A Current Review of Machine Learning and Deep Learning Models in Oral Cancer Diagnosis: Recent Technologies, Open Challenges, and Future Research Directions
Abstract
:1. Introduction
1.1. Objectives
1.2. Current Insights into Oral Cancer Diagnostics
1.3. Contribution of This Review
- We carried out a review of recent methods for detecting the early signs of oral cancer, including extreme learning machines, DBN, the deep generative model, and others. Furthermore, more traditional methods from the field of artificial intelligence, including random forest, ANN, DNN, KNN, and others were also included.
- We used an extensive tabular style to describe the studies on the use of ML and DL in OC. The summary includes information regarding the model, significant contributions, and model constraints.
- This review specifically addressed current issues and potential solutions for diagnosing and treating oral cancer.
- Table 2 compares the current review with earlier surveys or other review articles of a similar nature.
1.4. Survey Methodology
1.4.1. Search Strategy and the Literature Sources
1.4.2. Inclusion Criteria
1.4.3. Elimination Criteria
1.4.4. Results
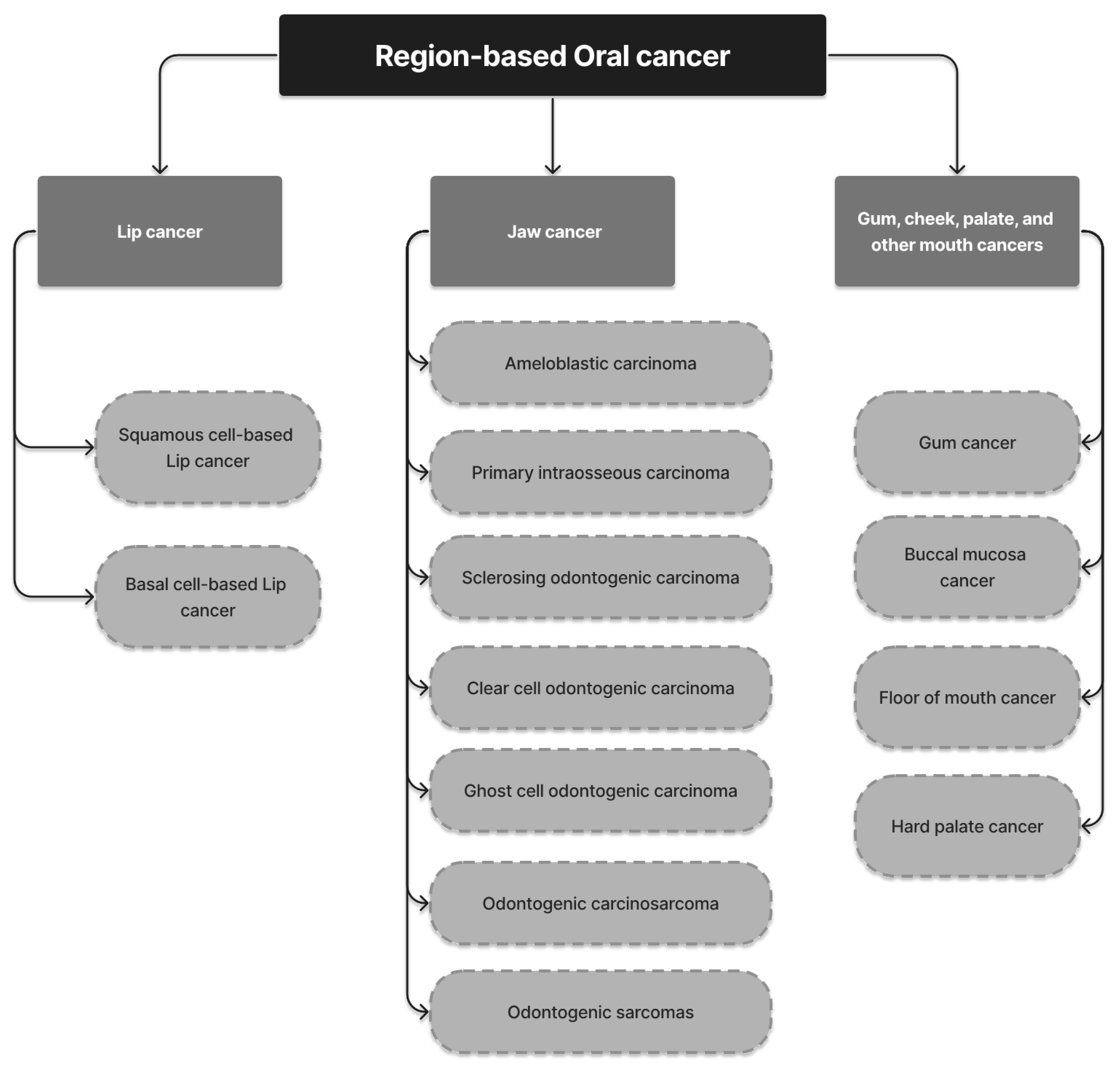
2. Region-Based Oral Cancer
2.1. Lip Cancer
2.1.1. Squamous-Cell-Based Lip Cancer
2.1.2. Basal-Cell-Based Lip Cancer
2.2. Jaw Cancer
2.2.1. Ameloblastic Carcinoma
2.2.2. Primary Intraosseous Carcinoma
2.2.3. Sclerosing Odontogenic Carcinoma
2.2.4. Clear Cell Odontogenic Carcinoma
2.2.5. Ghost Cell Odontogenic Carcinoma (GCOC)
2.2.6. Odontogenic Carcinosarcoma
2.2.7. Odontogenic Sarcomas
2.3. Gum, Cheek, Palate, and Other Mouth Cancers
2.3.1. Gum Cancer
2.3.2. Buccal Mucosa (Inner Cheek) Cancer
2.3.3. Floor of the Mouth Cancer
2.3.4. Hard Palate (Roof of the Mouth) Cancer
3. Recent Technologies in Oral Cancer Diagnosis
3.1. Visual Staining
3.2. Cytological Techniques
3.3. Optical Imaging
3.4. Saliva-Based Oral Cancer Diagnosis
3.5. Tomography
3.6. Tissue Auto-Fluorescence
3.7. Biopsy
3.8. Lab-On-Chip
4. Machine Learning and Deep Learning Models for Oral Cancer Diagnosis
4.1. Machine Learning Techniques
4.1.1. Artificial Neural Network
4.1.2. Naïve Bayes
4.1.3. Decision Tree
4.1.4. K-Nearest Neighbor
4.1.5. K-means Clustering
4.1.6. Random Forest
4.1.7. Support Vector Machine
4.1.8. Ensemble Models
4.1.9. Summary of the ML Model
4.1.10. Limitations of the ML Model
4.2. Deep Learning Techniques
4.2.1. Recurrent Neural Networks
4.2.2. Deep Autoencoder
4.2.3. Deep Neural Network
4.2.4. Deep Belief Network
4.2.5. Deep Convolutional Neural Network
4.2.6. Deep Generative Models
4.2.7. Deep Boltzmann Machine
4.2.8. Deep Reinforcement Learning
4.2.9. Extreme Learning Machine
4.2.10. Summary of DL Models
4.2.11. Limitation of DL Models
5. Open Challenges
5.1. Precision Medicine
5.1.1. Using Appropriate Datasets
5.1.2. Use of Bio-Inspired Computing Approaches
5.1.3. Difficulty in Achieving Accuracy
5.1.4. Choosing the Correct Features
5.1.5. Trustworthy AI
5.1.6. Data Privacy and Confidentiality
- Data breaches: AI-based disease diagnosis systems store large amounts of sensitive patient data, making them a target for cyberattacks. A data breach could result in the unauthorized access or disclosure of patient information, which could lead to serious privacy violations.
