KRAS Mutational Profiles among Colorectal Cancer Patients in the East Coast of Peninsular Malaysia
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. DNA Extraction from CRC FFPE Tissues
2.3. Detection of Mutations in Codons 12 and 13 of KRAS
2.4. Statistical Analysis
3. Results
3.1. Patient Clinical Characteristics
3.2. Mutation Characteristic of KRAS
3.3. Relationship between Clinicopathological Features of CRC and KRAS Mutations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef]
- Azizah, A.; Hashimah, B.; Nirmal, K.; Siti Zubaidah, A.; Puteri, N.; Nabihah, A.; Sukumaran, R.; Balqis, B.; Nadia, S.; Sharifah, S.; et al. Malaysia National Cancer Registry Report (MNCR); National Cancer Institute, Ministry of Health: Putrajaya, Malaysia, 2019. [Google Scholar]
- Tumors, K.; Drosten, M.; Barbacid, M. Review Targeting the MAPK Pathway. Cancer Cell 2020, 37, 543–550. [Google Scholar]
- Fang, J.Y.; Richardson, B.C. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Neumann, J.; Zeindl-eberhart, E.; Kirchner, T.; Jung, A. Pathology—Research and Practice Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol. Res. Pract. 2009, 205, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.M.; Kozuch, P.S. Impact of KRAS Mutations on Management of Colorectal Carcinoma. Pathol. Res. Int. 2011, 2011, 219309. [Google Scholar] [CrossRef] [PubMed]
- Brink, M.; De Goeij, A.F.; Weijenberg, M.P.; Roemen, G.M.; Lentjes, M.H.; Pachen, M.M.; Smits, K.M.; De Bruïne, A.P.; Goldbohm, R.A.; van den Brandt, P. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis 2003, 24, 703–710. [Google Scholar] [CrossRef]
- Meng, M.; Zhong, K.; Jiang, T.; Liu, Z.; Yee, H. Biomedicine & Pharmacotherapy The current understanding on the impact of KRAS on colorectal cancer. Biomed. Pharm. 2021, 140, 111717. [Google Scholar]
- Andreyev, H.J.; Norman, A.R.; Cunningham, D.; Oates, J.; Dix, B.R.; Iacopetta, B.J.; Young, J.; Walsh, T.; Ward, R.; Hawkins, N.; et al. Kirsten ras mutations in patients with colorectal cancer: The ‘RASCAL II’ study. Br. J. Cancer 2001, 85, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Hayama, T.; Hashiguchi, Y.; Okamoto, K.; Okada, Y.; Ono, K.; Shimada, R.; Ozawa, T.; Toyoda, T.; Tsuchiya, T.; Iinuma, H.; et al. G12V and G12C mutations in the gene KRAS are associated with a poorer prognosis in primary colorectal cancer. Int. J. Color. Dis. 2019, 34, 1491–1496. [Google Scholar] [CrossRef]
- Scott, A.; Goffredo, P.; Ginader, T.; Hrabe, J.; Gribovskaja-Rupp, I.; Kapadia, M.R.; Weigel, R.J.; Hassan, I. The Impact of KRAS Mutation on the Presentation and Prognosis of Non-Metastatic Colon Cancer: An Analysis from the National Cancer Database. J. Gastrointest. Surg. 2020, 24, 1402–1410. [Google Scholar] [CrossRef] [PubMed]
- Abdul Murad, N.A.; Othman, Z.; Khalid, M.; Abdul Razak, Z.; Hussain, R.; Nadesan, S.; Sagap, I.; Mohamed Rose, I.; Wan Ngah, W.Z.; Jamal, R. Missense Mutations in MLH1, MSH2, KRAS, and APC Genes in Colorectal Cancer Patients in Malaysia. Dig. Dis. Sci. 2012, 57, 2863–2872. [Google Scholar] [CrossRef] [PubMed]
- Yip, W.K.; Choo, C.W.; Leong, V.C.-S.; Leong, P.P.; Jabar, M.F.; Seow, H.F. Molecular alterations of Ras-Raf-mitogen-activated protein kinase and phosphatidylinositol 3-kinase-Akt signaling pathways in colorectal cancers from a tertiary hospital at Kuala Lumpur, Malaysia. APMIS 2013, 121, 954–966. [Google Scholar] [CrossRef] [PubMed]
- Othman, Z. Predominance of G to A codon 12 mutation K-ras gene in Dukes ’ B colorectal cancer. Singap. Med. J. 2012, 53, 26–31. [Google Scholar]
- Lee, C.B.; Kien, Y.W.; Dusa, N.; Mohtarrudin, N.; Fong, S.H. Identifying common mutations in colorectal cancer using a 7-gene panel by next generation sequencing. Malays. J. Med. Health Sci. 2019, 15, 95–102. [Google Scholar]
- Malaysia, D. Current Population Estimates, Malaysia. 2022. Available online: https://www.dosm.gov.my/v1/index.php?r=column/pdfPrev&id=dTZXanV6UUdyUEQ0SHNWOVhpSXNMUT09 (accessed on 5 October 2022).
