Phenotyping the Histopathological Subtypes of Non-Small-Cell Lung Carcinoma: How Beneficial Is Radiomics?
Abstract
:1. Introduction
1.1. Related Works
1.2. Research Motivation and Contribution
- Investigates the possibility of building a multiclass multicenter radiomics-based machine learning model, which is capable of differentiating between the NSCLC subtypes with the aim of improving treatment personalization;
- Evaluates the presence of batch effects in a multicenter study with the aim to assess the impact of feature harmonization on machine learning models;
- Evaluates the stability of the feature selection procedure by iterating the machine learning modeling pipeline 10 times with the aim of reducing the result variability;
- Shows that selected feature subsets are influenced by training/testing set splitting;
- Provides a scientific and critical analysis of the advantages and disadvantages of radiomics in the absence of well-defined standard guidelines with the aim of promoting the need for reproducible and repeatable radiomics studies.
2. Materials and Methods
2.1. Data Preparation and Image Segmentation
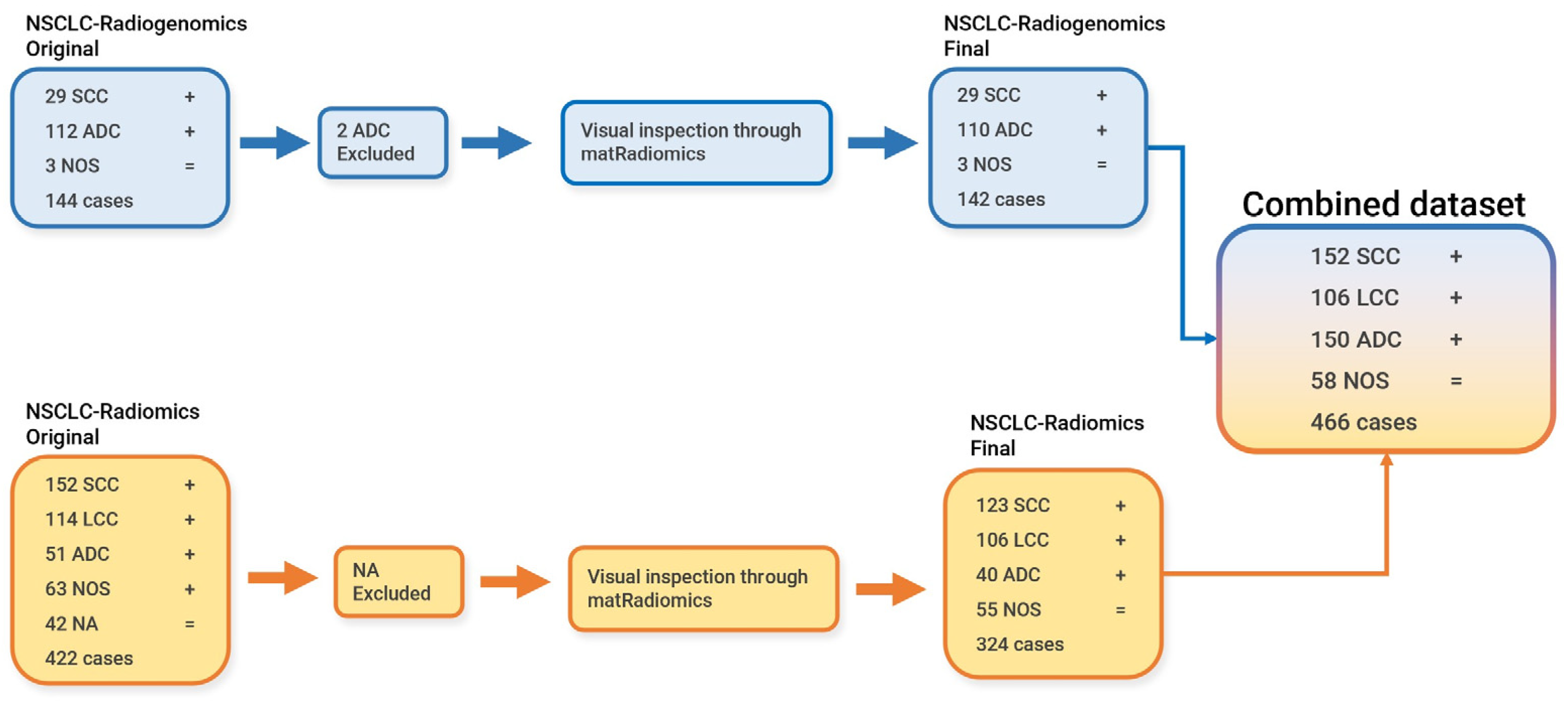
2.2. NSCLC-Radiomics Dataset
2.3. NSCLC-Radiogenomics
2.3.1. CT Images’ Pixel Spacing, Slice Thickness, Matrix Dimension, and Manufacturers
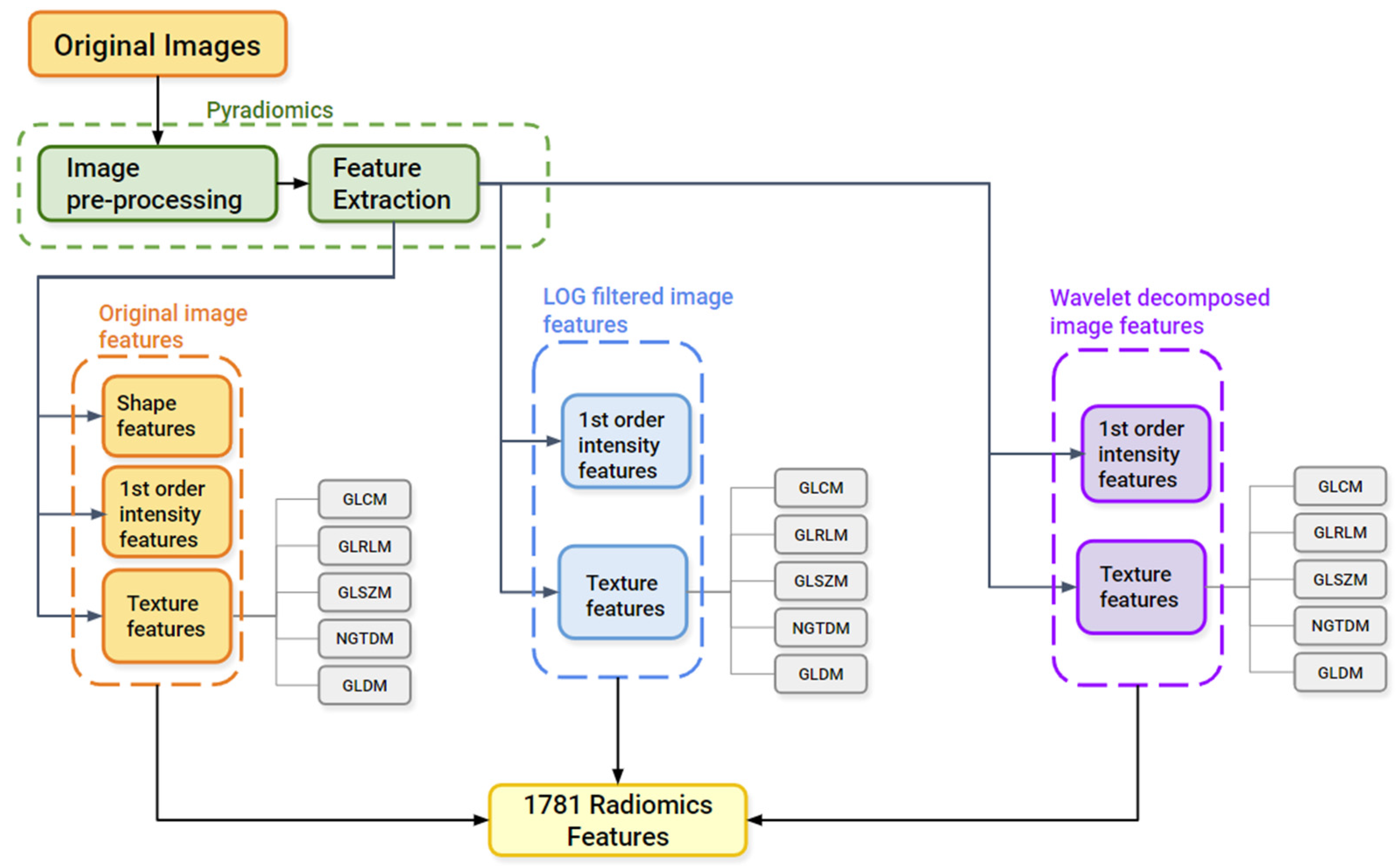
2.3.2. Image Pre-Processing and Feature Extraction
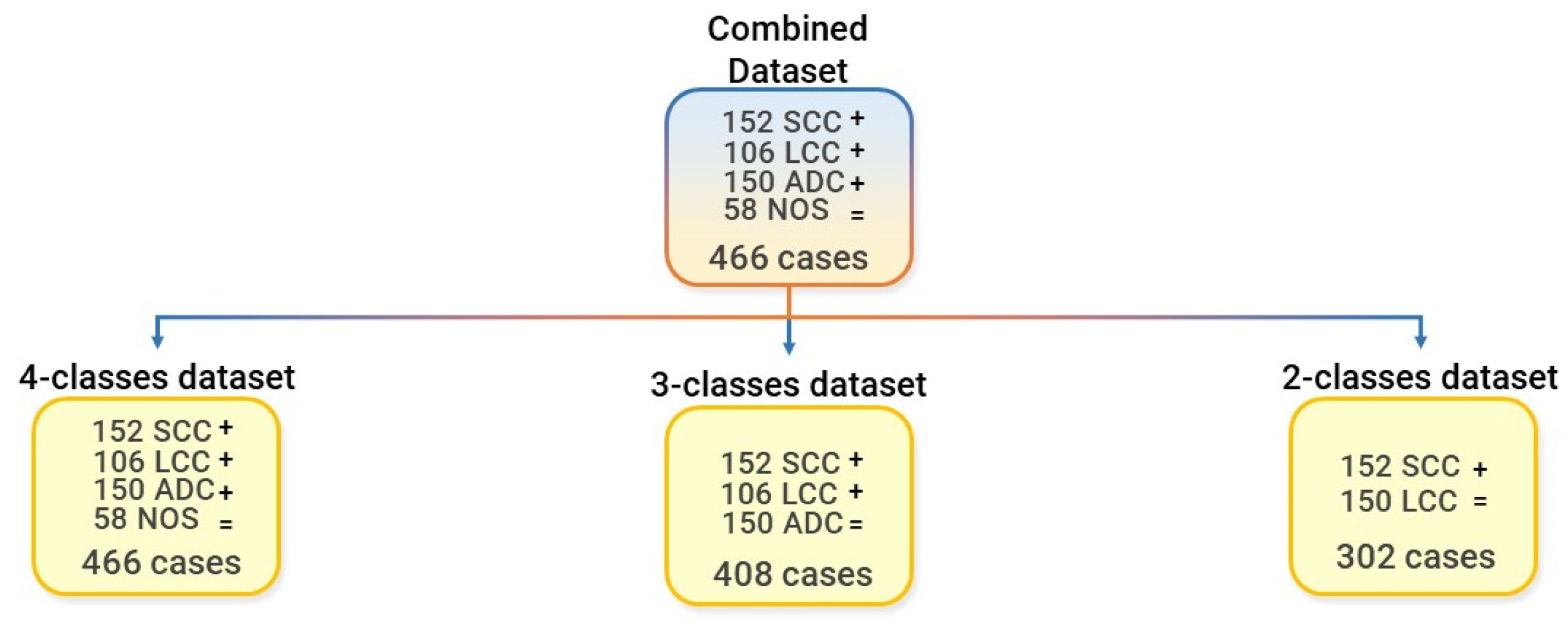
2.3.3. Dataset Preparation for Batch Analysis, Feature Harmonization, and for the Machine Learning Modeling Pipeline
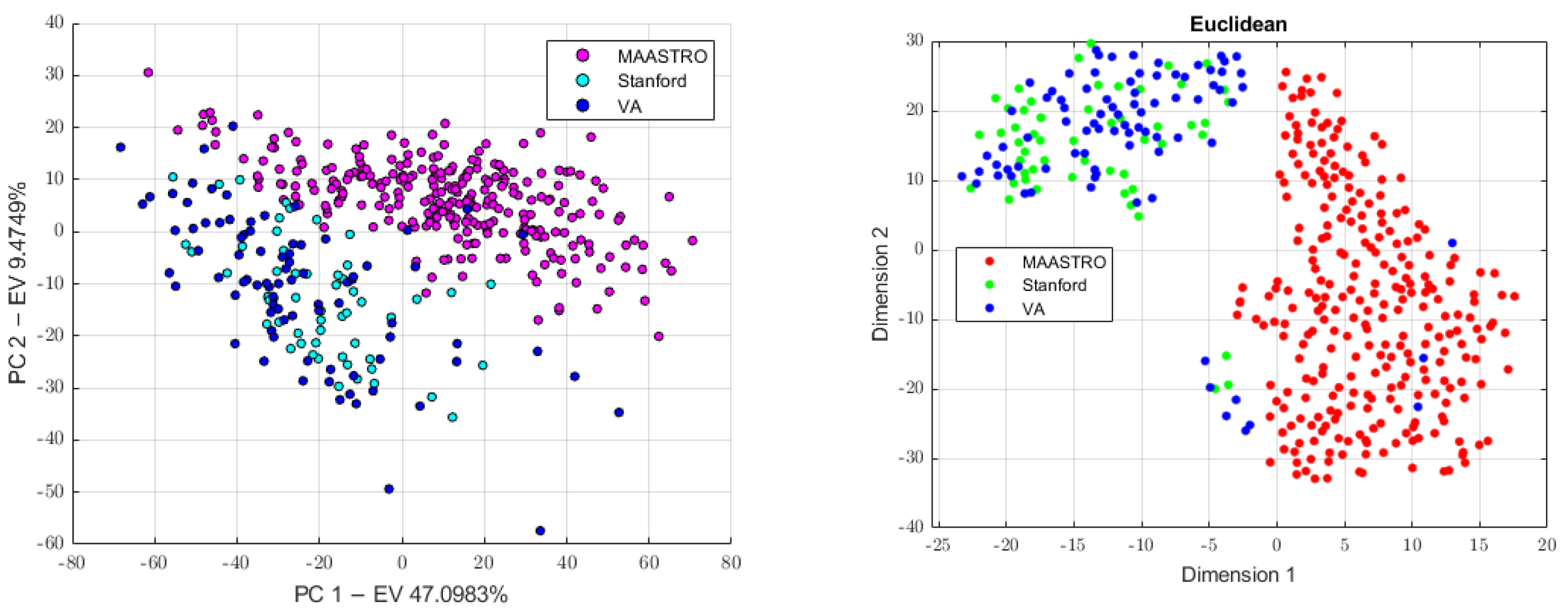
2.3.4. Batch Analysis and Feature Harmonization
2.3.5. Machine Learning Modeling Pipeline
2.4. Feature Selection and Feature Stability
2.5. Classification
Software Used for The Radiomics Analysis
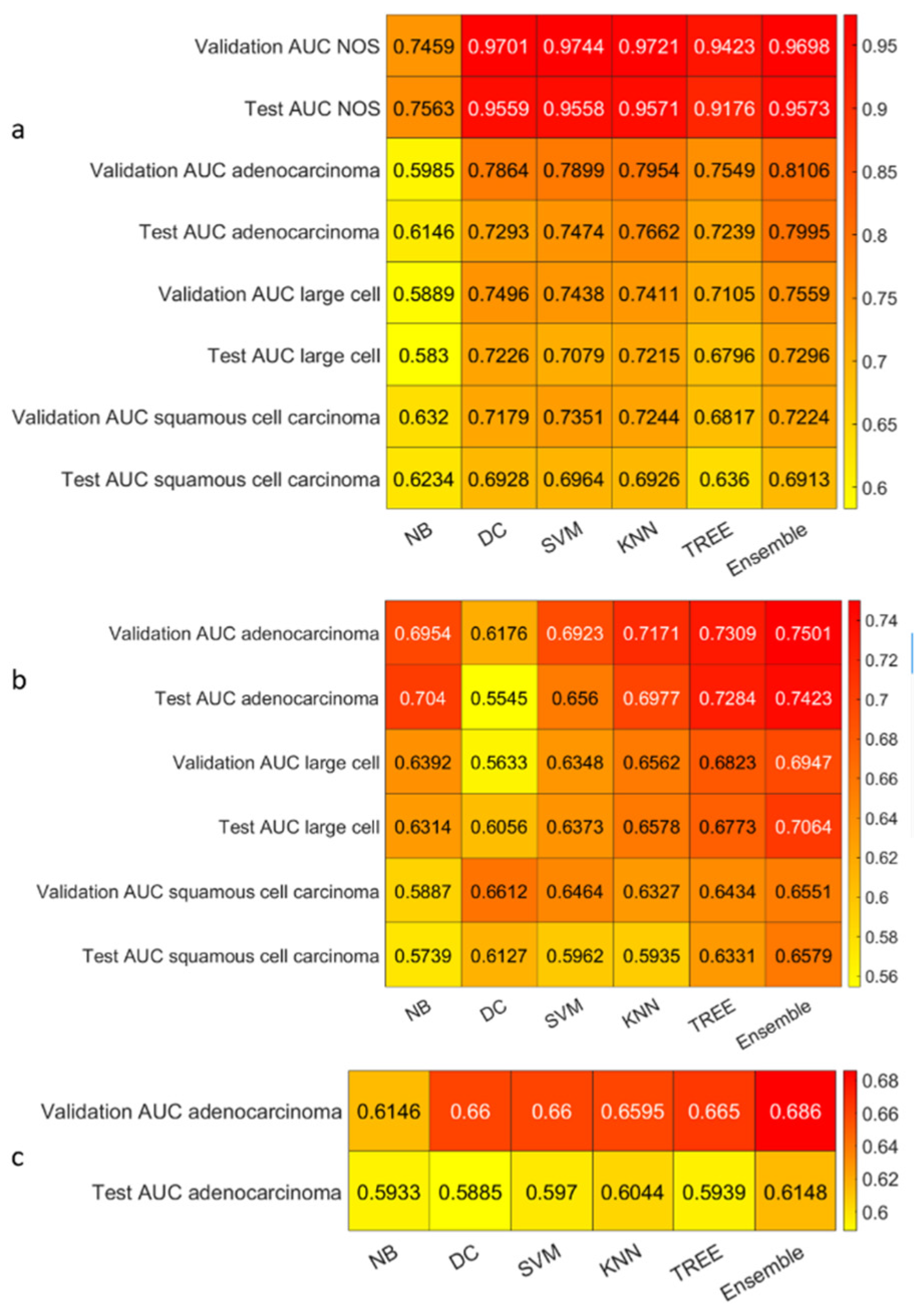
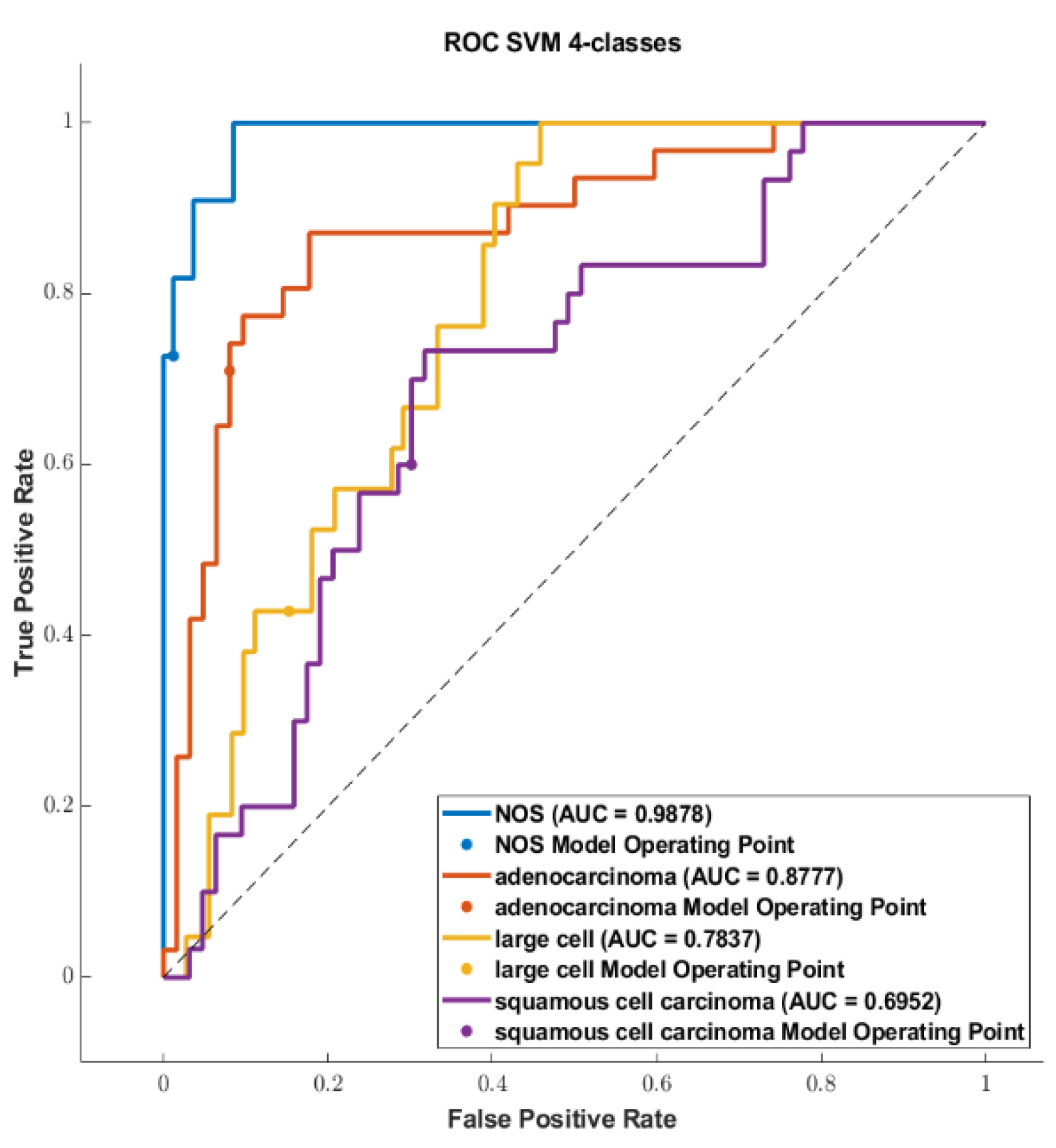
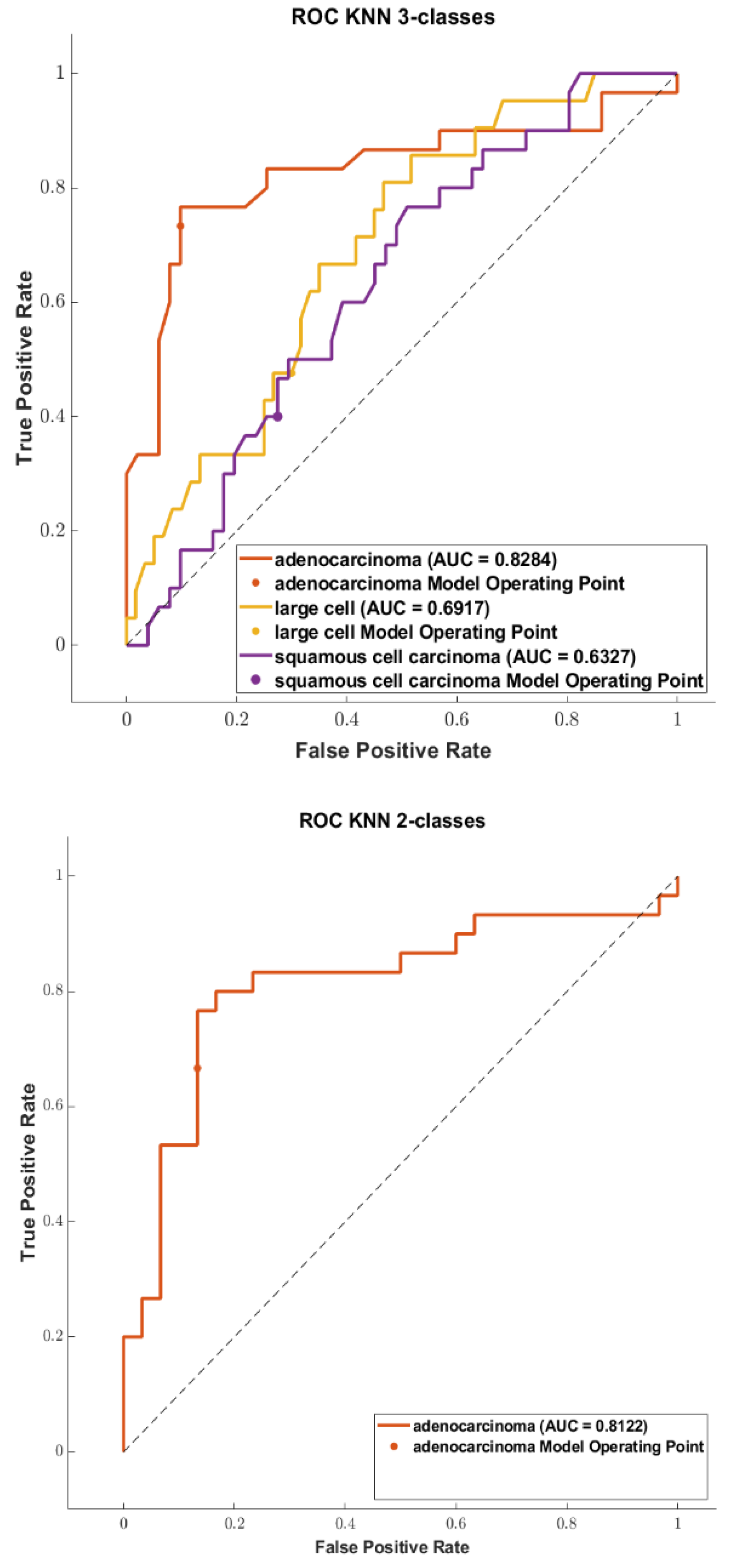
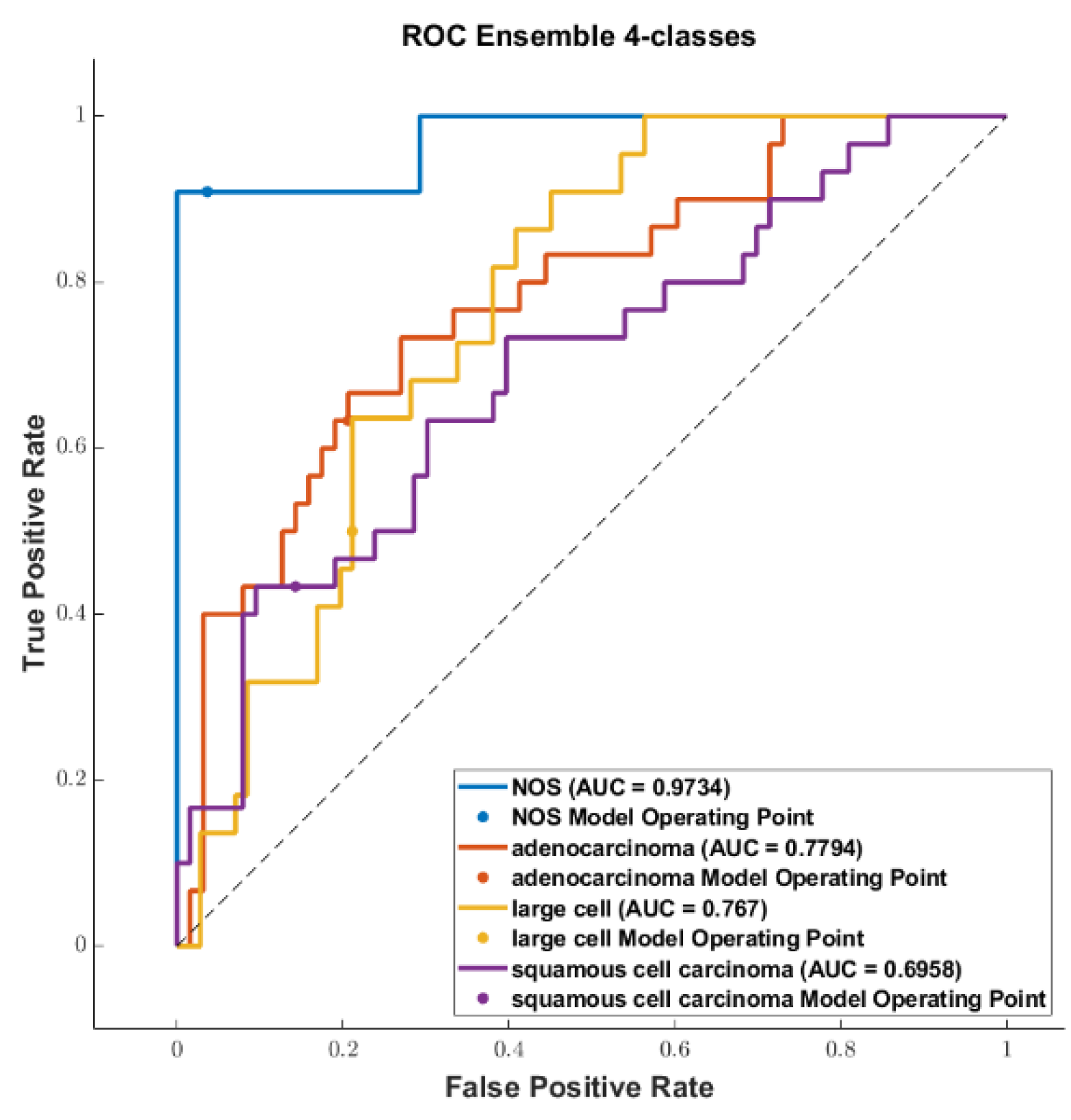
3. Results
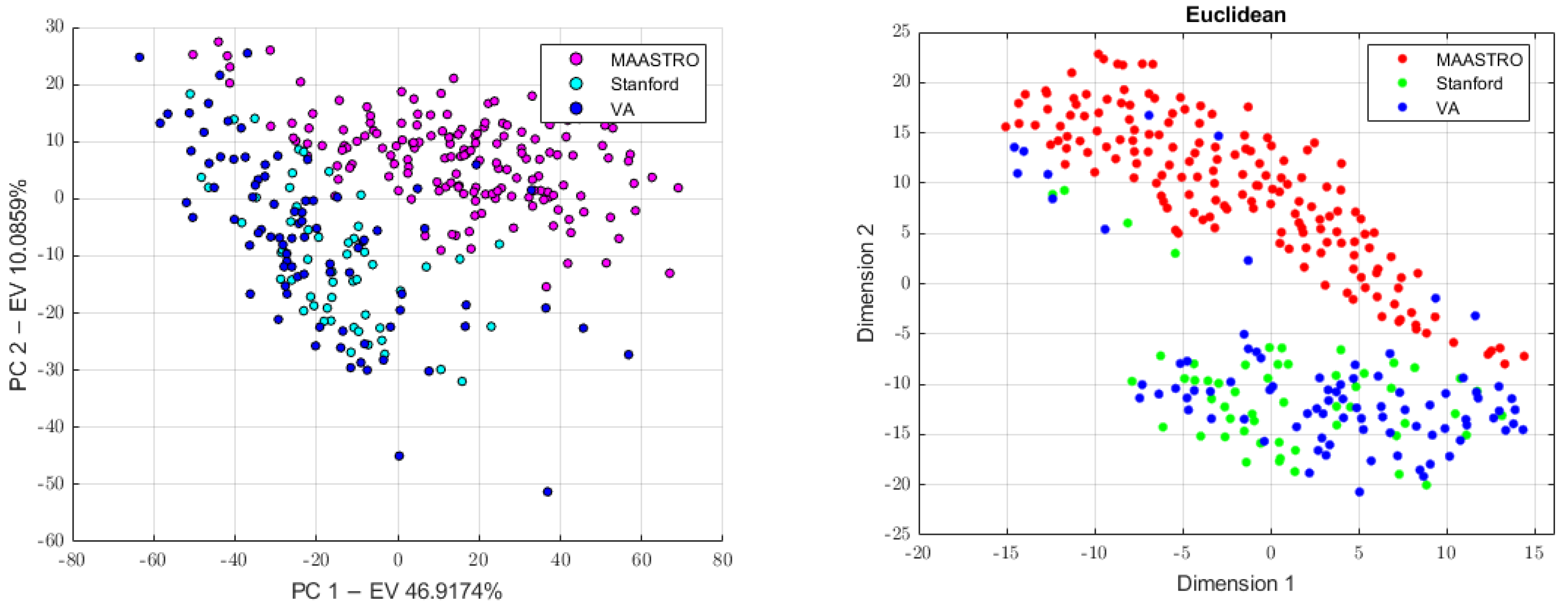
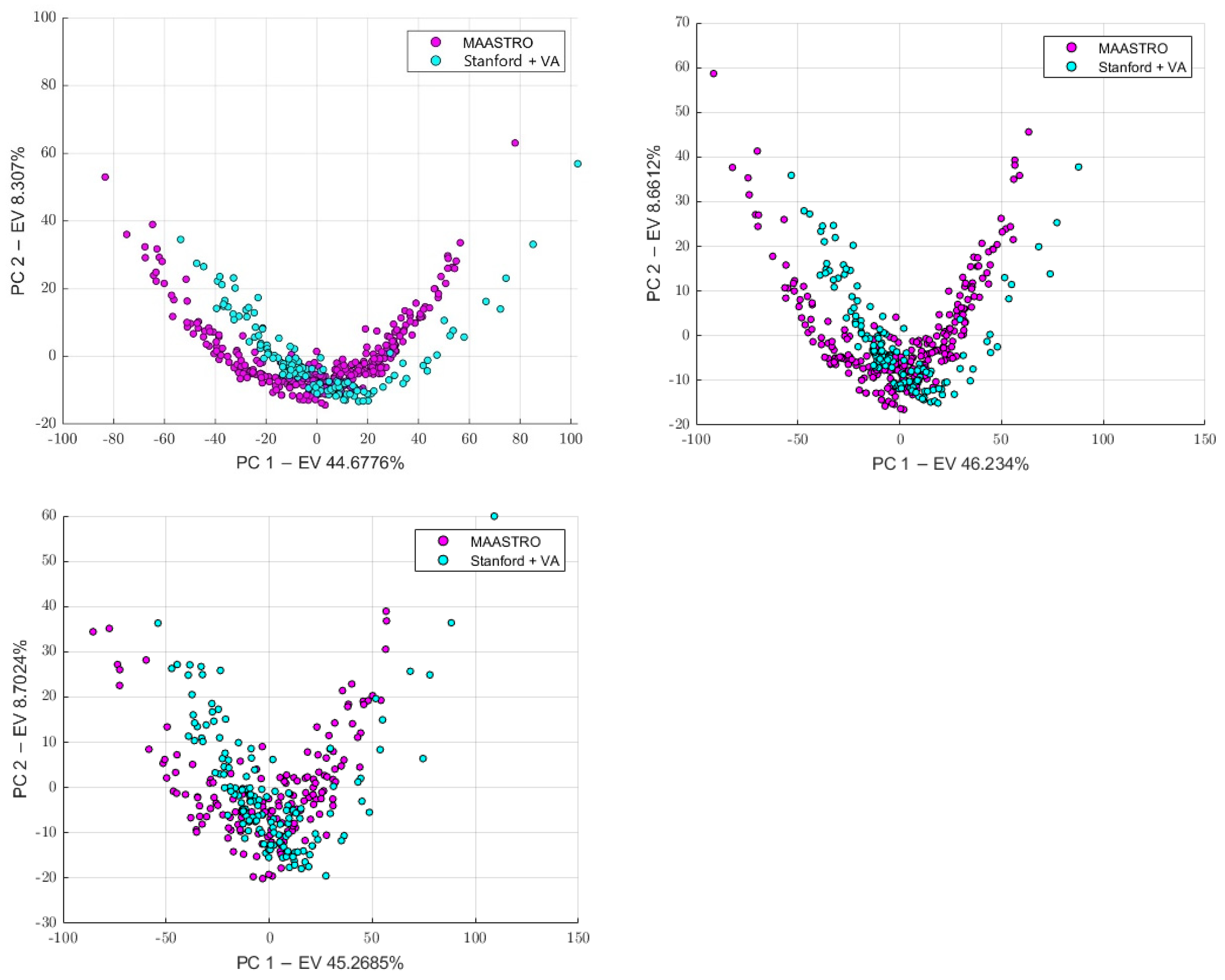
3.1. Batch Analysis and Feature Harmonization
3.2. Feature Selection and Stability
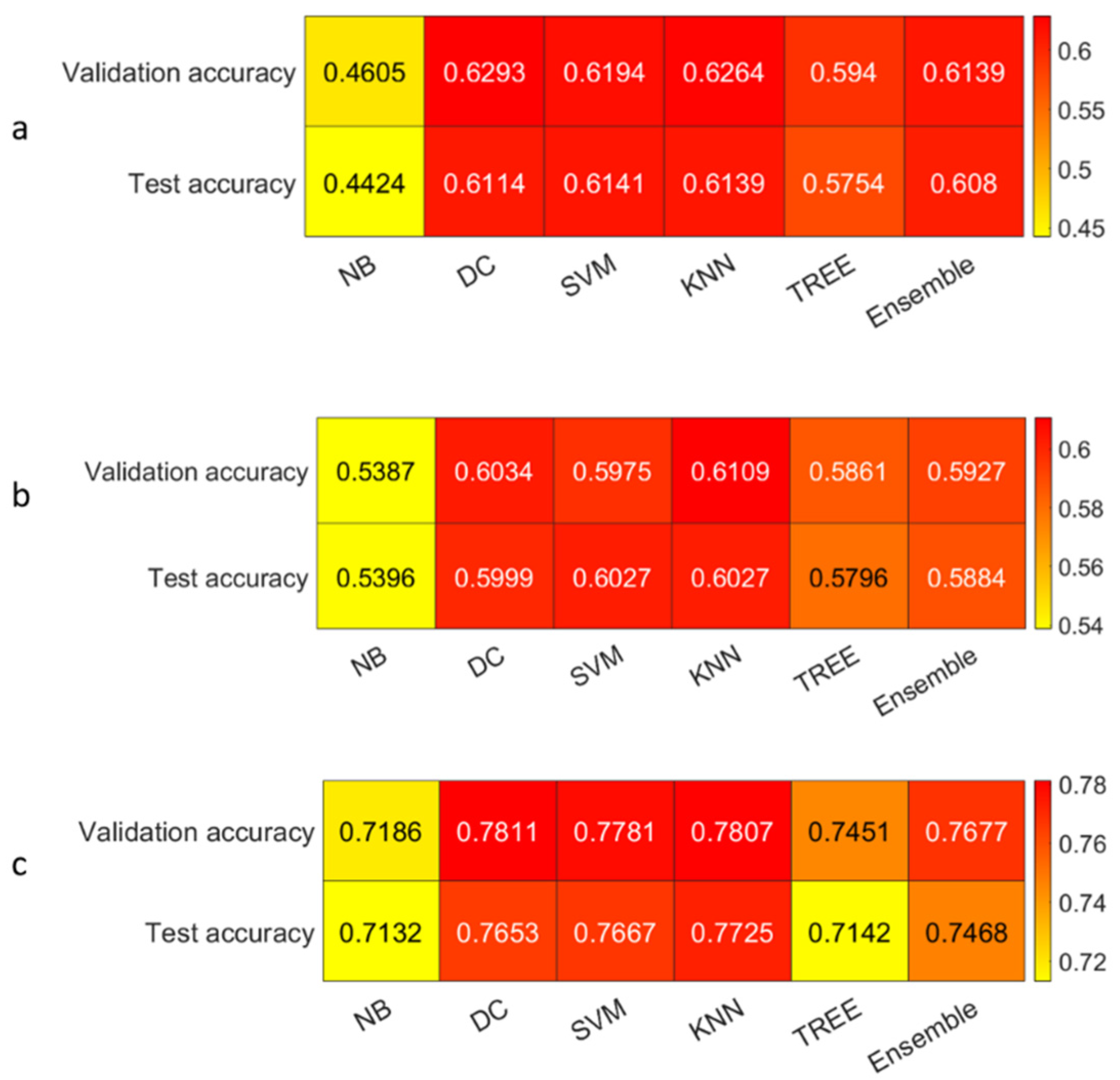
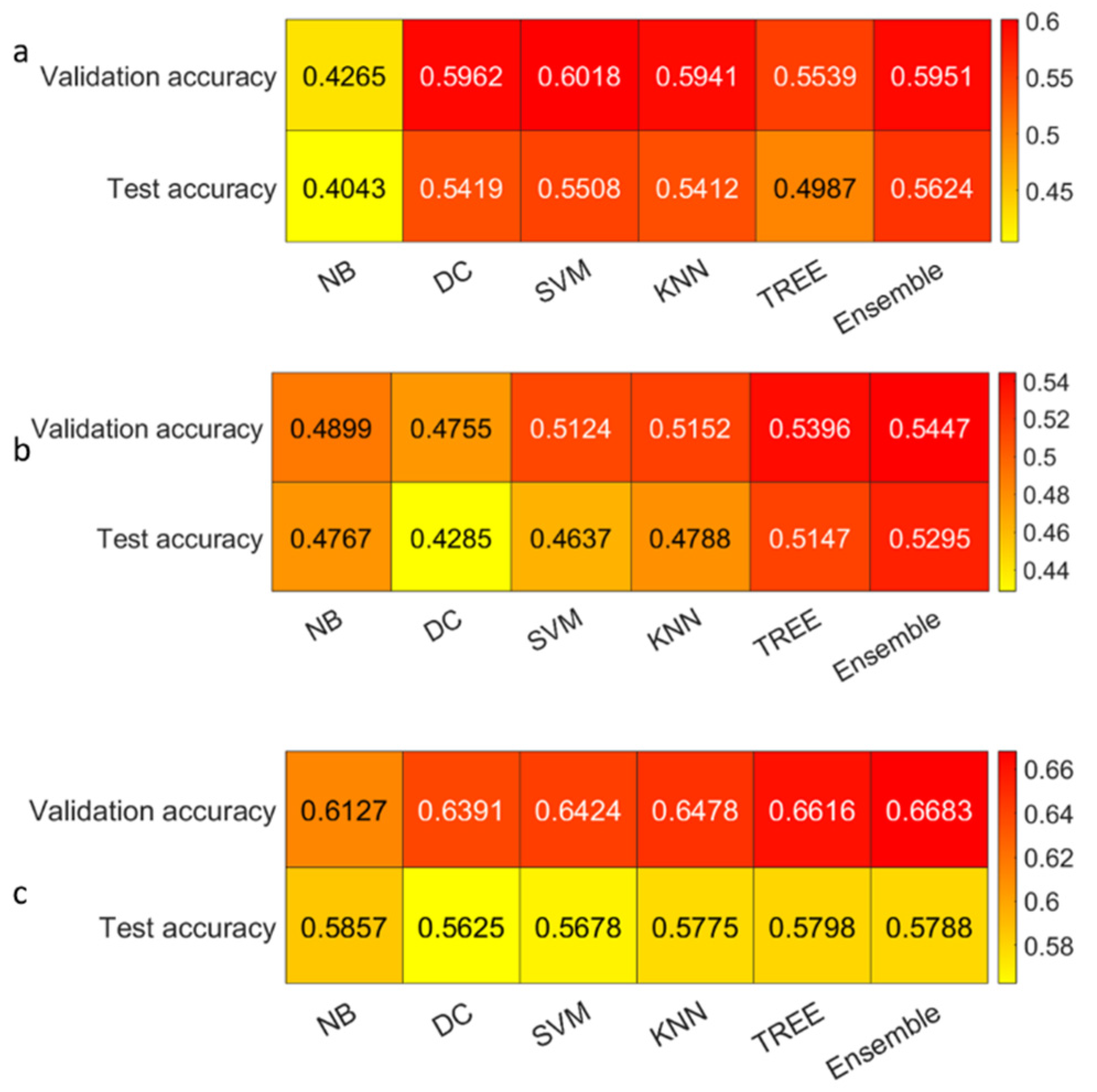
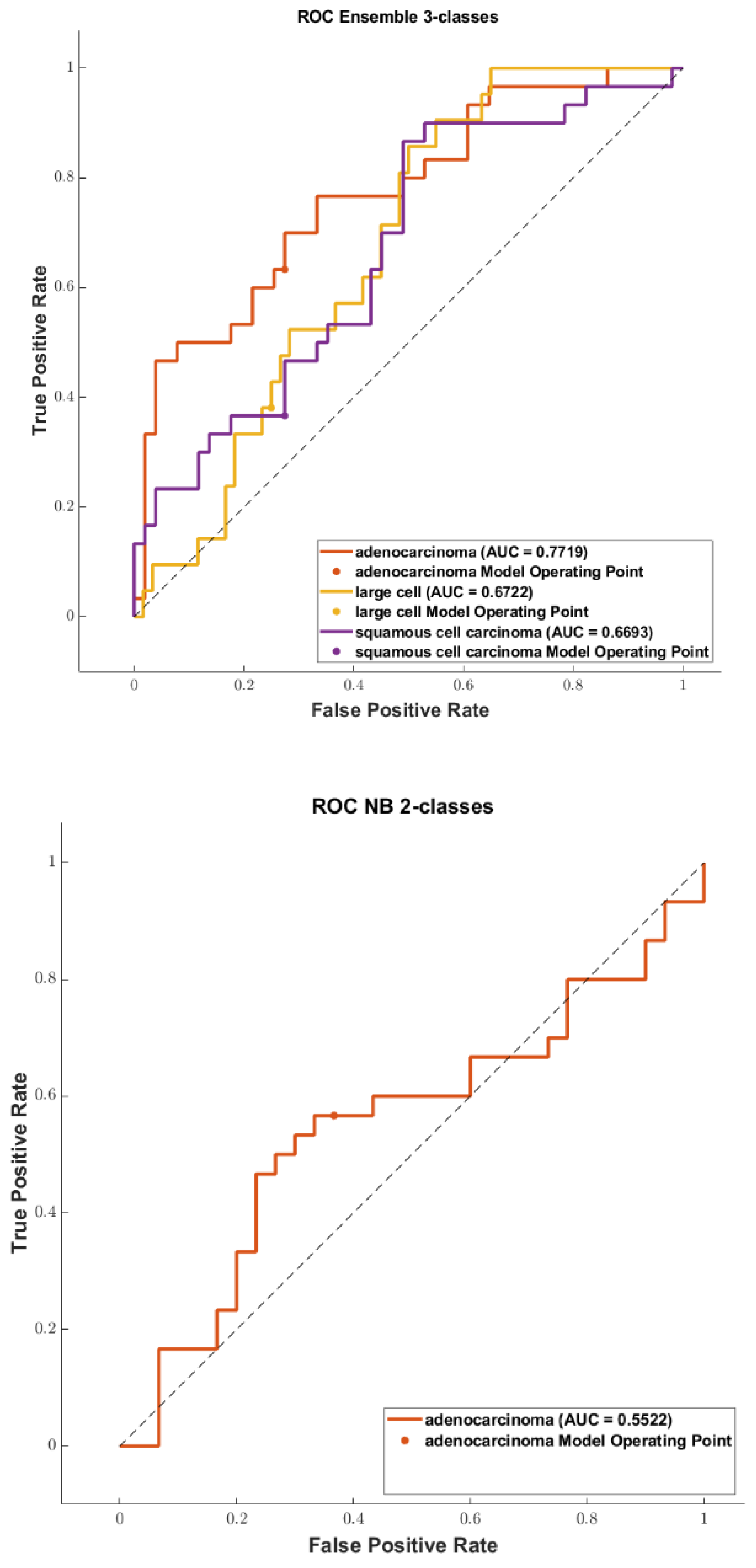
3.3. Classification
4. Discussion
4.1. Comparison of the Highest Accuracies and AUCs with Related Works
5. Limitation
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Extractor | Pre-Processing | Extracted Features |
|---|---|---|
| PyRadiomics v.3.0.1 | Voxel resampling: 1 × 1 × 1 mm3 Resampling interpolator: Sitk Linear. LoG filter: sigma (0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, and 5) Wavelet decomposition method: Haar Other parameters: PyRadiomics’ default. | GLCM GLSZM GLRLM NGTDM GLDM |
Appendix B
| Acronym | Stands for |
|---|---|
| CT | Computed tomography |
| NSCLC | Non-small-cell lung cancer |
| SCC | Squamous cell carcinoma |
| LCC | Large cell carcinoma |
| ADC | Adenocarcinoma |
| NOS | No other specified |
| IBSI | Image Biomarker Standardization Initiative |
| AUC | Area under curve |
| LoG | Laplacian of Gaussian |
| SCLC | Small-cell lung cancer |
| WHO | World Health Organization |
| GS | Gleason score |
| MRI | Magnetic resonance imaging |
| EDSS | Expanded disability status scale |
| DICOM | Digital imaging and communication in medicine |