- Data sharing: AI-based disease diagnosis systems often share patient data with other organizations, such as research institutions and other healthcare providers. This can raise concerns regarding the security and privacy of data, as well as the potential misuse of data.
- Data anonymization: AI-based disease diagnosis systems may use anonymized data to protect patient privacy. However, it is possible to re-identify patients from anonymized data, and there is a risk that the data could be used for unintended purposes.
- Data storage: AI-based disease diagnosis systems store large amounts of patient data. This data can be stored in multiple locations and can be vulnerable to hacking, data breaches, and data loss.
- Lack of transparency: AI-based disease diagnosis systems may lack transparency in the way they collect, store, and use patient data, which can make it difficult for patients to understand how their data are being used and control access to their data.
- Bias and discrimination: AI models can be affected by bias and discrimination, which can lead to inaccurate or unreliable results, especially for a certain population group.
6. Limitations of This Review
7. Future Research Directions
7.1. Integration with Other Diagnostic Tools
7.2. Handling Missing Data and Uncertainty
7.3. Personalized Medicine
7.4. Deep Learning Algorithm
7.5. Real-Time Analysis
7.6. Explainable AI
7.7. Automated Diagnosis
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sankaranarayanan, R.; Ramadas, K.; Amarasinghe, H.; Subramanian, S.; Johnson, N. Oral cancer: Prevention, early detection, and treatment. In Cancer: Disease Control Priorities, 3rd ed.; National Library of Medicine: Bethesda, MD, USA, 2015; Volume 3. [Google Scholar]
- Oral Cavity & Oropharyngeal Cancer Key Statistics 2021 (n.d.). Oral Cavity & Oropharyngeal Cancer Key Statistics. Available online: https://www.cancer.org/cancer/oral-cavity-and-oropharyngeal-cancer/about/key-statistics.html (accessed on 29 January 2023).
- Borse, V.; Konwar, A.N.; Buragohain, P. Oral cancer diagnosis and perspectives in India. Sens. Int. 2020, 1, 100046. [Google Scholar] [CrossRef]
- Rajaguru, H.; Prabhakar, S.K. Performance Comparison of Oral Cancer Classification with Gaussian Mixture Measures and Multi Layer Perceptron. IFMBE Proc. 2017, 61, 123–129. [Google Scholar] [CrossRef]
- GLOBOCAN 2020: New Global Cancer Data|UICC. GLOBOCAN 2020: New Global Cancer Data|UICC. 27 June 2022. Available online: https://www.uicc.org/news/globocan-2020-new-globalcan-cer%20zdata#:~:text=What%20is%20GLOBOCAN%3F,for%20all%20cancer%20sites%20combined (accessed on 29 January 2023).
- Lavanya, J.; Kavya, G.; Prasamya, N. Oral Cancer Diagnosis using Deep Learning for Early Detection. In Proceeedings of the 2022 International Conference on Electronics and Renewable Systems (ICEARS), 16–18 March 2022; IEEE: Tuticorin, India, 2022; pp. 1260–1268. [Google Scholar]
- Rivera, C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11884–11894. [Google Scholar] [PubMed]
- Le Campion, A.C.O.V.; Ribeiro, C.M.B.; Luiz, R.R.; Júnior, F.F.D.S.; Barros, H.C.S.; Santos, K.D.C.B.D.; Ferreira, S.J.; Gonçalves, L.S.; Ferreira, S.M.S. Low Survival Rates of Oral and Oropharyngeal Squamous Cell Carcinoma. Int. J. Dent. 2017, 2017, 5815493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, N.; Om, H. Extracting Significant Patterns for Oral Cancer Detection Using Apriori Algorithm. Intell. Inf. Manag. 2014, 6, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Song, B.; Sunny, S.; Uthoff, R.; Patrick, S.; Suresh, A.; Kolur, T.; Keerthi, G.; Anbarani, A.; Wilder-Smith, P.; Kuriakose, M.A.; et al. Automatic classification of dual-modalilty, smartphone-based oral dysplasia and malignancy images using deep learning. Biomed. Opt. Express 2018, 9, 5318–5329. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Hussain, E.; Mahanta, L.B. Automated classification of cells into multiple classes in epithelial tissue of oral squamous cell carcinoma using transfer learning and convolutional neural network. Neural Networks 2020, 128, 47–60. [Google Scholar] [CrossRef]
- Liu, X.; Faes, L.; Kale, A.U.; Wagner, S.K.; Fu, D.J.; Bruynseels, A.; Mahendiran, T.; Moraes, G.; Shamdas, M.; Kern, C.; et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health 2019, 1, e271–e297. [Google Scholar] [CrossRef]
- Yang, H.; Jo, E.; Kim, H.J.; Cha, I.-H.; Jung, Y.-S.; Nam, W.; Kim, J.-Y.; Kim, J.-K.; Kim, Y.H.; Oh, T.G.; et al. Deep Learning for Automated Detection of Cyst and Tumors of the Jaw in Panoramic Radiographs. J. Clin. Med. 2020, 9, 1939. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Aubreville, M.; Knipfer, C.; Oetter, N.; Jaremenko, C.; Rodner, E.; Denzler, J.; Bohr, C.; Neumann, H.; Stelzle, F.; Maier, A. Automatic Classification of Cancerous Tissue in Laserendomicroscopy Images of the Oral Cavity using Deep Learning. Sci. Rep. 2017, 7, 11979. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.K.; Kaur, M.; Manhas, J. Tissue Level Based Deep Learning Framework for Early Detection of Dysplasia in Oral Squamous Epithelium. J. Multimedia Inf. Syst. 2019, 6, 81–86. [Google Scholar] [CrossRef]
- López-Cortés, X.A.; Matamala, F.; Venegas, B.; Rivera, C. Machine-Learning Applications in Oral Cancer: A Sys-tematic Review. Appl. Sci. 2022, 12, 5715. [Google Scholar] [CrossRef]
- Sulochana, C.; Sumathi, M. A Systematic Review on Oral Cancer Diagnosis and Prognosis using Machine Learning Techniques. J. Algebraic Stat. 2022, 13, 3542–3550. [Google Scholar]
- Adeoye, J.; Tan, J.Y.; Choi, S.-W.; Thomson, P. Prediction models applying machine learning to oral cavity cancer outcomes: A systematic review. Int. J. Med Inform. 2021, 154, 104557. [Google Scholar] [CrossRef]