- Malapelle, U.; Bellevicine, C.; Salatiello, M.; de Luca, C.; Rispo, E.; Riccio, P.; Sparano, L.; De Stefano, A.; Carlomagno, C.; Maiello, F.M.; et al. Sanger sequencing in routine KRAS testing: A review of 1720 cases from a pathologist’s perspective. J. Clin. Pathol. 2012, 65, 940–944. [Google Scholar] [CrossRef] [PubMed]
- Korphaisarn, K.; Pongpaibul, A.; Roothumnong, E.; Pongsuktavorn, K.; Thamlikitkul, L.; Anekpuritanang, T.; Poungvarin, N.; Thongnoppakhun, W.; Pithukpakorn, M. High frequency of KRAS codon 146 and FBXW7 mutations in Thai patients with stage II-III colon cancer. Asian Pacific. J. Cancer Prev. 2019, 20, 2319. [Google Scholar] [CrossRef]
- Cefalì, M.; Epistolio, S.; Palmarocchi, M.C.; Frattini, M.; De Dosso, S. Research progress on KRAS mutations in colorectal cancer. J. Cancer Metastasis Treat. 2021, 7, 26. [Google Scholar] [CrossRef]
- Sanchez-Ibarra, H.E.; Jiang, X.; Gallegos-Gonzalez, E.Y.; Cavazos-González, A.C.; Chen, Y.; Morcos, F.; Barrera-Saldaña, H.A. KRAS, NRAS, and BRAF mutation prevalence, clinicopathological association, and their application in a predictive model in Mexican patients with metastatic colorectal cancer: A retrospective cohort study. PLoS ONE 2020, 15, e0235490. [Google Scholar] [CrossRef]
- Ghee, L.K. A review of colorectal cancer research in Malaysia. Med. J. Malays. 2014, 69, 23–32. [Google Scholar]
- Sharif, R.; Mohammad, N.M.A.; Xin, Y.J.; Hamid, N.H.A.; Shahar, S.; Ali, R.A.R. Dietary Risk Factors and Odds of Colorectal Adenoma in Malaysia: A Case Control Study. Nutr. Cancer 2022, 74, 2757–2768. [Google Scholar] [CrossRef] [PubMed]
- Institute for Public Health, National Institute of Health & Malaysia, Ministry of Health Malaysia. National Health and Morbidity Survey (NHMS) 2019: Vol. I: NCDs—Non-Communicable Diseases: Risk Factors and other Health Problems; Institute for Public Health, National Institutes of Health, Ministry of Health Malaysia: Selangor, Malaysia, 2020. Available online: https://iku.gov.my/images/IKU/Document/REPORT/NHMS2019/Report_NHMS2019-HCD-eBook_p.pdf (accessed on 26 September 2022).