| GLCM | Gray level co-occurrence matrix |
| GLRLM | Gray level run length matrix |
| GLSZM | Gray level size zone matrix |
| NGTDM | Neighboring gray tone difference matrix |
| GLDM | Gray level dependence matrix |
| HHH | High-high-high |
| HHL | High-high-low |
| HLH | High-low-high |
| LHH | Low-high-high |
| LLL | Low-low-low |
| LHL | Low-high-low |
| HLL | High-low-low |
| LLH | Low-low-high |
| 4-c | 4-classes dataset |
| 3-c | 3-classes dataset |
| 2-c | 2-classes dataset |
| PCA | Principal component analysis |
| PC1 | First principal component |
| PC2 | Second principal component |
| tSNE | t-distributed stochastic neighbor embedding |
| 4-c-h | 4-classes harmonized dataset |
| 3-c-h | 3-classes harmonized dataset |
| 2-c-h | 2-classes harmonized dataset |
| LASSO | Least absolute shrinkage and selection operator |
| DA | Discriminant analysis |
| KNN | K-nearest neighbors |
| SVM | Support vector machines |
| NB | Naïve Bayes |
| SMOTE | Synthetic minority over-sampling technique |
| GINI | Gini index |
| IG | Information gain |
| GR | Gain ratio |
| LS | Laplacian score |
| MDL | Minimum description length |
| SPEC | Spectral feature selection |
| ℓ2,1NR | ℓ2,1-norm regularization |
| RFS | robust feature selection |
| MCFS | Multi-cluster feature selection |
| CSS | Chi-square score |
| FS | Fisher score based on statistics |
| TS | T-score |
| mRMR | Redundancy maximum relevance feature selection |
| SFS | Sequential forward selection |
| RF | Random forest |
| GNB | Gaussian Naïve Bayes |
| AdaB | AdaBoost |
| XGBoost | Extreme gradient boosting |
| BAG | Bagging |
| DT | Decision tree |
| GDBT | Gradient boosting decision tree |
| LR | Logistic regression |
| MLP | Multilayer perceptron |
| LDA | Linear discriminant analysis |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Dalmartello, M.; La Vecchia, C.; Bertuccio, P.; Boffetta, P.; Levi, F.; Negri, E.; Malvezzi, M. European cancer mortality predictions for the year 2022 with focus on ovarian cancer. Ann. Oncol. 2022, 33, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef] [Green Version]
- Xing, P.; Zhu, Y.; Wang, L.; Hui, Z.; Liu, S.; Ren, J.; Zhang, Y.; Song, Y.; Liu, C.; Huang, Y.; et al. What are the clinical symptoms and physical signs for non-small cell lung cancer before diagnosis is made? A nation-wide multicenter 10-year retrospective study in China. Cancer Med. 2019, 8, 4055–4069. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.; Liu, S.V.; Subramaniam, D.S.; Giaccone, G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat. Rev. Clin. Oncol. 2015, 12, 511–526. [Google Scholar] [CrossRef]
- Cuocolo, R.; Cipullo, M.B.; Stanzione, A.; Ugga, L.; Romeo, V.; Radice, L.; Brunetti, A.; Imbriaco, M. Machine learning applications in prostate cancer magnetic resonance imaging. Eur. Radiol. Exp. 2019, 3, 35. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Comelli, A.; Stefano, A.; Coronnello, C.; Russo, G.; Vernuccio, F.; Cannella, R.; Salvaggio, G.; Lagalla, R.; Barone, S. Radiomics: A New Biomedical Workflow to Create a Predictive Model. In Proceedings of the Medical Image Understanding and Analysis; Papież, B.W., Namburete, A.I.L., Yaqub, M., Noble, J.A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 280–293. [Google Scholar]