- Sujir, N.; Ahmed, J.; Pai, K.; Denny, C.; Shenoy, N. Challenges in early diagnosis of oral cancer: Cases series. Acta Stomatol. Croat. Int. J. Oral Sci. Dent. Med. 2019, 53, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Goswami, C.P.; Nakshatri, H. PROGgene: Gene expression based survival analysis web application for multiple cancers. J. Clin. Bioinform. 2013, 3, 22. [Google Scholar] [CrossRef] [Green Version]
- Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef] [Green Version]
- Elfilali, A.; Lair, S.; Verbeke, C.; La Rosa, P.; Radvanyi, F.; Barillot, E. ITTACA: A new database for integrated tumor transcriptome array and clinical data analysis. Nucleic Acids Res. 2006, 34, D613–D616. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Xie, L.; Dang, Y.; Sun, X.; Xie, T.; Guo, J.; Han, Y.; Yan, Z.; Zhu, W.; Wang, Y.; et al. OSlms: A Web Server to Evaluate the Prognostic Value of Genes in Leiomyosarcoma. Front. Oncol. 2019, 9, 190. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Wang, F.; Lv, J.; Xin, J.; Xie, L.; Zhu, W.; Guo, X. Interactive online consensus survival tool for esoph-ageal squamous cell carcinoma prognosis analysis. Oncol. Lett. 2019, 18, 1199–1206. [Google Scholar] [PubMed]
- Ilhan, B.; Guneri, P.; Wilder-Smith, P. The contribution of artificial intelligence to reducing the diagnostic delay in oral cancer. Oral Oncol. 2021, 116, 105254. [Google Scholar] [CrossRef]
- Montero, P.H.; Patel, S.G. Cancer of the oral cavity. Surg. Oncol. Clin. 2015, 24, 491–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruccia, M.; Onesti, M.G.; Parisi, P.; Cigna, E.; Troccola, A.; Scuderi, N. Lip cancer: A 10-year retrospective epide-miological study. Anticancer. Res. 2012, 32, 1543–1546. [Google Scholar]
- de Morais, E.F.; Carlan, L.M.; de Farias Morais, H.G.; Pinheiro, J.C.; Martins, H.D.D.; Barboza, C.A.G.; de Almeida Freitas, R. Primary intraosseous squamous cell carcinoma involving the jaw bones: A systematic review and up-date. Head Neck Pathol. 2021, 15, 608–616. [Google Scholar] [CrossRef]
- Warnakulasuriya, S. Oral potentially malignant disorders: A comprehensive review on clinical aspects and man-agement. Oral Oncol. 2020, 102, 104550. [Google Scholar] [CrossRef] [PubMed]
- García-Pola, M.; Pons-Fuster, E.; Suárez-Fernández, C.; Seoane-Romero, J.; Romero-Méndez, A.; López-Jornet, P. Role of Artificial Intelligence in the Early Diagnosis of Oral Cancer. A Scoping Review. Cancers 2021, 13, 4600. [Google Scholar] [CrossRef]
- Avon, S.L.; McComb, J.; Clokie, C. Ameloblastic carcinoma: Case report and literature review. J. Can. Dent. Assoc. 2003, 69, 573–576. [Google Scholar]
- Alhabbab, R.; Johar, R. Lip cancer prevalence, epidemiology, diagnosis, and management: A review of the litera-ture. Adv. Oral Maxillofac. Surg. 2022, 24, 100276. [Google Scholar] [CrossRef]
- de Visscher, J.; Schaapveld, M.; Otter, R.; Visser, O.; van der Waal, I. Epidemiology of cancer of the lip in the Netherlands. Oral Oncol. 1998, 34, 421–426. [Google Scholar] [CrossRef]
- Wien, R.O. Oral Cancer; Shah, J.P., Johnson, N.W., Batsakis, J.G., Dunitz, M., Eds.; Thieme Medical Publishers, Inc.: New York, NY, USA, 2003; p. 496. [Google Scholar]
- Shah, J.P.; Gil, Z. Current concepts in management of oral cancer–surgery. Oral Oncol. 2009, 45, 394–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Types of Skin Cancer: Common, Rare and More Varieties. Cancer Treatment Centers of America. 1 June 2022. Available online: https://www.cancercenter.com/cancer-types/skin-cancer/types (accessed on 15 January 2023).
- Chakraborty, D.; Natarajan, C.; Mukherjee, A. Advances in oral cancer detection. Adv. Clin. Chem. 2019, 91, 181–200. [Google Scholar] [CrossRef]
- Manikandan, M.; Rao, A.K.D.M.; Arunkumar, G.; Manickavasagam, M.; Rajkumar, K.S.; Rajaraman, R.; Munirajan, A.K. Oral squamous cell carcinoma: MicroRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol. Cancer 2016, 15, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, J.; Sertel, O.; Shimada, H.; Boyer, K.; Saltz, J.; Gurcan, M. Computer-aided evaluation of neuroblastoma on whole-slide histology images: Classifying grade of neuroblastic differentiation. Pattern Recognit. 2009, 42, 1080–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santana, M.F.; Ferreira, L.C.L. Diagnostic errors in surgical pathology. J. Bras. Patol. Med. Lab. 2017, 53, 124–129. [Google Scholar] [CrossRef]
- Ghosh, A.; Chaudhuri, D.; Adhikary, S.; Chatterjee, K.; Roychowdhury, A.; Das, A.K.; Barui, A. Deep reinforced neural network model for cyto-spectroscopic analysis of epigenetic markers for automated oral cancer risk prediction. Chemom. Intell. Lab. Syst. 2022, 224, 104548. [Google Scholar] [CrossRef]
- Lo Muzio, L. Nevoid basal cell carcinoma syndrome (Gorlin syndrome). Orphanet J. Rare Dis. 2008, 3, 32. [Google Scholar] [CrossRef] [Green Version]
- Fania, L.; Didona, D.; Morese, R.; Campana, I.; Coco, V.; Di Pietro, F.R.; Ricci, F.; Pallotta, S.; Candi, E.; Abeni, D.; et al. Basal Cell Carcinoma: From Pathophysiology to Novel Therapeutic Approaches. Biomedicines 2020, 8, 449. [Google Scholar] [CrossRef]
- Marzuka, A.G.; Book, S.E. Basal cell carcinoma: Pathogenesis, epidemiology, clinical features, diagnosis, histo-pathology, and management. Yale J. Biol. Med. 2015, 88, 167–179. [Google Scholar]
- Furdova, A.; Lukacko, P. Periocular Basal Cell Carcinoma Predictors for Recurrence and Infiltration of the Orbit. J. Craniofacial Surg. 2017, 28, e84–e87. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.T.; Wu, A.; Figueira, E.; Huilgol, S.; Selva, D. Management of periorbital basal cell carcinoma with orbital invasion. Futur. Oncol. 2015, 11, 3003–3010. [Google Scholar] [CrossRef]
- Jaw Cancer Symptoms, Causes, Treatment & Survival Rate. Cancer Treatment Centers of America. 31 May 2022. Available online: https://www.cancercenter.com/cancer-types/oral-cancer/types/jaw-cancer (accessed on 6 December 2022).
