Validation of an Automated System for the Extraction of a Wide Dataset for Clinical Studies Aimed at Improving the Early Diagnosis of Candidemia
Abstract
1. Introduction
2. Methods
2.1. Setting and Objective
2.2. Definitions
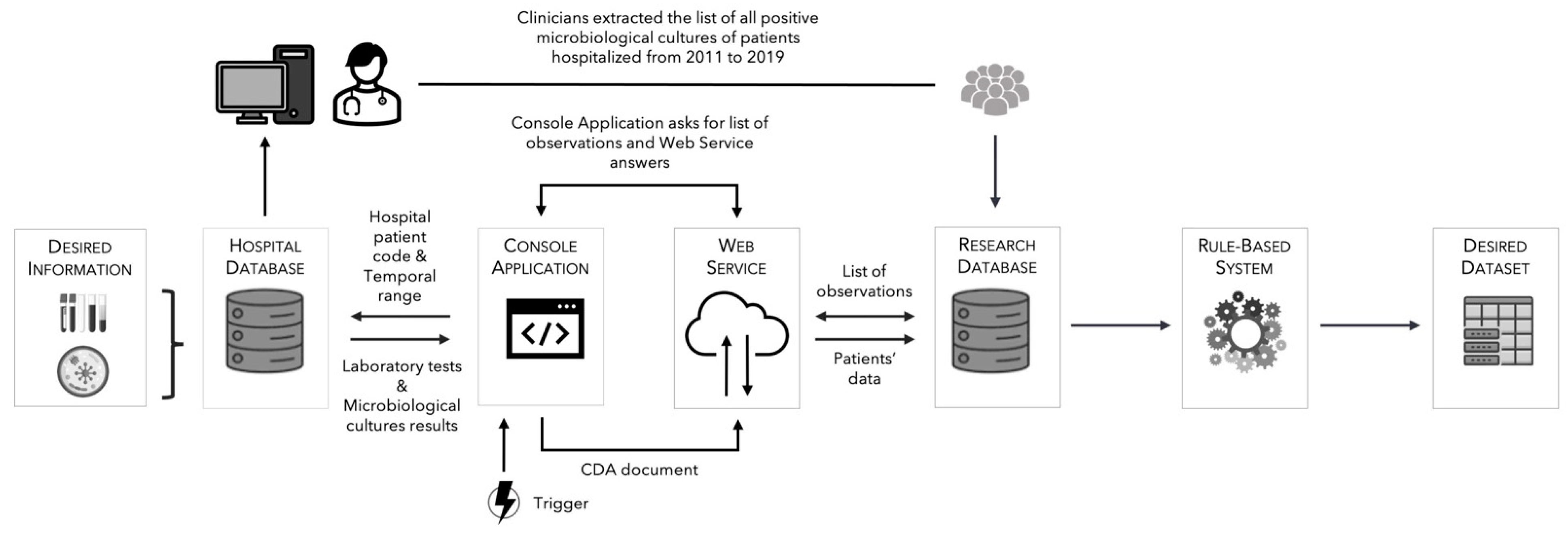
2.3. Data Collected for the Study
2.4. Manual Validation Procedure
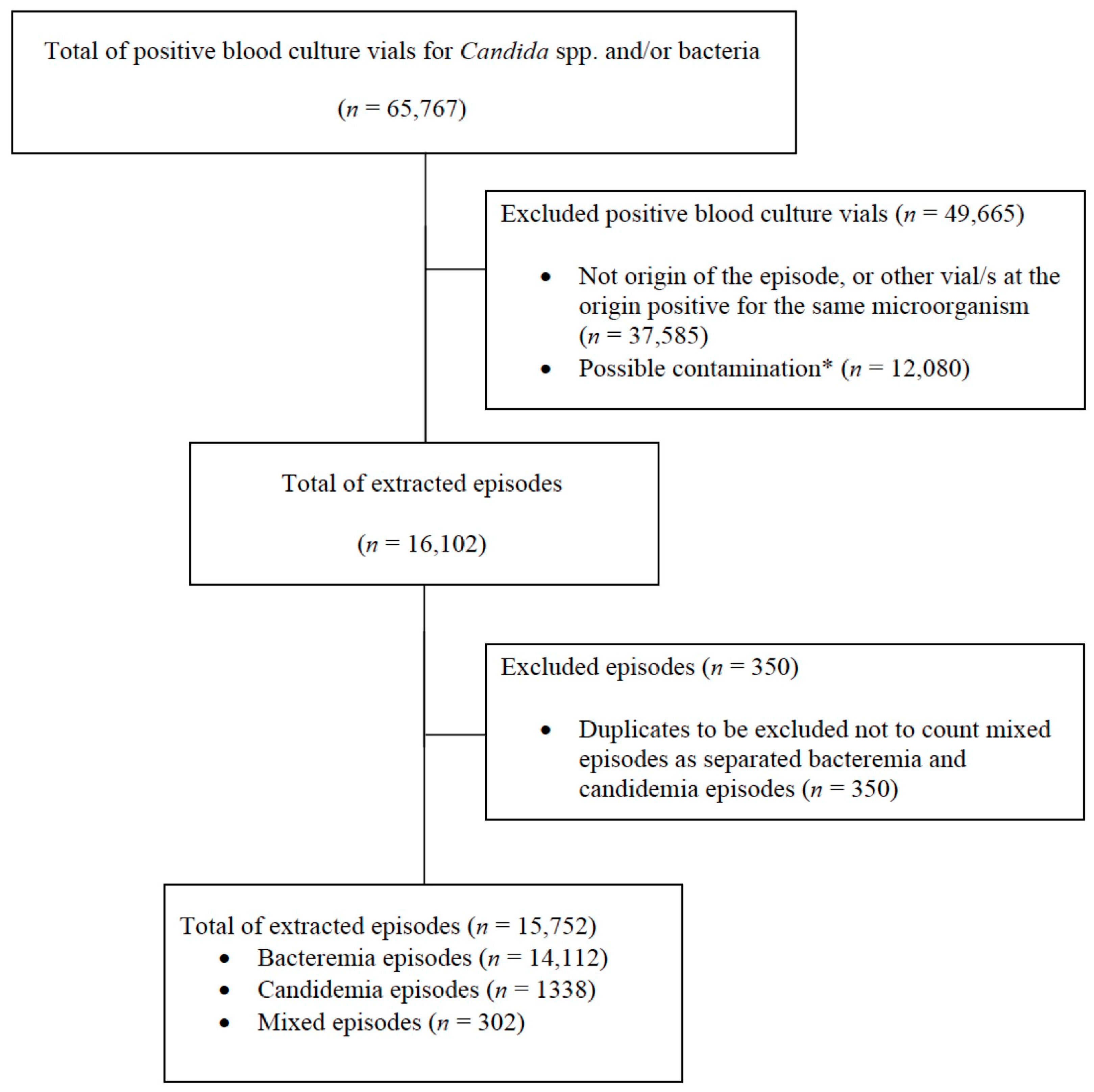
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef]
- Bouza, E.; Munoz, P. Epidemiology of candidemia in intensive care units. Int. J. Antimicrob. Agents 2008, 32 (Suppl. S2), S87–S91. [Google Scholar] [CrossRef]
- Bougnoux, M.E.; Kac, G.; Aegerter, P.; d’Enfert, C.; Fagon, J.Y.; CandiRea Study, G. Candidemia and candiduria in critically ill patients admitted to intensive care units in France: Incidence, molecular diversity, management and outcome. Intensiv. Care Med. 2008, 34, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Righi, E.; Ansaldi, F.; Merelli, M.; Trucchi, C.; De Pascale, G.; Diaz-Martin, A.; Luzzati, R.; Rosin, C.; Lagunes, L.; et al. A multicenter study of septic shock due to candidemia: Outcomes and predictors of mortality. Intensiv. Care Med. 2014, 40, 839–845. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Vena, A.; Trucchi, C.; Ansaldi, F.; Antonelli, M.; Adamkova, V.; Alicino, C.; Almyroudi, M.P.; Atchade, E.; et al. Incidence and outcome of invasive candidiasis in intensive care units (ICUs) in Europe: Results of the EUCANDICU project. Crit. Care 2019, 23, 219. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Prim. 2018, 4, 18026. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Vena, A.; Wolff, M. Diagnosis and Treatment of Candidemia in the Intensive Care Unit. Semin. Respir. Crit. Care Med. 2019, 40, 524–539. [Google Scholar] [CrossRef]
- Johnson, M.D.; Lewis, R.E.; Dodds Ashley, E.S.; Ostrosky-Zeichner, L.; Zaoutis, T.; Thompson, G.R.; Andes, D.R.; Walsh, T.J.; Pappas, P.G.; Cornely, O.A.; et al. Core Recommendations for Antifungal Stewardship: A Statement of the Mycoses Study Group Education and Research Consortium. J. Infect. Dis. 2020, 222, S175–S198. [Google Scholar] [CrossRef]
- Giacobbe, D.R.; Signori, A.; Tumbarello, M.; Ungaro, R.; Sarteschi, G.; Furfaro, E.; Mikulska, M.; Sanguinetti, M.; Posteraro, B.; Losito, A.R.; et al. Desirability of outcome ranking (DOOR) for comparing diagnostic tools and early therapeutic choices in patients with suspected candidemia. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Rautemaa-Richardson, R.; Rautemaa, V.; Al-Wathiqi, F.; Moore, C.B.; Craig, L.; Felton, T.W.; Muldoon, E.G. Impact of a diagnostics-driven antifungal stewardship programme in a UK tertiary referral teaching hospital. J. Antimicrob. Chemother. 2018, 73, 3488–3495. [Google Scholar] [CrossRef]