- Slattery, M.L.; Wolff, R.K.; Herrick, J.S. Diet, physical activity, and body size associations with rectal tumor mutations and epigenetic changes. Cancer Causes Control. 2010, 21, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- El Asri, A.; Zarrouq, B.; El Kinany, K.; Bouguenouch, L.; Ouldim, K.; El Rhazi, K. Associations between nutritional factors and KRAS mutations in colorectal cancer: A systematic review. BMC Cancer 2020, 20, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, Y.; Cai, S.; Yang, C.; Lin, Z.; Zhou, L.; Liu, L.; Cheng, X.; Zeng, W. Not all mutations of KRAS predict poor prognosis in patients with colorectal cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 957. [Google Scholar]
- Osterlund, E.; Ristimäki, A.; Kytölä, S.; Kuopio, T.; Heervä, E.; Muhonen, T.; Halonen, P.; Kallio, R.; Soveri, L.-M.; Sundström, J.; et al. KRAS-G12C Mutation in One Real-Life and Three Population-Based Nordic Cohorts of Metastatic Colorectal Cancer. Front. Oncol. 2022, 12, 274. [Google Scholar] [CrossRef]
- Fakih, M.G.; Kopetz, S.; Kuboki, Y.; Kim, T.W.; Munster, P.N.; Krauss, J.C.; Falchook, G.S.; Han, S.W.; Heinemann, V.; Muro, K.; et al. Sotorasib for previously treated colorectal cancers with KRAS G12C mutation (CodeBreaK100): A prespecified analysis of a single-arm, phase 2 trial. Lancet Oncol. 2022, 23, 115–124. [Google Scholar] [CrossRef]
- Zlobec, I.; Bihl, M.P.; Schwarb, H.; Terracciano, L.; Lugli, A. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int. J. Cancer 2010, 127, 367–380. [Google Scholar]
- Zihui Yong, Z.; Ching, G.T.H.; Ching, M.T.C. Metastatic Profile of Colorectal Cancer: Interplay Between Primary Tumor Location and KRAS Status. J. Surg. Res. 2020, 246, 325–334. [Google Scholar] [CrossRef]
- Xie, M.Z.; Li, J.L.; Cai, Z.M.; Li, K.Z.; Hu, B.L. Impact of primary colorectal Cancer location on the KRAS status and its prognostic value. BMC Gastroenterol. 2019, 19, 46. [Google Scholar] [CrossRef]
- Bylsma, L.C.; Gillezeau, C.; Garawin, T.A.; Kelsh, M.A.; Fryzek, J.P.; Sangaré, L.; Lowe, K.A. Prevalence of RAS and BRAF mutations in metastatic colorectal cancer patients by tumor sidedness: A systematic review and meta-analysis. Cancer Med. 2020, 9, 1044–1057. [Google Scholar] [CrossRef]
- Paschke, S.; Jafarov, S.; Staib, L.; Kreuser, E.-D.; Maulbecker-Armstrong, C.; Roitman, M.; Holm, T.; Harris, C.C.; Link, K.-H.; Kornmann, M. Are colon and rectal cancer two different tumor entities? A proposal to abandon the term colorectal cancer. Int. J. Mol. Sci. 2018, 19, 2577. [Google Scholar] [CrossRef]
- Mukund, K.; Syulyukina, N.; Ramamoorthy, S.; Subramaniam, S. Right and left-sided colon cancers-specificity of molecular mechanisms in tumorigenesis and progression. BMC Cancer 2020, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Baran, B.; Ozupek, N.M.; Tetik, N.Y.; Acar, E.; Bekcioglu, O.; Baskin, Y. Difference between left-sided and right-sided colorectal cancer: A focused review of literature. Gastroenterol Res 2018, 11, 264. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.R.; Kuk, J.K.; Kim, T.; Shin, E.J. Comparison of oncological outcomes of right-sided colon cancer versus left-sided colon cancer after curative resection. Medicine 2017, 96, e8241. [Google Scholar] [CrossRef] [PubMed]
- Hardiman, K.M. Update on Sporadic Colorectal Cancer Genetics. Clin. Colon Rectal Surg. 2018, 31, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Ooi, C.Y.; Hanafi, N.S.; Liew, S.M. Knowledge and practice of colorectal cancer screening in an urban setting: Cross-sectional survey of primary care physicians in government clinics in Malaysia. Singap. Med. J. 2019, 60, 596. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Health Malaysia. National Strategic Plan for Colorectal Cancer (2021–2025). 2021. Available online: https://www.moh.gov.my/moh/resources/Penerbitan/Rujukan/NCD/Kanser/National_Strategic_Plan_for_Colorectal_Cancer_(NSPCRC)_2021-2025.pdf (accessed on 18 September 2022).