- Israel, O.; Kuten, A. Early detection of cancer recurrence: 18F-FDG PET/CT can make a difference in diagnosis and patient care. J. Nucl. Med. 2007, 48, 28S–35S. [Google Scholar] [PubMed]
- Soyele, O.O.; Adebiyi, K.E.; Adesina, O.M.; Ladeji, A.M.; Aborisade, A.; Olatunji, A.; Adeola, H.A. Ameloblastic carcinoma: A clinicopathologic analysis of cases seen in a Nigerian Teaching Hospital and review of literature. Pan Afr. Med J. 2018, 31, 208. [Google Scholar] [CrossRef] [PubMed]
- Moro, A.; Foresta, E.; Gasparini, G.; Pelo, S.; Forcione, M.; Cristallini, E.G.; Toraldo, M.; Lorenzo, C.; Falchi, M.; Saponaro, G. Ameloblastic carcinoma of the maxilla: A case report and an updated review of the literature. Oncol. Lett. 2016, 12, 4339–4350. [Google Scholar] [CrossRef] [Green Version]
- Effiom, O.A.; Ogundana, O.M.; Akinshipo, A.O.; Akintoye, S.O. Ameloblastoma: Current etiopathological con-cepts and management. Oral Dis. 2018, 24, 307–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angiero, F.; Borloni, R.; Macchi, M.; Stefani, M. Ameloblastic carcinoma of the maxillary sinus. Anticancer. Res. 2009, 28, 3847–3854. [Google Scholar]
- Mubeen, K.; Shakya, H.K.; Jigna, V.R. Ameloblastic carcinoma of mandible. A rare case report with review of literature. J. Clin. Exp. Dent. 2010, 2, e83–e87. [Google Scholar]
- Wu, J.Y.; Chi, L.H.; Pemg, B.Y.; Lin, Y.H. Ameloblastic carcinoma of the maxilla-case report. J. Dent. Sci. 2007, 2, 164–170. [Google Scholar]
- Datta, R.; Winston, J.S.; Diaz-Reyes, G.; Loree, T.R.; Myers, L.; Kuriakose, M.; Rigual, N.R.; Hicks, W.L. Ameloblastic carcinoma: Report of an aggressive case with multiple bony metastases. Am. J. Otolaryngol. 2003, 24, 64–69. [Google Scholar] [CrossRef]
- Uzawa, N.; Suzuki, M.; Miura, C.; Tomomatsu, N.; Izumo, T.; Harada, K. Primary ameloblastic carcinoma of the maxilla: A case report and literature review. Oncol. Lett. 2014, 9, 459–467. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, H.; Katase, N.; Hara, M.; Asaumi, J.I.; Yanagi, Y.; Unetsubo, T.; Nagatsuka, H. Ameloblastic carcino-ma: A case report with radiological features of computed tomography and magnetic resonance imaging and positron emis-sion tomography. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2011, 112, e40–e47. [Google Scholar] [CrossRef]
- Saxena, C.; Aggarwal, P.; Wadhwan, V.; Bansal, V. Primary intraosseous squamous cell carcinoma in odontogenic keratocyst: A rare entity. J. Oral Maxillofac. Pathol. 2015, 19, 406. [Google Scholar] [CrossRef]
- Sengupta, S.; Vij, H.; Vij, R. Primary intraosseous carcinoma of the mandible: A report of two cases. J. Oral Maxillofac. Pathol. 2010, 14, 69–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nokovitch, L.; Bodard, A.-G.; Corradini, N.; Crozes, C.; Guyennon, A.; Deneuve, S. Pediatric case of squamous cell carcinoma arising from a keratocystic odontogenic tumor. Int. J. Pediatr. Otorhinolaryngol. 2018, 112, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Abdelkarim, A.Z.; Elzayat, A.M.; Syed, A.Z.; Lozanoff, S. Delayed diagnosis of a primary intraosseous squamous cell carcinoma: A case report. Imaging Sci. Dent. 2019, 49, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Todorovic, E.; Berthelet, E.; O’Connor, R.; Durham, J.S.; Tran, E.; Martin, M.; Hayes, M.M.; Ng, T.L. Sclerosing Odontogenic Carcinoma with Local Recurrence: Case Report and Review of Literature. Head Neck Pathol. 2018, 13, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Speight, P.M.; Takata, T. New tumour entities in the 4th edition of the World Health Organization Classification of Head and Neck tumours: Odontogenic and maxillofacial bone tumours. Virchows Arch. 2017, 472, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.H.; Yeo, J.F.; Pang, B.N.K.; Petersson, F. An intraosseous sclerosing odontogenic tumor predominantly composed of epithelial cells: Relation to (so-called) sclerosing odontogenic carcinoma and epithelial-rich central odontogenic fibroma. Oral Surgery, Oral Med. Oral Pathol. Oral Radiol. 2014, 118, e119–e125. [Google Scholar] [CrossRef]
- Hussain, O.; Rendon, A.T.; Orr, R.L.; Speight, P.M. Sclerosing odontogenic carcinoma in the maxilla: A rare primary intraosseous carcinoma. Oral Surgery, Oral Med. Oral Pathol. Oral Radiol. 2013, 116, e283–e286. [Google Scholar] [CrossRef]
- Koutlas, I.G.; Allen, C.M.; Warnock, G.R.; Manivel, J.C. Sclerosing odontogenic carcinoma: A previously unre-ported variant of a locally aggressive odontogenic neoplasm without apparent metastatic potential. Am. J. Surg. Pathol. 2008, 32, 1613–1619. [Google Scholar] [CrossRef]
- Richardson, M.S.; Muller, S. Malignant Odontogenic Tumors: An Update on Selected Tumors. Head Neck Pathol. 2014, 8, 411–420. [Google Scholar] [CrossRef] [Green Version]
- Wood, A.; Young, F.; Morrison, J.; Conn, B.I. Sclerosing odontogenic carcinoma presenting on the hard palate of a 43-year-old female: A case report. Oral Surgery, Oral Med. Oral Pathol. Oral Radiol. 2016, 122, e204–e208. [Google Scholar] [CrossRef] [PubMed]
- Ide, F.; Mishima, K.; Saito, I.; Kusama, K. Diagnostically Challenging Epithelial Odontogenic Tumors: A Selective Review of 7 Jawbone Lesions. Head Neck Pathol. 2009, 3, 18–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yancoskie, A.E.; Sreekantaiah, C.; Jacob, J.; Rosenberg, A.; Edelman, M.; Antonescu, C.R.; Fantasia, J.E. EWSR1 and ATF1 rearrangements in clear cell odontogenic carcinoma: Presentation of a case. Oral Surgery, Oral Med. Oral Pathol. Oral Radiol. 2014, 118, e115–e118. [Google Scholar] [CrossRef] [PubMed]
- Labrador, A.J.P.; Marin, N.R.G.; Valdez, L.H.M.; Valentina, M.P.; Sanchez, K.B.T.; Ibazetta, K.A.R.; Johan, B.; Cesar, A.V.; Wright, J.M. Clear Cell Odontogenic Carcinoma a Systematic Review. Head Neck Pathol. 2021, 16, 838–848. [Google Scholar] [CrossRef]
- Santana, T.; de Andrade, F.L.; de Sousa Melo, M.C.; da Rocha, G.B.L.; Trierveiler, M. Clear cell odontogenic car-cinoma harboring the EWSR1–ATF1 fusion gene: Report of a rare case. Head Neck Pathol. 2020, 14, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Ellis, G.L. Clear cell neoplasms in salivary glands: Clearly a diagnostic challenge. Ann. Diagn. Pathol. 1998, 2, 61–78. [Google Scholar] [CrossRef]
- Guastaldi, F.P.S.; Faquin, W.C.; Gootkind, F.; Hashemi, S.; August, M.; Iafrate, A.J.; Troulis, M.J. Clear cell odontogenic carcinoma: A rare jaw tumor. A summary of 107 reported cases. Int. J. Oral Maxillofac. Surg. 2019, 48, 1405–1410. [Google Scholar] [CrossRef]
- Jain, A.; Shetty, D.C.; Juneja, S.; Narwal, N. Molecular characterization of clear cell lesions of head and neck. J. Clin. Diagn. Res. 2016, 10, ZE18. [Google Scholar] [CrossRef]
- Kumar, M.; Fasanmade, A.; Barrett, A.W.; Mack, G.; Newman, L.; Hyde, N.C. Metastasising clear cell odontogenic carcinoma: A case report and review of the literature. Oral Oncol. 2002, 39, 190–194. [Google Scholar] [CrossRef]