- Bharati, S.; Podder, P.; Mondal, M.R.H. Hybrid deep learning for detecting lung diseases from X-ray images. Inform. Med. Unlocked 2020, 20, 100391. [Google Scholar] [CrossRef] [PubMed]
- Afshar, P.; Mohammadi, A.; Plataniotis, K.N.; Oikonomou, A.; Benali, H. From Handcrafted to Deep-Learning-Based Cancer Radiomics: Challenges and Opportunities. IEEE Signal Process. Mag. 2019, 36, 132–160. [Google Scholar] [CrossRef] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging-”how-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Johnson, W.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Fortin, J.-P.; Cullen, N.; Sheline, Y.I.; Taylor, W.D.; Aselcioglu, I.; Cook, P.A.; Adams, P.; Cooper, C.; Fava, M.; McGrath, P.J.; et al. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 2018, 167, 104–120. [Google Scholar] [CrossRef]
- Alongi, P.; Stefano, A.; Comelli, A.; Laudicella, R.; Scalisi, S.; Arnone, G.; Barone, S.; Spada, M.; Purpura, P.; Bartolotta, T.V.; et al. Radiomics analysis of 18F-Choline PET/CT in the prediction of disease outcome in high-risk prostate cancer: An explorative study on machine learning feature classification in 94 patients. Eur. Radiol. 2021, 31, 4595–4605. [Google Scholar] [CrossRef] [PubMed]
- Cutaia, G.; La Tona, G.; Comelli, A.; Vernuccio, F.; Agnello, F.; Gagliardo, C.; Salvaggio, L.; Quartuccio, N.; Sturiale, L.; Stefano, A.; et al. Radiomics and Prostate MRI: Current Role and Future Applications. J. Imaging 2021, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Pasini, G.; Bini, F.; Russo, G.; Comelli, A.; Marinozzi, F.; Stefano, A. matRadiomics: A Novel and Complete Radiomics Framework, from Image Visualization to Predictive Model. J. Imaging 2022, 8, 221. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Cannella, R.; La Grutta, L.; Midiri, M.; Bartolotta, T.V. New advances in radiomics of gastrointestinal stromal tumors. World J. Gastroenterol. 2020, 26, 4729–4738. [Google Scholar] [CrossRef]
- Russo, G.; Stefano, A.; Comelli, A.; Savoca, G.; Richiusa, S.; Sabini, M.; Cosentino, S.; Alongi, P.; Ippolito, M. Radiomics features of 11[C]-MET PET/CT in primary brain tumors: Preliminary results on grading discrimination using a machine learning model. Phys. Med. 2021, 62, S44–S45. [Google Scholar] [CrossRef]
- Alongi, P.; Laudicella, R.; Panasiti, F.; Stefano, A.; Comelli, A.; Giaccone, P.; Arnone, A.; Minutoli, F.; Quartuccio, N.; Cupidi, C.; et al. Radiomics Analysis of Brain [18F]FDG PET/CT to Predict Alzheimer’s Disease in Patients with Amyloid PET Positivity: A Preliminary Report on the Application of SPM Cortical Segmentation, Pyradiomics and Machine-Learning Analysis. Diagnostics 2022, 12, 933. [Google Scholar] [CrossRef]
- Shu, Z.; Cui, S.; Wu, X.; Xu, Y.; Huang, P.; Pang, P.; Zhang, M. Predicting the progression of Parkinson’s disease using conventional MRI and machine learning: An application of radiomic biomarkers in whole-brain white matter. Magn. Reson. Med. 2021, 85, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Shoeibi, A.; Khodatars, M.; Jafari, M.; Ghassemi, N.; Moridian, P.; Alizadehsani, R.; Ling, S.H.; Khosravi, A.; Alinejad-Rokny, H.; Lam, H.; et al. Diagnosis of brain diseases in fusion of neuroimaging modalities using deep learning: A review. Inf. Fusion 2023, 93, 85–117. [Google Scholar] [CrossRef]