- Yuen, S.T.; Davies, H.; Chan, T.L.; Ho, J.W.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Tsui, W.W.; Chan, A.S.; et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002, 62, 6451–6455. [Google Scholar]
- Artale, S.; Sartore-Bianchi, A.; Veronese, S.M.; Gambi, V.; Sarnataro, C.S.; Gambacorta, M.; Lauricella, C.; Siena, S. Mutations of KRAS and BRAF in Primary and Matched Metastatic Sites of Colorectal Cancer. J. Clin. Oncol. 2008, 26, 4217–4219. [Google Scholar] [CrossRef]
- Cejas, P.; López-Gómez, M.; Aguayo, C.; Madero, R.; Carpeño, J.D.C.; Belda-Iniesta, C.; Barriuso, J.; Garcia, V.M.; Larrauri, J.; Lopez, R.; et al. KRAS mutations in primary colorectal cancer tumors and related metastases: A potential role in prediction of lung metastasis. PLoS ONE 2009, 4, e8199. [Google Scholar] [CrossRef]
- Wojciechowicz, D.C.; Park, P.Y.; Datta, R.V.; Paty, P.B. CEA is the major PHA-L-reactive glycoprotein in colon carcinoma cell lines and tumors: Relationship between K-ras activation and β1-6 branching of N-linked carbohydrate on CEA. Biochem. Biophys. Res. Commun. 2000, 273, 147–153. [Google Scholar] [CrossRef]
- Selcukbiricik, F.; Bilici, A.; Tural, D.; Erdamar, S.; Soyluk, O.; Buyukunal, E.; Demirelli, F.; Serdengecti, S. Are high initial CEA and CA 19-9 levels associated with the presence of K-ras mutation in patients with metastatic colorectal cancer? Tumor Biol. 2013, 34, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Cao, Y.; Yang, J.; Li, H.; Wu, K.; Wang, J.; Peng, T.; Cai, K. Serum Tumor Markers Combined with Clinicopathological Characteristics for Predicting MMR and KRAS Status in 2279 Chinese Colorectal Cancer Patients: A Retrospective Analysis. Front. Oncol. 2021, 11, 582244. [Google Scholar] [CrossRef] [PubMed]
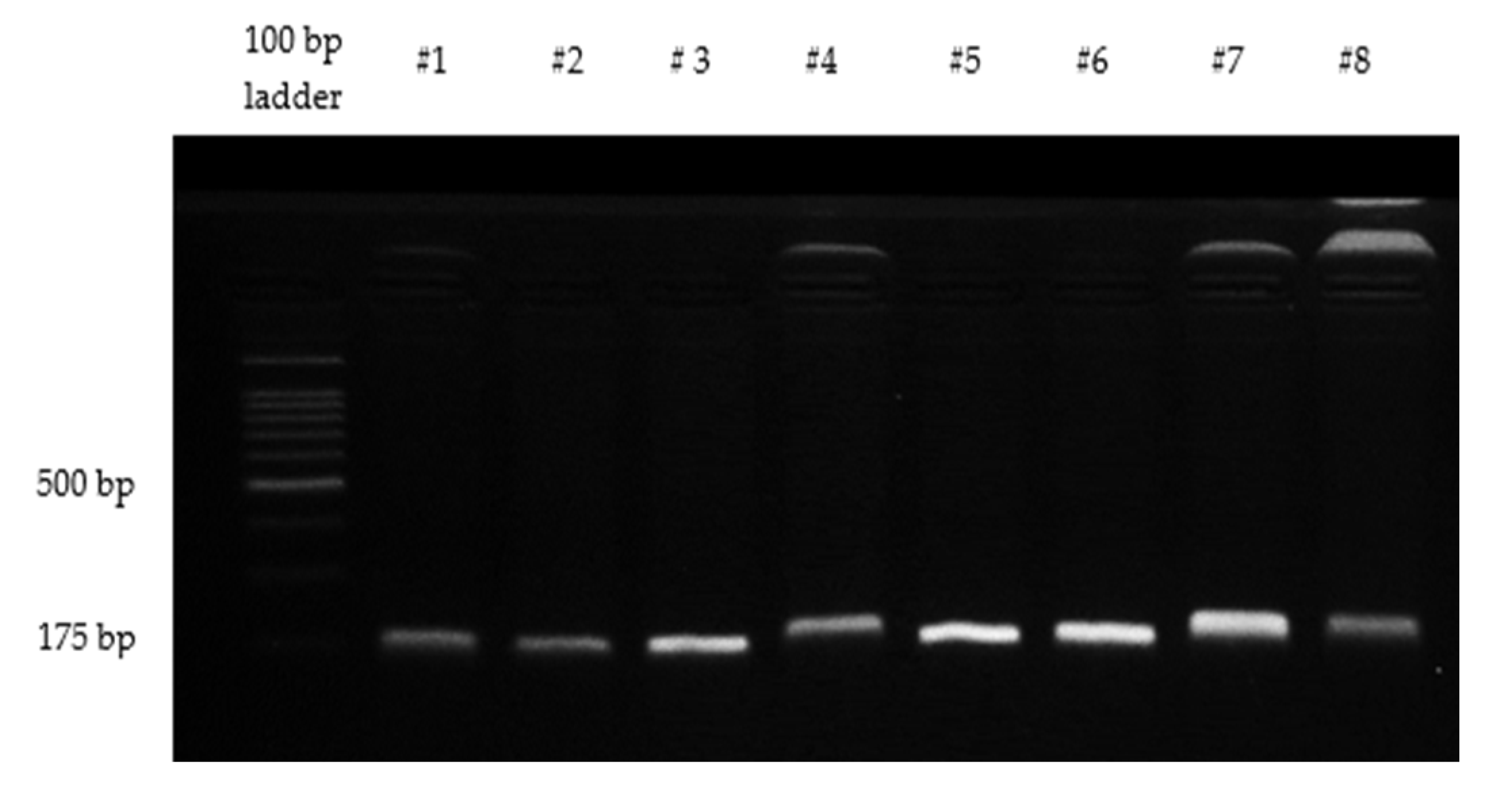

| Features | KRAS Status | p-Value | |