- Chera, B.S.; Villaret, D.B.; Orlando, C.A.; Mendenhall, W.M. Clear cell odontogenic carcinoma of the maxilla: A case report and literature review. Am. J. Otolaryngol. 2008, 29, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, S.; Kumar, R.; Sarkar, C.; Ralte, M.; Sharma, M.C. Clear cell odontogenic carcinoma: A diagnostic dilemma. Pathol. Oncol. Res. 2002, 8, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, R.; Ray, J.; Swain, N. Clear cell odontogenic carcinoma of maxilla: A case report and mini review. J. Oral Maxillofac. Pathol. 2013, 17, 89–94. [Google Scholar] [CrossRef] [Green Version]
- Loyola, A.M.; Cardoso, S.V.; de Faria, P.R.; Servato, J.P.S.; de Paulo, L.F.B.; Eisenberg, A.L.A.; Dias, F.L.; Gomes, C.C.; Gomez, R.S. Clear cell odontogenic carcinoma: Report of 7 new cases and systematic review of the current knowledge. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 2015, 120, 483–496. [Google Scholar] [CrossRef]
- Kwon, I.J.; Kim, S.M.; Amponsah, E.K.; Myoung, H.; Lee, J.H.; Lee, S.K. Mandibular clear cell odontogenic car-cinoma. World J. Surg. Oncol. 2015, 13, 284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panda, S.; Sahoo, S.R.; Srivastav, G.; Padhiary, S.; Dhull, K.S.; Aggarwal, S. Pathogenesis and Nomenclature of Odontogenic Carcinomas: Revisited. J. Oncol. 2014, 2014, 197425. [Google Scholar] [CrossRef] [Green Version]
- Ghita, I.; Nagai, M.Y.; Lubek, J.E.; Stashek, K.M.; Basile, J.R.; Price, J.B.; Papadimitriou, J.C.; Dyalram, D.; Younis, R.H. Ghost Cell Odontogenic Carcinoma Arising in a Previous Calcifying Odontogenic Cyst: A Case Report and Review of Literature. Head Neck Pathol. 2022, 16, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Montes, C.; Gorlin, R.J.; Shear, M.; Prae’torius, F.; Mosqueda-Taylor, A.; Altini, M.; Meneses-García, A. International collaborative study on ghost cell odontogenic tumours: Calcifying cystic odontogenic tumour, dentinogenic ghost cell tumour and ghost cell odontogenic carcinoma. J. Oral Pathol. Med. 2008, 37, 302–308. [Google Scholar] [CrossRef]
- Lee, S.K.; Kim, Y.-S. Current Concepts and Occurrence of Epithelial Odontogenic Tumors: II. Calcifying Epithelial Odontogenic Tumor Versus Ghost Cell Odontogenic Tumors Derived from Calcifying Odontogenic Cyst. Korean J. Pathol. 2014, 48, 175–187. [Google Scholar] [CrossRef]
- Alekhya, B.; Majumdar, S.; Uppala, D.; Sreekanth, K. Odontogenic carcinosarcoma—A rare case report with review of literature. J. Oral Maxillofac. Pathol. 2022, 26, 51. [Google Scholar] [CrossRef]
- da Silva, K.D.; Flores, I.L.; Etges, A.; Vasconcelos, A.C.U.; Mesquita, R.A.; Gomes, A.P.N.; Tarquinio, S.B.C. Unusual osteolytic lesion of the jaw. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2017, 124, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Marin, C.; Dave, M.; Hunter, K.D. Malignant Odontogenic Tumours: A Systematic Review of Cases Reported in Literature. Front. Oral Heal. 2021, 2, 82. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.-K.; Pae, S.-P.; Cho, H.-Y.; Seo, J.-H.; Lee, D.-H.; Park, I.-S. Odontogenic carcinosarcoma of the mandible: A case report and review. J. Korean Assoc. Oral Maxillofac. Surg. 2015, 41, 139–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, I.R.; Pindborg, J.J.; Shear, M. The World Health Organization histological typing of odontogenic tumours. Introducing the second edition. Eur. J. Cancer Part B: Oral Oncol. 1993, 29, 169–171. [Google Scholar] [CrossRef] [PubMed]
- Bregni, R.C.; Taylor, A.M.; García, A.M. Ameloblastic fibrosarcoma of the mandible: Report of two cases and review of the literature. J. Oral Pathol. Med. 2001, 30, 316–320. [Google Scholar] [CrossRef]
- Chrcanovic, B.R.; Brennan, P.A.; Rahimi, S.; Gomez, R.S. Ameloblastic fibroma and ameloblastic fibrosar-coma: A systematic review. J. Oral Pathol. Med. 2017, 47, 315–325. [Google Scholar] [CrossRef]
- Ramani, P.; Krishnan, R.P.; Karunagaran, M.; Muthusekhar, M.R. Odontogenic sarcoma: First report after new who nomenclature with systematic review. J. Oral Maxillofac. Pathol. 2020, 24, 157–163. [Google Scholar] [CrossRef]
- Buccal Mucosa Cancer: Symptoms, Causes & Treatment. Cleveland Clinic. Available online: https://my.clevelandclinic.org/health/diseases/23423-buccal-mucosa-inner-cheek-cancer (accessed on 3 January 2023).
- Sánchez-Alarcón, J.; Milić, M.; Gómez-Arroyo, S.; Montiel-González, J.M.R.; Valencia-Quintana, J.M.R.M.-G.A.R. Assessment of DNA Damage by Comet Assay in Buccal Epithelial Cells: Problems, Achievement, Perspectives. In Environmental Health Risk-Hazardous Factors to Living Species; InTech: Rijeka, Croatia, 2016. [Google Scholar] [CrossRef] [Green Version]
- Yanuaryska, R.D. Comet Assay Assessment of DNA Damage in Buccal Mucosa Cells Exposed to X-Rays via Panoramic Radiography. J. Dent. Indones. 2018, 25, 53–57. [Google Scholar] [CrossRef]
- Sieczka, E.; Datta, R.; Singh, A.; Loree, T.; Rigual, N.; Orner, J.; Hicks, W., Jr. Cancer of the buccal mucosa: Are mar-gins and T-stage accurate predictors of local control? Am. J. Otolaryngol. 2001, 22, 395–399. [Google Scholar] [CrossRef]
- Hicks, W.L., Jr.; Loree, T.R.; Garcia, R.I.; Maamoun, S.; Marshall, D.; Orner, J.B.; Shedd, D.P. Squamous cell car-cinoma of the floor of mouth: A 20-year review. Head Neck J. Sci. Spec. Head Neck 1997, 19, 400–405. [Google Scholar] [CrossRef]
- Luryi, A.L.; Chen, M.M.; Mehra, S.; Roman, S.A.; Sosa, J.A.; Judson, B.L. Positive Surgical Margins in Early Stage Oral Cavity Cancer: An Analysis of 20,602 Cases. Otolaryngol. Neck Surg. 2014, 151, 984–990. [Google Scholar] [CrossRef] [PubMed]
- Delclos, L.; Lindberg, R.; Fletcher, G. Squamous cell carcinoma of the oral tongue and floor of mouth. Evaluation of interstitial radium therapy. Am. J. Roentgenol. 1976, 126, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Krause, C.J.; Lee, J.G.; McCabe, B.F. Carcinoma of the oral cavity: A comparison of therapeutic modali-ties. Arch. Otolaryngol. 1973, 97, 354–358. [Google Scholar] [CrossRef] [PubMed]
- E Marks, J.; Lee, F.; Freeman, R.B.; Zivnuska, F.R.; Ogura, J.H. Carcinoma of the oral tongue: A study of patient selection and treatment results. Laryngoscope 1981, 91, 1548–1559. [Google Scholar] [CrossRef]
- Mendenhall, W.M.; Van Cise, W.S.; Bova, F.J.; Million, R.R. Analysis of time-dose factors in squamous cell car-cinoma of the oral tongue and floor of mouth treated with radiation therapy alone. Int. J. Radiat. Oncol. Biol. Phys. 1981, 7, 1005–1011. [Google Scholar] [CrossRef]
- Hammouda, Y.; Halily, S.; Oukessou, Y.; Rouadi, S.; Abada, R.; Roubal, M.; Mahtar, M. Malignant tumors of the hard palate: Report of 4 cases and review of the literature. Int. J. Surg. Case Rep. 2020, 78, 228–234. [Google Scholar] [CrossRef]
- Hard Palate Cancer (n.d.). Hard Palate Cancer | Memorial Sloan Kettering Cancer Center. Available online: https://www.mskcc.org/cancer-care/types/mouth/types-mouth/hard-palate (accessed on 27 January 2023).