- Shoeibi, A.; Khodatars, M.; Jafari, M.; Moridian, P.; Rezaei, M.; Alizadehsani, R.; Khozeimeh, F.; Gorriz, J.M.; Heras, J.; Panahiazar, M.; et al. Applications of deep learning techniques for automated multiple sclerosis detection using magnetic resonance imaging: A review. Comput. Biol. Med. 2021, 136, 104697. [Google Scholar] [CrossRef] [PubMed]
- Nepi, V.; Pasini, G.; Bini, F.; Marinozzi, F.; Russo, G.; Stefano, A. MRI-Based Radiomics Analysis for Identification of Features Correlated with the Expanded Disability Status Scale of Multiple Sclerosis Patients. In Proceedings of the Image Analysis and Processing; Mazzeo, P.L., Frontoni, E., Sclaroff, S., Distante, C., Eds.; ICIAP 2022 Workshops; Springer International Publishing: Cham, Switzerland, 2022; pp. 362–373. [Google Scholar] [CrossRef]
- Gao, A.; Yang, H.; Wang, Y.; Zhao, G.; Wang, C.; Wang, H.; Zhang, X.; Zhang, Y.; Cheng, J.; Yang, G.; et al. Radiomics for the Prediction of Epilepsy in Patients With Frontal Glioma. Front. Oncol. 2021, 11, 725926. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jiang, X.; Xu, H.; Zhang, D.; Liu, L.-N.; Xia, Y.-X.; Xu, D.-K.; Wu, H.-J.; Cheng, G.; Shi, Y.-H. Preoperative prediction of mediastinal lymph node metastasis in non-small cell lung cancer based on 18F-FDG PET/CT radiomics. Clin. Radiol. 2023, 78, 8–17. [Google Scholar] [CrossRef]
- Wu, W.; Parmar, C.; Grossmann, P.; Quackenbush, J.; Lambin, P.; Bussink, J.; Mak, R.; Aerts, H.J.W.L. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front. Oncol. 2016, 6, 71. [Google Scholar] [CrossRef] [Green Version]
- Haga, A.; Takahashi, W.; Aoki, S.; Nawa, K.; Yamashita, H.; Abe, O.; Nakagawa, K. Classification of early stage non-small cell lung cancers on computed tomographic images into histological types using radiomic features: Interobserver delineation variability analysis. Radiol. Phys. Technol. 2018, 11, 27–35. [Google Scholar] [CrossRef]
- Han, Y.; Ma, Y.; Wu, Z.; Zhang, F.; Zheng, D.; Liu, X.; Tao, L.; Liang, Z.; Yang, Z.; Li, X.; et al. Histologic subtype classification of non-small cell lung cancer using PET/CT images. Eur. J. Nucl. Med. 2020, 48, 350–360. [Google Scholar] [CrossRef]
- Yang, F.; Chen, W.; Wei, H.; Zhang, X.; Yuan, S.; Qiao, X.; Chen, Y.-W. Machine Learning for Histologic Subtype Classification of Non-Small Cell Lung Cancer: A Retrospective Multicenter Radiomics Study. Front. Oncol. 2021, 10, 608598. [Google Scholar] [CrossRef]
- Song, F.; Song, X.; Feng, Y.; Fan, G.; Sun, Y.; Zhang, P.; Li, J.; Liu, F.; Zhang, G. Radiomics feature analysis and model research for predicting histopathological subtypes of non-small cell lung cancer on CT images: A multi-dataset study. Med. Phys. 2023; early view. [Google Scholar] [CrossRef]
- Liu, J.; Cui, J.; Liu, F.; Yuan, Y.; Guo, F.; Zhang, G. Multi-subtype classification model for non-small cell lung cancer based on radiomics: SLS model. Med. Phys. 2019, 46, 3091–3100. [Google Scholar] [CrossRef]
- Khodabakhshi, Z.; Mostafaei, S.; Arabi, H.; Oveisi, M.; Shiri, I.; Zaidi, H. Non-small cell lung carcinoma histopathological subtype phenotyping using high-dimensional multinomial multiclass CT radiomics signature. Comput. Biol. Med. 2021, 136, 104752. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, K.; Rajpal, A.; Rajpal, S.; Agarwal, M.; Kumar, V.; Kumar, N. An explainable AI-driven biomarker discovery framework for Non-Small Cell Lung Cancer classification. Comput. Biol. Med. 2023, 153, 106544. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Wee, L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; et al. Data From NSCLC-Radiomics 2019. The Cancer Imaging Archive. Available online: https://doi.org/10.7937/k9/tcia.2015.pf0m9rei (accessed on 1 January 2023).