- Yera, H.; Sendid, B.; Francois, N.; Camus, D.; Poulain, D. Contribution of serological tests and blood culture to the early diagnosis of systemic candidiasis. Eur. J. Clin. Microbiol. Infect. Dis. 2001, 20, 864–870. [Google Scholar] [CrossRef]
- White, P.L.; Archer, A.E.; Barnes, R.A. Comparison of non-culture-based methods for detection of systemic fungal infections, with an emphasis on invasive Candida infections. J. Clin. Microbiol. 2005, 43, 2181–2187. [Google Scholar] [CrossRef]
- Wei, S.; Wu, T.; Wu, Y.; Ming, D.; Zhu, X. Diagnostic accuracy of Candida albicans germ tube antibody for invasive candidiasis: Systematic review and meta-analysis. Diagn. Microbiol. Infect. Dis. 2019, 93, 339–345. [Google Scholar] [CrossRef]
- Walker, B.; Powers-Fletcher, M.V.; Schmidt, R.L.; Hanson, K.E. Cost-Effectiveness Analysis of Multiplex PCR with Magnetic Resonance Detection versus Empiric or Blood Culture-Directed Therapy for Management of Suspected Candidemia. J. Clin. Microbiol. 2016, 54, 718–726. [Google Scholar] [CrossRef]
- Rouze, A.; for the S-TAFE Study Group; Loridant, S.; Poissy, J.; Dervaux, B.; Sendid, B.; Cornu, M.; Nseir, S. Biomarker-based strategy for early discontinuation of empirical antifungal treatment in critically ill patients: A randomized controlled trial. Intensiv. Care Med. 2017, 43, 1668–1677. [Google Scholar] [CrossRef] [PubMed]
- Raineri, S.M.; Cortegiani, A.; Vitale, F.; Iozzo, P.; Giarratano, A. Procalcitonin for the diagnosis of invasive candidiasis: What is the evidence? J. Intensiv. Care 2017, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Tumbarello, M.; De Pascale, G.; Liberto, E.; Vallecoccia, M.S.; De Carolis, E.; Di Gravio, V.; Trecarichi, E.M.; Sanguinetti, M.; Antonelli, M. (1,3)-beta-d-Glucan-based antifungal treatment in critically ill adults at high risk of candidaemia: An observational study. J. Antimicrob. Chemother. 2016, 71, 2262–2269. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; De Pascale, G.; Tumbarello, M.; Torelli, R.; Pennisi, M.A.; Bello, G.; Maviglia, R.; Fadda, G.; Sanguinetti, M.; Antonelli, M. Early diagnosis of candidemia in intensive care unit patients with sepsis: A prospective comparison of (1-->3)-beta-D-glucan assay, Candida score, and colonization index. Crit. Care 2011, 15, R249. [Google Scholar] [CrossRef]
- Paphitou, N.I.; Ostrosky-Zeichner, L.; Rex, J.H. Rules for identifying patients at increased risk for candidal infections in the surgical intensive care unit: Approach to developing practical criteria for systematic use in antifungal prophylaxis trials. Med. Mycol. 2005, 43, 235–243. [Google Scholar] [CrossRef]