- Amisha; Malik, P.; Pathania, M.; Rathaur, V.K. Overview of artificial intelligence in medicine. J. Fam. Med. Prim. Care 2019, 8, 2328–2331. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Bossuyt, P.M.; Reitsma, J.B.; E Bruns, D.; A Gatsonis, C.; Glasziou, P.P.; Irwig, L.; Lijmer, J.G.; Moher, D.; Rennie, D.; de Vet, H.C.W.; et al. STARD 2015: An Updated List of Essential Items for Reporting Diagnostic Accuracy Studies. Clin. Chem. 2015, 61, 1446–1452. [Google Scholar] [CrossRef] [Green Version]
- Sreeshyla, H.S.; Sudheendra, U.; Shashidara, R. Vital tissue staining in the diagnosis of oral precancer and cancer: Stains, technique, utility, and reliability. Clin. Cancer Investig. J. 2014, 3, 141. [Google Scholar] [CrossRef]
- Nagaraju, K.; Prasad, S.; Ashok, L. Diagnostic efficiency of toluidine blue with Lugol′s iodine in oral premalignant and malignant lesions. Indian J. Dent. Res. 2010, 21, 218–223. [Google Scholar] [CrossRef]
- Mehrotra, R.; Singh, M.; Thomas, S.; Nair, P.; Pandya, S.; Nigam, N.S.; Shukla, P. A cross-sectional study evaluating chemiluminescence and autofluorescence in the detection of clinically innocuous precancerous and cancerous oral le-sions. J. Am. Dent. Assoc. 2010, 141, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Vigneswaran, N.; Gillenwater, A.; Richards-Kortum, R. Advances in fluorescence imaging techniques to detect oral cancer and its precursors. Futur. Oncol. 2010, 6, 1143–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messadi, D.V. Diagnostic aids for detection of oral precancerous conditions. Int. J. Oral Sci. 2013, 5, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Nagler, R.M. Saliva as a tool for oral cancer diagnosis and prognosis. Oral Oncol. 2009, 45, 1006–1010. [Google Scholar] [CrossRef] [PubMed]
- Bahar, G.; Feinmesser, R.; Shpitzer, T.; Popovtzer, A.; Nagler, R.M. Salivary analysis in oral cancer patients: DNA and protein oxidation, reactive nitrogen species, and antioxidant profile. Cancer 2007, 109, 54–59. [Google Scholar] [CrossRef]
- Li, Y.; John, M.A.R.S.; Wong, D.T.; Zhou, X.; Kim, Y.; Sinha, U.; Jordan, R.C.K.; Eisele, D.; Abemayor, E.; Elashoff, D.; et al. Salivary Transcriptome Diagnostics for Oral Cancer Detection. Clin. Cancer Res. 2004, 10, 8442–8450. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Hou, D.; Wen, X.; Xin, M.; Li, Z.; Wu, L.; Pathak, J.L. Gold nanomaterials for oral cancer diagnosis and therapy: Advances, challenges, and prospects. Mater. Today Bio 2022, 15, 100333. [Google Scholar] [CrossRef]
- Zuluaga, A.F.; Follen, M.; Boiko, I.; Malpica, A.; Richards-Kortum, R. Optical coherence tomography: A pilot study of a new imaging technique for noninvasive examination of cervical tissue. Am. J. Obstet. Gynecol. 2005, 193, 83–88. [Google Scholar] [CrossRef]
- Assayag, O.; Antoine, M.; Sigal-Zafrani, B.; Riben, M.; Harms, F.; Burcheri, A.; Boccara, C. Large field, high reso-lution fullfield optical coherence tomography: A pre-clinical study of human breast tissue and cancer assessment. Technol. Cancer Res. Treat. 2014, 13, 455–468. [Google Scholar]
- Iftimia, N.; Iyer, A.K.; Hammer, D.X.; Lue, N.; Mujat, M.; Pitman, M.; Ferguson, R.D.; Amiji, M. Fluorescence-guided optical coherence tomography imaging for colon cancer screening: A preliminary mouse study. Biomed. Opt. Express 2011, 3, 178–191. [Google Scholar] [CrossRef] [Green Version]
- Tests for Oral Cavity (Mouth) and Oropharyngeal (Throat) Cancers (n.d.). Tests for Oral Cavity (Mouth) and Oropharyngeal (Throat) Cancers. Available online: https://www.cancer.org/cancer/oral-cavity-and-oropharyngeal-cancer/detection-diagnosis-staging/how-diagnosed.html (accessed on 6 December 2022).
- Pandya, D.; Nagarajappa, A.K.; Reddy, S.; Bhasin, M. Lab-on-a-chip-oral cancer diagnosis at your door step. J. Int. Oral Health 2015, 7, 122. [Google Scholar]
- Daniel, G.S.T.; Thiruppathy, M.; Aswath, N.; Narayanan, S.R. Lab on a Chip: Conquer Disease at the Earliest. J. Pharm. Bioallied Sci. 2018, 10, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Skandarajah, A.; Sunny, S.P.; Gurpur, P.; Reber, C.D.; D’Ambrosio, M.V.; Raghavan, N.; James, B.L.; Ramanjinappa, R.D.; Suresh, A.; Kandasarma, U.; et al. Mobile microscopy as a screening tool for oral cancer in India: A pilot study. PLoS ONE 2017, 12, e0188440. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.-F.; Chen, Y.-J.; Tsai, F.-T.; Li, W.-C.; Hsu, M.-L.; Wang, D.-H.; Yang, C.-C. Current Insights into Oral Cancer Diagnostics. Diagnostics 2021, 11, 1287. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.; Kumar, V.; Saini, S. Texture analysis based segmentation and classification of oral cancer lesions in color images using ANN. In Proceedings of the 2013 IEEE International Conference on Signal Processing, Computing and Control (ISPCC), Solan, India, 26–28 September 2013; pp. 1–5. [Google Scholar] [CrossRef]
- Singh, B.K.; Verma, K.; Panigrahi, L.; Thoke, A.S. Integrating radiologist feedback with computer aided diagnos-tic systems for breast cancer risk prediction in ultrasonic images: An experimental investigation in machine learning para-digm. Expert Syst. Appl. 2017, 90, 209–223. [Google Scholar] [CrossRef]
- Nanditha, B.R.; MP, G.K.A.S. Oral Cancer Detection using Machine Learning and Deep Learning Techniques. Int. J. Cur. Res. Rev. 2022, 14, 64. [Google Scholar]
- Alhazmi, A.; Alhazmi, Y.; Makrami, A.; Masmali, A.; Salawi, N.; Masmali, K.; Patil, S. Application of artificial in-telligence and machine learning for prediction of oral cancer risk. J. Oral Pathol. Med. 2021, 50, 444–450. [Google Scholar] [CrossRef]
- Sunny, S.; Baby, A.; James, B.L.; Balaji, D.; Rana, M.H.; Gurpur, P.; Skandarajah, A.; D’Ambrosio, M.; Ramanjinappa, R.D.; Mohan, S.P.; et al. A smart tele-cytology point-of-care platform for oral cancer screening. PLoS ONE 2019, 14, e0224885. [Google Scholar] [CrossRef] [Green Version]
- Tseng, W.-T.; Chiang, W.-F.; Liu, S.-Y.; Roan, J.; Lin, C.-N. The Application of Data Mining Techniques to Oral Cancer Prognosis. J. Med. Syst. 2015, 39, 59. [Google Scholar] [CrossRef]
- Dharani, R.; Revathy, S. DEEPORCD: Detection of Oral Cancer using Deep Learning. J. Physics: Conf. Ser. 2021, 1911, 012006. [Google Scholar] [CrossRef]
