrosine Kinase Inhibitors: Risk of Inadequate Detection with Current Amplicon-Based NGS Panels. J. Thorac. Oncol. 2017, 12, 1582–1587. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, O.; Wright, M.C.; O’Brien, C.; Geoghegan, O.; Leonard, N.; Nicholson, S.; Cuffe, S.; Fabre, A.; Jochum, W.; Joerger, M.; et al. Cost-Efficient and Easy to Perform PCR-Based Assay to Identify Met Exon 14 Skipping in Formalin-Fixed Paraf-fin-Embedded (FFPE) Non-Small Cell Lung Cancer (NSCLC) Samples. Diagnostics 2019, 18, 13. [Google Scholar]
- Kwon, D.; Koh, J.; Kim, S.; Go, H.; Kim, Y.A.; Keam, B.; Kim, T.M.; Kim, D.W.; Jeon, Y.K.; Chung, D.H. MET exon 14 skipping mutation in triple-negative pulmonary adenocarcinomas and pleomorphic carcinomas: An analysis of intratumoral MET status hetero-geneity and clinicopathological characteristics. Lung Cancer 2017, 106, 131–137. [Google Scholar]
- Liu, X.; Jia, Y.; Stoopler, M.B.; Shen, Y.; Cheng, H.; Chen, J.; Mansukhani, M.; Koul, S.; Halmos, B.; Borczuk, A.C. Next-Generation Se-quencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. J. Clin. Oncol. 2016, 34, 794–802. [Google Scholar]
- Descarpentries, C.; Lepretre, F.; Escande, F.; Kherrouche, Z.; Figeac, M.; Sebda, S.; Baldacci, S.; Grégoire, V.; Jamme, P.; Copin, M.-C.; et al. Optimization of Routine Testing for MET Exon 14 Splice Site Mutations in NSCLC Patients. J. Thorac. Oncol. 2018, 13, 1873–1883. [Google Scholar] [CrossRef]
- Benayed, R.; Offin, M.; Mullaney, K.; Sukhadia, P.; Rios, K.; Desmeules, P.; Ptashkin, R.; Won, H.; Chang, J.; Halpenny, D.; et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin. Cancer Res. 2019, 25, 4712–4722. [Google Scholar] [CrossRef]
- Low, S.-K.; Ariyasu, R.; Uchibori, K.; Hayashi, R.; Chan, H.T.; Chin, Y.M.; Akita, T.; Harutani, Y.; Kiritani, A.; Tsugitomi, R.; et al. Rapid genomic profiling of circulating tumor DNA in non-small cell lung cancer using Oncomine Precision Assay with GenexusTM integrated sequencer. Transl. Lung Cancer Res. 2022, 11, 711–721. [Google Scholar] [CrossRef]
- Pisapia, P.; Pepe, F.; Sgariglia, R.; Nacchio, M.; Russo, G.; Conticelli, F.; Girolami, I.; Eccher, A.; Bellevicine, C.; Vigliar, E.; et al. Next generation sequencing in cytology. Cytopathology 2021, 32, 588–595. [Google Scholar] [CrossRef]
- Ilié, M.; Hofman, V.; Bontoux, C.; Heeke, S.; Lespinet-Fabre, V.; Bordone, O.; Lassalle, S.; Lalvée, S.; Tanga, V.; Allegra, M.; et al. Setting Up an Ultra-Fast Next-Generation Sequencing Approach as Reflex Testing at Diagnosis of Non-Squamous Non-Small Cell Lung Cancer; Experience of a Single Center (LPCE, Nice, France). Cancers 2022, 14, 2258. [Google Scholar] [CrossRef]


| RT-PCR-Based Technologies | ||||||
|---|---|---|---|---|---|---|
| Center | Extraction Kit | RNA Amount (ng/µL) | Platform | Assay | Results | Cq Value |
| 1 | MagCore Total RNA FFPE One-step kit | NA * | EasyPGX | EasyPGX ready ALK/ROS1/RET/MET | METΔ exon 14 | 30.7 |
| 2 | MagCore Total RNA FFPE One-step kit | 10.0 | EasyPGX | EasyPGX ready ALK/ROS1/RET/MET | METΔ exon 14 | 31.4 |
| 3 | HGH Pure MIRNA isolation kit | 19.5 | EasyPGX | EasyPGX ready ALK/ROS1/RET/MET | METΔ exon 14 | 27.1 |
| 4 | RNeasy FFPE Kit | 40.7 | EasyPGX | EasyPGX ready ALK/ROS1/RET/MET | METΔ exon 14 | 29.7 |
| 5 | Maxwell RSC RNA FFPE Kit | 8.7 | EasyPGX | EasyPGX ready ALK/ROS1/RET/MET | METΔ exon 14 | 32.0 |
| NGS-based technologies | ||||||
| Center | Extraction kit | RNA amount (ng/µL) | Platform | Assay | Results | Read count |
| 6 | Maxwell RSC RNA FFPE Kit | 4.3 | iSeql100™ | Myriapod®NGS Cancer Panel RNA | METΔ exon 14 | 160 |
| 7 | Magmax FFPE DNA/RNA ultra kit | 18.0 | S5™ | Oncomine Focus Assay | METΔ exon 14 | NR ** |
| 8 | Rneasy FFPE Kit | 51.3 | MiSeq | Myriapod®NGS Cancer Panel RNA | METΔ exon 14 | 176 |
| 9 | Rneasy FFPE Kit | 18.3 | S5™ | Oncomine Focus Assay | METΔ exon 14 | 7526 |
| 10 | Rneasy FFPE Kit | 49.0 | Genexus | Oncomine Precision Assay | METΔ exon 14 | 2194 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bironzo, P.; Pepe, F.; Russo, G.; Pisapia, P.; Gragnano, G.; Aquino, G.; Bessi, S.; Buglioni, S.; Bartoccini, F.; Ferrero, G.; et al. An Italian Multicenter Perspective Harmonization Trial for the Assessment of MET Exon 14 Skipping Mutations in Standard Reference Samples. Diagnostics 2023, 13, 629. https://doi.org/10.3390/diagnostics13040629
Bironzo P, Pepe F, Russo G, Pisapia P, Gragnano G, Aquino G, Bessi S, Buglioni S, Bartoccini F, Ferrero G, et al. An Italian Multicenter Perspective Harmonization Trial for the Assessment of MET Exon 14 Skipping Mutations in Standard Reference Samples. Diagnostics. 2023; 13(4):629. https://doi.org/10.3390/diagnostics13040629
Chicago/Turabian StyleBironzo, Paolo, Francesco Pepe, Gianluca Russo, Pasquale Pisapia, Gianluca Gragnano, Gabriella Aquino, Silvia Bessi, Simonetta Buglioni, Federico Bartoccini, Giuseppina Ferrero, and et al. 2023. "An Italian Multicenter Perspective Harmonization Trial for the Assessment of MET Exon 14 Skipping Mutations in Standard Reference Samples" Diagnostics 13, no. 4: 629. https://doi.org/10.3390/diagnostics13040629