- Bakr, S.; Gevaert, O.; Echegaray, S.; Ayers, K.; Zhou, M.; Shafiq, M.; Zheng, H.; Zhang, W.; Leung, A.; Kadoch, M.; et al. Data for NSCLC Radiogenomics Collection 2017. The Cancer Imaging Archive. Available online: http://doi.org/10.7937/K9/TCIA.2017.7hs46erv (accessed on 1 January 2023).
- Grove, O.; Berglund, A.E.; Schabath, M.B.; Aerts, H.J.W.L.; Dekker, A.; Wang, H.; Velazquez, E.R.; Lambin, P.; Gu, Y.; Balagurunathan, Y.; et al. Quantitative Computed Tomographic Descriptors Associate Tumor Shape Complexity and Intratumor Heterogeneity with Prognosis in Lung Adenocarcinoma. PLoS ONE 2015, 10, e0118261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Wang, S.; Li, T.; Lu, J.; HuangFu, Y.; Wang, D. A Large-Scale CT and PET/CT Dataset for Lung Cancer Diagnosis. 2020. Available online: https://doi.org/10.7937/TCIA.2020.NNC2-0461 (accessed on 1 January 2023).
- Wee, L.; Aerts, H.J.; Kalendralis, P.; Dekker, A. Data from NSCLC-Radiomics-Interobserver1 2019. Available online: https://doi.org/10.7937/tcia.2019.cwvlpd26 (accessed on 1 January 2023).
- Kirk, S.; Lee, Y.; Kumar, P.; Filippini, J.; Albertina, B.; Watson, M.; Rieger-Christ, K.; Lemmerman, J. Radiology Data from The Cancer Genome Atlas Lung Squamous Cell Carcinoma [TCGA-LUSC] Collection. 2016. Available online: https://doi.org/10.7937/k9/tcia.2016.tygkkfmq (accessed on 1 January 2023).
- Albertina, B.; Watson, M.; Holback, C.; Jarosz, R.; Kirk, S.; Lee, Y.; Rieger-Christ, K.; Lemmerman, J. Radiology Data from The Cancer Genome Atlas Lung Adenocarcinoma [TCGA-LUAD]. 2016. Available online: https://doi.org/10.7937/k9/tcia.2016.jgnihep5 (accessed on 1 January 2023).
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Data From NSCLC-Radiomics-Genomics. 2015. Available online: https://doi.org/10.7937/k9/tcia.2015.l4fret6z (accessed on 1 January 2023).
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orlhac, F.; Frouin, F.; Nioche, C.; Ayache, N.; Buvat, I. Validation of A Method to Compensate Multicenter Effects Affecting CT Radiomics. Radiology 2019, 291, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.-H.; Park, H. Classification of low-grade and high-grade glioma using multi-modal image radiomics features. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Republic of Korea, 11–15 July 2017; pp. 3081–3084. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, X.-L.; Zhang, T.; Wang, J.; Zhang, T.; Tian, R. Use of radiomics based on 18F-FDG PET/CT and machine learning methods to aid clinical decision-making in the classification of solitary pulmonary lesions: An innovative approach. Eur. J. Nucl. Med. 2021, 48, 2904–2913. [Google Scholar] [CrossRef]
- Bertolini, M.; Trojani, V.; Botti, A.; Cucurachi, N.; Galaverni, M.; Cozzi, S.; Borghetti, P.; La Mattina, S.; Pastorello, E.; Avanzo, M.; et al. Novel Harmonization Method for Multi-Centric Radiomic Studies in Non-Small Cell Lung Cancer. Curr. Oncol. 2022, 29, 5179–5194. [Google Scholar] [CrossRef]