- Romeo, V.; Cuocolo, R.; Ricciardi, C.; Ugga, L.; Cocozza, S.; Verde, F.; Stanzione, A.; Napolitano, V.; Russo, D.; Improta, G.; et al. Prediction of Tumor Grade and Nodal Status in Oropharyngeal and Oral Cavity Squamous-cell Carcinoma Using a Radiomic Approach. Anticancer. Res. 2019, 40, 271–280. [Google Scholar] [CrossRef]
- Exarchos, K.P.; Goletsis, Y.; Fotiadis, D.I. Multiparametric decision support system for the prediction of oral can-cer reoccurrence. IEEE Trans. Inf. Technol. Biomed. 2011, 16, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Anuradha, K.; Sankaranarayanan, K. Identification of Suspicious Regions to Detect Oral Cancers At An Earlier Stage-A Literature Survey. Int. J. Adv. Eng. Technol. 2012, 3, 84. [Google Scholar]
- Alkhadar, H.; Macluskey, M.; White, S.; Ellis, I.; Gardner, A. Comparison of machine learning algorithms for the prediction of five-year survival in oral squamous cell carcinoma. J. Oral Pathol. Med. 2021, 50, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Alabi, R.O.; Elmusrati, M.; Sawazaki-Calone, I.; Kowalski, L.P.; Haglund, C.; Coletta, R.D.; Leivo, I. Compari-son of supervised machine learning classification techniques in prediction of locoregional recurrences in early oral tongue cancer. Int. J. Med. Inform. 2020, 136, 104068. [Google Scholar] [CrossRef] [PubMed]
- Alabi, R.O.; Elmusrati, M.; Sawazaki-Calone, I.; Kowalski, L.P.; Haglund, C.; Coletta, R.D.; Almangush, A. Ma-chine learning application for prediction of locoregional recurrences in early oral tongue cancer: A Web-based prognostic tool. Virchows Arch. 2019, 475, 489–497. [Google Scholar] [CrossRef] [Green Version]
- Hung, M.; Park, J.; Hon, E.S.; Bounsanga, J.; Moazzami, S.; Ruiz-Negrón, B.; Wang, D. Artificial intelligence in den-tistry: Harnessing big data to predict oral cancer survival. World J. Clin. Oncol. 2020, 11, 918. [Google Scholar] [CrossRef]
- Lavanya, L.; Chandra, J. Oral cancer analysis using machine learning techniques. Int. J. Eng. Res. Technol 2019, 12, 596–601. [Google Scholar]
- Suji, R.J.; Rajagopalan, S.P. An automatic oral cancer classification using data mining techniques. Int. J. Adv. Res. Comput. Commun. Eng. 2013, 2, 3759–3765. [Google Scholar]
- Siddalingappa, R.; Kanagaraj, S. K-nearest-neighbor algorithm to predict the survival time and classification of various stages of oral cancer: A machine learning approach. F1000Research 2022, 11, 70. [Google Scholar] [CrossRef]
- Chu, C.S.; Lee, N.P.; Adeoye, J.; Thomson, P.; Choi, S. Machine learning and treatment outcome prediction for oral cancer. J. Oral Pathol. Med. 2020, 49, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Harnale, S.; Maktedar, D.D. Oral cancer detection: Hybrid method of KFCM clustering. Int. J. Recent Technol. Eng. 2020, 8, 2287–2292. [Google Scholar] [CrossRef]
- Vidhu, R.; Kiruthika, S. A new feature selection method for oral cancer using data mining techniques. Int. J. Adv. Res. Comput. Commun. Eng. 2016, 5. [Google Scholar] [CrossRef]
- Pilling, M.J.; Henderson, A.; Shanks, J.H.; Brown, M.D.; Clarke, N.W.; Gardner, P. Infrared spectral histopathol-ogy using haematoxylin and eosin (H&E) stained glass slides: A major step forward towards clinical translation. Analyst 2017, 142, 1258–1268. [Google Scholar]
- Das, D.K.; Chakraborty, C.; Sawaimoon, S.; Maiti, A.K.; Chatterjee, S. Automated identification of keratinization and keratin pearl area from in situ oral histological images. Tissue Cell 2015, 47, 349–358. [Google Scholar] [CrossRef]
- James, B.L.; Sunny, S.P.; Heidari, A.E.; Ramanjinappa, R.D.; Lam, T.; Tran, A.V.; Kuriakose, M.A. Validation of a point-of-care optical coherence tomography device with machine learning algorithm for detection of oral potentially ma-lignant and malignant lesions. Cancers 2021, 13, 3583. [Google Scholar] [CrossRef]
- Jain, D.K.; Dubey, S.B.; Choubey, R.K.; Sinhal, A.; Arjaria, S.K.; Jain, A.; Wang, H. An approach for hyperspectral image classification by optimizing SVM using self organizing map. J. Comput. Sci. 2018, 25, 252–259. [Google Scholar] [CrossRef]
- Mohd, F.; Abu Bakar, Z.; Noor, N.M.M.; Rajion, Z.A.; Saddki, N. A Hybrid Selection Method Based on HCELFS and SVM for the Diagnosis of Oral Cancer Staging. In Advanced Computer and Communication Engineering Technology: Proceedings of the 1st International Conference on Communication and Computer Engineering; Springer International Publishing: New York, NY, USA, 2014; pp. 821–831. [Google Scholar] [CrossRef]
- Chu, C.S.; Lee, N.P.; Ho, J.W.; Choi, S.W.; Thomson, P.J. Deep learning for clinical image analyses in oral squa-mous cell carcinoma: A review. JAMA Otolaryngol.—Head Neck Surg. 2021, 147, 893–900. [Google Scholar] [CrossRef]
- Patil, S.; Awan, K.; Arakeri, G.; Seneviratne, C.J.; Muddur, N.; Malik, S.; Ferrari, M.; Rahimi, S.; Brennan, P.A. Machine learning and its potential applications to the genomic study of head and neck cancer—A systematic review. J. Oral Pathol. Med. 2019, 48, 773–779. [Google Scholar] [CrossRef]
- Sharma, N.; Om, H. Using MLP and SVM for predicting survival rate of oral cancer patients. Netw. Model. Anal. Heal. Inform. Bioinform. 2014, 3, 1–10. [Google Scholar] [CrossRef]
- Pavani, V.; Babu, I.R. Using fog and Replication Techniques Inorder to Enhance Cloud Data Security. J. Crit. Rev. 2020, 7, 202–207. [Google Scholar]
- Liu, Y.; Li, Y.; Fu, Y.; Liu, T.; Liu, X.; Zhang, X.; Fu, J.; Guan, X.; Chen, T.; Chen, X.; et al. Quantitative prediction of oral cancer risk in patients with oral leukoplakia. Oncotarget 2017, 8, 46057–46064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yang, J.; Wei, C.; Zhou, G.; Wu, L.; Gao, Q.; He, X.; Shi, J.; Mei, Y.; Liu, Y.; et al. A personalized computational model predicts cancer risk level of oral potentially malignant disorders and its web application for promotion of non-invasive screening. J. Oral Pathol. Med. 2019, 49, 417–426. [Google Scholar] [CrossRef]
- Nanditha, B.R.; Geetha, A.; Chandrashekar, H.S.; Dinesh, M.S.; Murali, S. An Ensemble Deep Neural Network Approach for Oral Cancer Screening. Int. J. Online Biomed. Eng. 2021, 17, 121–134. [Google Scholar]