|---|---|---|---|
| Wild-Type n (%) | Mutant n (%) | ||
| Total | 21 (63.6) | 12 (36.4) | |
| Gender Male Female | 11 (73.3) 10 (55.6) | 4 (26.7) 8 (44.4) | 0.469 |
| Age (years) ≤60 >60 | 9 (56.3) 12 (70.6) | 7 (43.8) 5 (29.4) | 0.392 |
| Ethnicity Malay Chinese | 17 (60.7) 4 (80) | 11 (39.3) 1 (20) | 0.630 |
| Tumor site Right colon Left colon Rectum | 3 (100) 15 (60) 3 (60) | 0 (0) 10 (40) 2 (40) | 0.420 |
| Differentiation Well Moderate Poor | 4 (57.1) 16 (64.0) - | 3 (42.9) 9 (36.0) - | >0.950 |
| AJCC Stage I or II III IV | 7 (70) 6 (60) 8 (61.5) | 3 (30) 4 (40) 5 (38.5) | >0.950 |
| CEA (ng/mL) ≤5 >5 | 7 (77.8) 14 (58.3) | 2 (22.2) 10 (41.7) | 0.429 |
| Mutation | Base Change | Frequency, n (%) |
|---|---|---|
| G12D | c.35G > A | 6 (50) |
| G12V | c.35G > T | 3 (25) |
| G12S | c.34G > A | 1 (8.3) |
| G13D | c.38G > A | 2 (16.7) |
| Case | KRAS Mutation Status | Primary CRC Location | Corresponding Metastatic Site(s) |
|---|---|---|---|
| 3 | G12D | Left colon | Liver, Lung, Spine |
| 5 | G12V | Left colon | Lung |
| 6 | G12D | Left colon | Liver, Lung |
| 9 | G13D | Left colon | Lung, Adrenal |
| 11 | G13D | Left colon | Lung |
| 14 | Wild-type | Left colon | Liver |
| 16 | Wild-type | Left colon | Lung |
| 19 | Wild-type | Left colon | Liver, Lung |
| 23 | Wild-type | Left colon | Liver, Peritoneum |
| 24 | Wild-type | Rectum | Liver |
| 26 | Wild-type | Right colon | Liver, Lung |
| 28 | Wild-type | Left colon | Liver |
| 30 | Wild-type | Rectum | Liver, Lung |
| Case | Age (Years) | Race | Tumor | Initial CEA (ng/mL) | KRAS Mutations, Amino Acid Changes | ||
|---|---|---|---|---|---|---|---|
| Site | Differentiation | Stage | |||||
| 1 | 30 | Malay | Rectum | Well | I | 0.3 | c.35G > A, G12D |
| 2 | 72 | Malay | Left side | Well | III | 3.8 | c.34G > A, G12S |
| 3 | 51 | Chinese | Left side | Moderate | IV | 914.8 | c.35G > A, G12D |