- Comelli, A.; Stefano, A.; Bignardi, S.; Russo, G.; Sabini, M.G.; Ippolito, M.; Barone, S.; Yezzi, A. Active contour algorithm with discriminant analysis for delineating tumors in positron emission tomography. Artif. Intell. Med. 2019, 94, 67–78. [Google Scholar] [CrossRef]
- Artzi, M.; Bressler, I.; Ben Bashat, D. Differentiation between glioblastoma, brain metastasis and subtypes using radiomics analysis. J. Magn. Reson. Imaging 2019, 50, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Comelli, A.; Stefano, A.; Russo, G.; Bignardi, S.; Sabini, M.G.; Petrucci, G.; Ippolito, M.; Yezzi, A. K-nearest neighbor driving active contours to delineate biological tumor volumes. Eng. Appl. Artif. Intell. 2019, 81, 133–144. [Google Scholar] [CrossRef]
- Licari, L.; Salamone, G.; Campanella, S.; Carfì, F.; Fontana, T.; Falco, N.; Tutino, R.; De Marco, P.; Comelli, A.; Cerniglia, D.; et al. Use of the KSVM-based system for the definition, validation and identification of the incisional hernia recurrence risk factors. Il Giornale di Chirurgia. G. Di Chir. J. Surg. 2019, 40, 32–38. [Google Scholar]
- Comelli, A.; Stefano, A.; Bignardi, S.; Coronnello, C.; Russo, G.; Sabini, M.G.; Ippolito, M.; Yezzi, A. Tissue Classification to Support Local Active Delineation of Brain Tumors. In Proceedings of the Medical Image Understanding and Analysis; Zheng, Y., Williams, B.M., Chen, K., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 3–14. [Google Scholar]
- Demircioğlu, A. The effect of preprocessing filters on predictive performance in radiomics. Eur. Radiol. Exp. 2022, 6, 40. [Google Scholar] [CrossRef]
- Stefano, A.; Leal, A.; Richiusa, S.; Trang, P.; Comelli, A.; Benfante, V.; Cosentino, S.; Sabini, M.G.; Tuttolomondo, A.; Altieri, R.; et al. Robustness of PET Radiomics Features: Impact of Co-Registration with MRI. Appl. Sci. 2021, 11, 10170. [Google Scholar] [CrossRef]
- Abd Elrahman, S.M.; Abraham, A. A review of class imbalance problem. J. Netw. Innov. Comput. 2013, 1, 332–340. [Google Scholar]
- Schwier, M.; van Griethuysen, J.; Vangel, M.G.; Pieper, S.; Peled, S.; Tempany, C.; Aerts, H.J.W.L.; Kikinis, R.; Fennessy, F.M.; Fedorov, A. Repeatability of Multiparametric Prostate MRI Radiomics Features. Sci. Rep. 2019, 9, 9441. [Google Scholar] [CrossRef] [Green Version]
- LaRue, R.T.H.M.; Van Timmeren, J.E.; De Jong, E.E.C.; Feliciani, G.; Leijenaar, R.T.H.; Schreurs, W.M.J.; Sosef, M.N.; Raat, F.H.P.J.; Van Der Zande, F.H.R.; Das, M.; et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: A comprehensive phantom study. Acta Oncol. 2017, 56, 1544–1553. [Google Scholar] [CrossRef]
- Tavolara, T.E.; Gurcan, M.N.; Niazi, M.K.K. Contrastive Multiple Instance Learning: An Unsupervised Framework for Learning Slide-Level Representations of Whole Slide Histopathology Images without Labels. Cancers 2022, 14, 5778. [Google Scholar] [CrossRef]




| Work | Dataset | Modality | Feature Extraction | M/B/H |
|---|---|---|---|---|
| [30] (2016) | 1 public dataset [38] (192) and lung 2 (152) | CT | Pre-processing: not all specified Extractor: Matlab 2012 Extracted Features: shape, 1st-order statistics, texture features (440) | No/no/no |
| [31] (2017) | Private dataset (40) | CT | Pre-processing: all specified Extractor: Mvalliers package Extracted Features: shape, 1st-order statistics, texture features (476) | No/no/no |