- Ostrosky-Zeichner, L.; Sable, C.; Sobel, J.; Alexander, B.D.; Donowitz, G.; Kan, V.; Kauffman, C.A.; Kett, D.; Larsen, R.A.; Morrison, V.; et al. Multicenter retrospective development and validation of a clinical prediction rule for nosocomial invasive candidiasis in the intensive care setting. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 271–276. [Google Scholar] [CrossRef]
- Mikulska, M.; Giacobbe, D.R.; Furfaro, E.; Mesini, A.; Marchese, A.; Del Bono, V.; Viscoli, C. Lower sensitivity of serum (1,3)-beta-d-glucan for the diagnosis of candidaemia due to Candida parapsilosis. Clin. Microbiol Infect. 2016, 22. [Google Scholar] [CrossRef]
- Martinez-Jimenez, M.C.; Munoz, P.; Valerio, M.; Alonso, R.; Martos, C.; Guinea, J.; Bouza, E. Candida biomarkers in patients with candidaemia and bacteraemia. J. Antimicrob. Chemother. 2015, 70, 2354–2361. [Google Scholar] [CrossRef]
- Leon, C.; Ruiz-Santana, S.; Saavedra, P.; Almirante, B.; Nolla-Salas, J.; Alvarez-Lerma, F.; Garnacho-Montero, J.; Leon, M.A.; Group, E.S. A bedside scoring system (“Candida score”) for early antifungal treatment in nonneutropenic critically ill patients with Candida colonization. Crit. Care Med. 2006, 34, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Giannella, M.; Paolucci, M.; Roncarati, G.; Vandi, G.; Pascale, R.; Trapani, F.; Bartoletti, M.; Tedeschi, S.; Ambretti, S.; Lewis, R.; et al. Potential role of T2Candida in the management of empirical antifungal treatment in patients at high risk of candidaemia: A pilot single-centre study. J. Antimicrob. Chemother. 2018, 73, 2856–2859. [Google Scholar] [CrossRef] [PubMed]
- Giacobbe, D.R.; Mikulska, M.; Tumbarello, M.; Furfaro, E.; Spadaro, M.; Losito, A.R.; Mesini, A.; De Pascale, G.; Marchese, A.; Bruzzone, M.; et al. Combined use of serum (1,3)-beta-D-glucan and procalcitonin for the early differential diagnosis between candidaemia and bacteraemia in intensive care units. Crit. Care 2017, 21, 176. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Andersen, J.S.; Holten, M.K.; Krarup, K.B.; Reiter, N.; Schierbeck, J.; Helleberg, M. Diagnostic Performance of T2Candida Among ICU Patients With Risk Factors for Invasive Candidiasis. Open Forum. Infect. Dis. 2019, 6, ofz136. [Google Scholar] [CrossRef]
- Mikulska, M.; Magnasco, L.; Signori, A.; Sepulcri, C.; Dettori, S.; Tutino, S.; Vena, A.; Miletich, F.; Ullah, N.; Morici, P.; et al. Sensitivity of Serum Beta-D-Glucan in Candidemia According to Candida Species Epidemiology in Critically Ill Patients Admitted to the Intensive Care Unit. J. Fungi. 2022, 8, 921. [Google Scholar] [CrossRef]