| 4 | 58 | Malay | Left side | Moderate | III | 12.6 | c.35G > A, G12D |
| 5 | 71 | Malay | Left side | Moderate | IV | 28.1 | c.35G > T, G12V |
| 6 | 39 | Malay | Left side | Moderate | IV | 166.9 | c.35G > A, G12D |
| 7 | 82 | Malay | Left side | Well | I | 12.2 | c.35G > A, G12D |
| 8 | 40 | Malay | Left side | Moderate | III | 66.5 | c.35G > A, G12D |
| 9 | 60 | Malay | Left side | Moderate | IV | 173.2 | c.38G > A, G13D |
| 10 | 83 | Malay | Rectum | Moderate | III | 25.0 | c.35G > T, G12V |
| 11 | 78 | Malay | Left side | Moderate | IV | 276.1 | c.38G > A, G13D |
| 12 | 59 | Malay | Left side | Moderate | II | 27.0 | c.35G > T, G12V |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasbullah, H.H.; Sulong, S.; Che Jalil, N.A.; Abdul Aziz, A.A.; Musa, N.; Musa, M. KRAS Mutational Profiles among Colorectal Cancer Patients in the East Coast of Peninsular Malaysia. Diagnostics 2023, 13, 822. https://doi.org/10.3390/diagnostics13050822
Hasbullah HH, Sulong S, Che Jalil NA, Abdul Aziz AA, Musa N, Musa M. KRAS Mutational Profiles among Colorectal Cancer Patients in the East Coast of Peninsular Malaysia. Diagnostics. 2023; 13(5):822. https://doi.org/10.3390/diagnostics13050822
Chicago/Turabian StyleHasbullah, Hidayati Husainy, Sarina Sulong, Nur Asyilla Che Jalil, Ahmad Aizat Abdul Aziz, Nurfadhlina Musa, and Marahaini Musa. 2023. "KRAS Mutational Profiles among Colorectal Cancer Patients in the East Coast of Peninsular Malaysia" Diagnostics 13, no. 5: 822. https://doi.org/10.3390/diagnostics13050822
APA StyleHasbullah, H. H., Sulong, S., Che Jalil, N. A., Abdul Aziz, A. A., Musa, N., & Musa, M. (2023). KRAS Mutational Profiles among Colorectal Cancer Patients in the East Coast of Peninsular Malaysia. Diagnostics, 13(5), 822. https://doi.org/10.3390/diagnostics13050822