| [32] (2021) | Private dataset (1419) | PET + CT | Pre-processing: not all specified Extractor: PyRadiomics Extracted Features: 1st-order statistics, texture Features (688) | No/no/no |
| [33] (2021) | Private dataset (302) and 2 public datasets [38] (203), [39] (140) | CT | Pre-processing: not all specified Extractor: PyRadiomics Extracted Features: shape, 1st-order statistics, texture features (788) | Yes/no/no |
| [34] (2023) | 8 public datasets [38,39,40,41,42,43,44,45] (868). | CT | Pre- processing: not all specified, interpolator specified Extractor: PyRadiomics Extracted Features: shape, 1st-order statistics, texture features (1409). | Yes/no/ no |
| ours | 2 public datasets: [38,39] (302) | CT | Pre-processing: all specified Extractor: PyRadiomics Extracted Features: shape, 1st-order statistics, texture features (1433) | Yes/Yes/Yes |
| Work | Dataset | Modality | Feature Extraction | M/B/H |
|---|---|---|---|---|
| [35] (2019) | 2 public datasets [38] (278), [45] (71) | CT | Pre-processing: not all specified. Extractor: not specified Extracted Features: shape, 1st-order statistics, texture features (440) | No/no/no |
| [36] (2021) | 1 public dataset: [38] (354) | CT | Pre-processing: not all specified, Extractor: PyRadiomics Extracted Features: shape, 1st-order statistics, texture features (1433) | No/no/no |
| ours | 2 public datasets: [38,39] (466) | CT | Pre-processing: all specified Extractor: PyRadiomics Extracted Features: shape, 1st-order statistics, texture features (1433) | Yes/Yes/Yes |
| Datasets | Pixel Spacing [x, y] (n°) | Slice Thickness [z] (n°) | Matrix Dimension [x, y] (n)° |
|---|---|---|---|
| NSCLC-Radiomics | [0.97656250, 9765625] (247) | 3 (247) | [512 × 512] (247) |
| NSCLC-Radiomics | [0.9770, 0.9770] (77) | 3 (77) | [512 × 512] (77) |
| NSCLC-Radiogenomics | In a range between [0.589844, 0.976562] (142) | In a range between [0.625, 3] (142) | [512 × 512] (142) |
| Size | 4-c-nh | 4-c-h | 3-c-nh | 3-c-h | 2-c-nh | 2-c-h |
|---|---|---|---|---|---|---|
| Minimum | 16 | 14 | 9 | 2 | 8 | 1 |
| Maximum | 28 | 22 | 23 | 14 | 22 | 9 |
| 4-c-nh | 4-c-h | 3-c-nh | 3-c-h | 2-c-nh | 2-c-h | ||
|---|---|---|---|---|---|---|---|
| Amount | 11 | 9 | 4 | 2 | 2 | 1 | |
| Shape | 3 | 1 | 0 | 0 | 0 | 0 | |
| Class | First order | 1 | 1 | 1 | 0 | 0 | 0 |
| Texture | 7 | 7 | 3 | 2 | 2 | 1 | |
| Original | 3 | 1 | 0 | 0 | 0 | 0 | |
| Image Type | LoG | 4 | 1 | 0 | 0 | 0 | 1 |
| Wavelet | 4 | 7 | 4 | 2 | 2 | 0 |
| Work | ML Methods | Training and Testing Sets | Validation and Testing Schemes | Results |
|---|---|---|---|---|
| [30] (2016) | Selection: correlation + GINI, IG, GR, MDL, DKM, ReliefF + variants. Classification: RF, NB, and KNN | Training: 192 (public dataset) Testing: 152 (Lung 2) | External testing | ReliefFdistance + NB: Test AUC 0.72 |
| [31] (2017) | Selection: univariate analysis p < 0.05 + interobserver variation analysis p < 0.1 + cross correlation analysis r < 0.7. Classification: NB | Training: 28 (private dataset) Testing: 12 (private dataset) | Training/testing sets splitting: 30 times repeated (70/30) | Test accuracy: 0.656 Test AUC: 0.725 |