- Yoo, J.; Kim, S.H.; Hur, S.; Ha, J.; Huh, K.; Cha, W.C. Candidemia Risk Prediction (CanDETEC) Model for Patients With Malignancy: Model Development and Validation in a Single-Center Retrospective Study. JMIR Med. Inf. 2021, 9, e24651. [Google Scholar] [CrossRef]
- Ripoli, A.; Sozio, E.; Sbrana, F.; Bertolino, G.; Pallotto, C.; Cardinali, G.; Meini, S.; Pieralli, F.; Azzini, A.M.; Concia, E.; et al. Personalized machine learning approach to predict candidemia in medical wards. Infection 2020, 48, 749–759. [Google Scholar] [CrossRef]
- Ngiam, K.Y.; Khor, I.W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019, 20, e262–e273. [Google Scholar] [CrossRef] [PubMed]
- Giacobbe, D.R.; Signori, A.; Del Puente, F.; Mora, S.; Carmisciano, L.; Briano, F.; Vena, A.; Ball, L.; Robba, C.; Pelosi, P.; et al. Early Detection of Sepsis With Machine Learning Techniques: A Brief Clinical Perspective. Front. Med. 2021, 8, 617486. [Google Scholar] [CrossRef]
- Beam, A.L.; Kohane, I.S. Big Data and Machine Learning in Health Care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef] [PubMed]
- Mora, S.; Giacobbe, D.R.; Russo, C.; Diana, E.; Signori, A.; Carmisciano, L.; Bassetti, M.; Giacomini, M. A Wide Database for Future Studies Aimed at Improving Early Recognition of Candidemia. Stud. Health Technol. Inf. 2021, 281, 1081–1082. [Google Scholar] [CrossRef]
- Giannini, B.; Riccardi, N.; Cenderello, G.; Di Biagio, A.; Dentone, C.; Giacomini, M. From Liguria HIV Web to Liguria Infectious Diseases Network: How a Digital Platform Improved Doctors’ Work and Patients’ Care. AIDS Res. Hum. Retrovir. 2018, 34, 239–240. [Google Scholar] [CrossRef]
- Mora, S.; Giannini, B.; Di Biagio, A.; Cenderello, G.; Nicolini, L.A.; Taramasso, L.; Dentone, C.; Bassetti, M.; Giacomini, M. Ten Years of Medical Informatics and Standards Support for Clinical Research in an Infectious Diseases Network. Appl. Clin. Inf. 2023, 14, 16–27. [Google Scholar] [CrossRef]
- Gazzarata, R.; Giannini, B.; Giacomini, M. A SOA-Based Platform to Support Clinical Data Sharing. J. Healthc. Eng. 2017, 2017, 2190679. [Google Scholar] [CrossRef]
- Kim, S.H.; Yoon, Y.K.; Kim, M.J.; Sohn, J.W. Risk factors for and clinical implications of mixed Candida/bacterial bloodstream infections. Clin. Microbiol. Infect. 2013, 19, 62–68. [Google Scholar] [CrossRef]
- ECDC. Healthcare-Associated Infections Acquired in Intensive Care Units. Annual Epidemiological Report for 2017. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2017-HAI.pdf (accessed on 30 October 2022).