| [32] (2021) | Selection: LS, ReliefF, SPEC, ℓ2,1NR, RFS, MCFS, CSS, FS, TS, and GINI. Classification: AdaB, BAG, DT, NB, KNN, LR, MLP, LDA, and SVM | Training: 1136 (private dataset) Testing: 283 (private dataset) | Training/testing sets splitting: (80/20). Validation: 10-fold cross validation | ℓ2,1NR + LDA: test accuracy 0.794 ℓ2,1NR + LDA: test AUC 0.863 |
| [33] (2021) | Selection: mRMR, SFS, and LASSO Classification: LR, SVM, and RF | Since the ratio of training/testing split is not specified (merged dataset), the number of samples used for training and testing is unknown | Training/testing sets splitting: ratio not specified Validation: 5-fold cross validation | Test accuracy 0.74 Test AUC 0.78 |
| [34] (2023) | Selection: wrapper ℓ2,1 norm minimization + 10-fold cross validated LR Classification: LR, SVM, RF, MLP, KNN, GNB, GBDT, AdaB, BAG, and XGBoost | Training: from 560 (5 merged datasets) to 940 after SMOTE. Testing: 140 (5 merged datasets) + external testing (168-3 datasets) | Training/Testing sets splitting: (80/20) Validation: 10-fold cross validation External testing | Bagging-AdaBoost-LR (ensemble): test accuracy 0.766 Bagging-AdaBoost-SVM (Ensemble): test AUC 0.815 |
| ours | Selection: Kruskal Wallis + LASSO Classification: NB, DA, KNN, SVM, TREE, and ensemble | Training: 242 (2 datasets merged) Testing: 60 (2 datasets merged) | Training/Testing sets splitting: 10 times repeated (80/20) Validation: 10 times repeated 5-fold cross validation | KNN: test accuracy 0.7725 KNN: test AUC 0.821 |
| Work | ML Methods | Training and Testing Sets | Validation and Testing Schemes | Results |
|---|---|---|---|---|
| [35] (2019) | Selection: wrapper ℓ2,1 norm minimization + SVM Classification: SVM | Training: from 279 (public dataset) to 760 after SMOTE Testing: 70 (public dataset) | Training/Testing sets splitting: (80/20) Validation: 10-fold cross validation | NO SMOTE test accuracy 0.67 SMOTE Test accuracy 0.86 |
| [36] (2021) | Selection: Wrapper algorithm, multivariate adaptive regression splines Classification: Multinomial logistic regression | Training: 354 (public dataset) | Validation: 1000 bootstrapping | Validation accuracy: 0.865 Validation AUC: 0.747 |
| ours | Selection: Kruskal Wallis + LASSO Classification: NB, DA, KNN, SVM, TREE, and ensemble | Training: 373 (2 datasets merged) Testing: 93 (2 datasets merged) | Training/Testing sets splitting: 10 times repeated (80/20) Validation: 10 times repeated 5-fold cross validation | DC: validation accuracy 0.6293 SVM: test accuracy 0.6141 DC: validation AUC 0.826 (averaged on 4 classes) KNN: test AUC 0.831 (averaged on 4 classes) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasini, G.; Stefano, A.; Russo, G.; Comelli, A.; Marinozzi, F.; Bini, F. Phenotyping the Histopathological Subtypes of Non-Small-Cell Lung Carcinoma: How Beneficial Is Radiomics? Diagnostics 2023, 13, 1167. https://doi.org/10.3390/diagnostics13061167
Pasini G, Stefano A, Russo G, Comelli A, Marinozzi F, Bini F. Phenotyping the Histopathological Subtypes of Non-Small-Cell Lung Carcinoma: How Beneficial Is Radiomics? Diagnostics. 2023; 13(6):1167. https://doi.org/10.3390/diagnostics13061167
Chicago/Turabian StylePasini, Giovanni, Alessandro Stefano, Giorgio Russo, Albert Comelli, Franco Marinozzi, and Fabiano Bini. 2023. "Phenotyping the Histopathological Subtypes of Non-Small-Cell Lung Carcinoma: How Beneficial Is Radiomics?" Diagnostics 13, no. 6: 1167. https://doi.org/10.3390/diagnostics13061167