- van Dijk, W.B.; Fiolet, A.T.L.; Schuit, E.; Sammani, A.; Groenhof, T.K.J.; van der Graaf, R.; de Vries, M.C.; Alings, M.; Schaap, J.; Asselbergs, F.W.; et al. Text-mining in electronic healthcare records can be used as efficient tool for screening and data collection in cardiovascular trials: A multicenter validation study. J. Clin. Epidemiol. 2021, 132, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Sirgo, G.; Esteban, F.; Gómez, J.; Moreno, G.; Rodríguez, A.; Blanch, L.; Guardiola, J.J.; Gracia, R.; De Haro, L.; Bodí, M. Validation of the ICU-DaMa tool for automatically extracting variables for minimum dataset and quality indicators: The importance of data quality assessment. Int. J. Med. Inform. 2018, 112, 166–172. [Google Scholar] [CrossRef]
- Kozak, M.; Krzanowski, W.; Cichocka, I.; Hartley, J. The effects of data input errors on subsequent statistical inference. J. Appl. Stat. 2015, 42, 2030–2037. [Google Scholar] [CrossRef]
- Hammond, K.W.; Helbig, S.T.; Benson, C.C.; Brathwaite-Sketoe, B.M. Are electronic medical records trustworthy? Observations on copying, pasting and duplication. AMIA Annu. Symp. Proc. 2003, 2003, 269–273. [Google Scholar]
- Mora, S.; Attene, J.; Gazzarata, R.; Giacobbe, D.R.; Blobel, B.; Parruti, G.; Giacomini, M. A NLP Pipeline for the Automatic Extraction of a Complete Microorganism’s Picture from Microbiological Notes. J. Pers. Med. 2022, 12, 1424. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giacobbe, D.R.; Mora, S.; Signori, A.; Russo, C.; Brucci, G.; Campi, C.; Guastavino, S.; Marelli, C.; Limongelli, A.; Vena, A.; et al. Validation of an Automated System for the Extraction of a Wide Dataset for Clinical Studies Aimed at Improving the Early Diagnosis of Candidemia. Diagnostics 2023, 13, 961. https://doi.org/10.3390/diagnostics13050961
Giacobbe DR, Mora S, Signori A, Russo C, Brucci G, Campi C, Guastavino S, Marelli C, Limongelli A, Vena A, et al. Validation of an Automated System for the Extraction of a Wide Dataset for Clinical Studies Aimed at Improving the Early Diagnosis of Candidemia. Diagnostics. 2023; 13(5):961. https://doi.org/10.3390/diagnostics13050961
Chicago/Turabian StyleGiacobbe, Daniele Roberto, Sara Mora, Alessio Signori, Chiara Russo, Giorgia Brucci, Cristina Campi, Sabrina Guastavino, Cristina Marelli, Alessandro Limongelli, Antonio Vena, and et al. 2023. "Validation of an Automated System for the Extraction of a Wide Dataset for Clinical Studies Aimed at Improving the Early Diagnosis of Candidemia" Diagnostics 13, no. 5: 961. https://doi.org/10.3390/diagnostics13050961
APA StyleGiacobbe, D. R., Mora, S., Signori, A., Russo, C., Brucci, G., Campi, C., Guastavino, S., Marelli, C., Limongelli, A., Vena, A., Mikulska, M., Marchese, A., Di Biagio, A., Giacomini, M., & Bassetti, M. (2023). Validation of an Automated System for the Extraction of a Wide Dataset for Clinical Studies Aimed at Improving the Early Diagnosis of Candidemia. Diagnostics, 13(5), 961. https://doi.org/10.3390/diagnostics13050961