- Xiao, Y.; Wu, J.; Lin, Z.; Zhao, X. A deep learning-based multi-model ensemble method for cancer prediction. Comput. Methods Programs Biomed. 2018, 153, 1–9. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010, 1. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Xie, L.; Abouelenien, M. A regularized ensemble framework of deep learning for cancer detection from multi-class, imbalanced training data. Pattern Recognit. 2018, 77, 160–172. [Google Scholar] [CrossRef]
- Tong, L.-I.; Chang, Y.-C.; Lin, S.-H. Determining the optimal re-sampling strategy for a classification model with imbalanced data using design of experiments and response surface methodologies. Expert Syst. Appl. 2011, 38, 4222–4227. [Google Scholar] [CrossRef]
- Krishnan, M.M.R.; Chakraborty, C.; Paul, R.R.; Ray, A.K. Hybrid segmentation, characterization and classifica-tion of basal cell nuclei from histopathological images of normal oral mucosa and oral submucous fibrosis. Expert Syst. Appl. 2012, 39, 1062–1077. [Google Scholar] [CrossRef]
- Das, D.K.; Bose, S.; Maiti, A.K.; Mitra, B.; Mukherjee, G.; Dutta, P.K. Automatic identification of clinically relevant regions from oral tissue histological images for oral squamous cell carcinoma diagnosis. Tissue Cell 2018, 53, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Campisi, G.; Calvino, F.; Carinci, F.; Matranga, D.; Carella, M.; Mazzotta, M.; Lo Muzio, L. Peri-tumoral inflam-matory cell infiltration in OSCC: A reliable marker of local recurrence and prognosis? An investigation using artificial neural networks. Int. J. Immunopathol. Pharmacol. 2011, 24 (Suppl. S2), 113–120. [Google Scholar] [CrossRef]
- Duran-Sierra, E.; Cheng, S.; Cuenca, R.; Ahmed, B.; Ji, J.; Yakovlev, V.V.; Jo, J.A. Machine-learning assisted dis-crimination of precancerous and cancerous from healthy oral tissue based on multispectral autofluorescence lifetime imag-ing endoscopy. Cancers 2021, 13, 4751. [Google Scholar] [CrossRef]
- Shan, J.; Jiang, R.; Chen, X.; Zhong, Y.; Zhang, W.; Xie, L.; Cheng, J.; Jiang, H. Machine Learning Predicts Lymph Node Metastasis in Early-Stage Oral Tongue Squamous Cell Carcinoma. J. Oral Maxillofac. Surg. 2020, 78, 2208–2218. [Google Scholar] [CrossRef] [PubMed]
- McRae, M.P.; Modak, S.S.; Bs, G.W.S.; Dds, D.A.T.; Kerr, A.R.; Thornhill, M.H.; Redding, S.W.; Vigneswaran, N.; Kang, S.; Christodoulides, N.J.; et al. Point-of-care oral cytology tool for the screening and assessment of potentially malignant oral lesions. Cancer Cytopathol. 2019, 128, 207–220. [Google Scholar] [CrossRef] [Green Version]
- Aravinth, M. Oral Cancer Detection Using RNN. IRJET, 09(09), Article e-ISSN: 2395-0056. Available online: https://www.irjet.net/archives/V9/i9/IRJET-V9I941.pdf (accessed on 24 September 2022).
- Zhang, Z.; Zhao, Y.; Liao, X.; Shi, W.; Li, K.; Zou, Q.; Peng, S. Deep learning in omics: A survey and guideline. Briefings Funct. Genom. 2018, 18, 41–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ding, Z.; Fu, Y. Feature selection guided auto-encoder. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, California USA, 4–9 February 2017; Volume 31. [Google Scholar]
- Zhang, X.; Zhang, J.; Sun, K.; Yang, X.; Dai, C.; Guo, Y. Integrated multi-omics analysis using varia-tional autoencoders: Application to pan-cancer classification. In 2019 IEEE International Conference on Bioinformatics and Bio-Medicine (BIBM), San Diego, CA, USA, 18–21 November 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 765–769. [Google Scholar]
- Simidjievski, N.; Bodnar, C.; Tariq, I.; Scherer, P.; Andres-Terre, H.; Shams, Z.; Jamnik, M.; Liò, P. Variational Autoencoders for Cancer Data Integration: Design Principles and Computational Practice. Front. Genet. 2019, 10, 1205. [Google Scholar] [CrossRef] [Green Version]
- Sheet, S.; Ghosh, A.; Ghosh, R.; Chakrabarti, A. Identification of Cancer Mediating Biomarkers using Stacked De-noising Autoencoder Model-An Application on Human Lung Data. Procedia Comput. Sci. 2020, 167, 686–695. [Google Scholar] [CrossRef]
- Antonio, V.A.A.; Ono, N.; Saito, A.; Sato, T.; Amin, A.U.; Kanaya, S. Classification of lung adenocarcinoma transcriptome subtypes from pathological images using deep convolutional networks. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1905–1913. [Google Scholar] [CrossRef] [Green Version]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the European Conference on Computer Vision; Springer: Zurich, Switzerland, 2014; pp. 818–833. [Google Scholar]
- Joshi, P.; Alsadoon, O.H.; Alsadoon, A.; AlSallami, N.; Rashid, T.A.; Prasad, P.; Haddad, S. Deep learning for size and microscope feature extraction and classification in Oral Cancer: Enhanced convolution neural network. Multimed. Tools Appl. 2022, 82, 6197–6220. [Google Scholar] [CrossRef]
- Kiruthika, S.; RahmathNisha, S. Automated Oral Cancer Detection and Classification using very Deep Convolutional Neural Network Algorithm. Test Manag. Eng. 2020, 83, 20019–20027. [Google Scholar]
- Welikala, R.A.; Remagnino, P.; Lim, J.H.; Chan, C.S.; Rajendran, S.; Kallarakkal, T.G.; Zain, R.B.; Jayasinghe, R.D.; Rimal, J.; Kerr, A.R.; et al. Automated Detection and Classification of Oral Lesions Using Deep Learning for Early Detection of Oral Cancer. IEEE Access 2020, 8, 132677–132693. [Google Scholar] [CrossRef]
- Jeyaraj, P.R.; Samuel Nadar, E.R. Computer-assisted medical image classification for early diagnosis of oral can-cer employing deep learning algorithm. J. Cancer Res. Clin. Oncol. 2019, 145, 829–837. [Google Scholar] [CrossRef]
- Matsuo, K.; Purushotham, S.; Jiang, B.; Mandelbaum, R.S.; Takiuchi, T.; Liu, Y.; Roman, L.D. Survival outcome prediction in cervical cancer: Cox models vs deep-learning model. Am. J. Obstet. Gynecol. 2018, 220, 381.e1–381.e14. [Google Scholar] [CrossRef] [PubMed]
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.; Kluger, Y. DeepSurv: Personalized treatment recom-mender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Shams, W.K.; Htike, Z.Z. Oral cancer prediction using gene expression profiling and machine learning. Int. J. Appl. Eng. Res 2017, 12, 4893–4898. [Google Scholar]
- Fu, Q.; Chen, Y.; Li, Z.; Jing, Q.; Hu, C.; Liu, H.; Bao, J.; Hong, Y.; Shi, T.; Li, K.; et al. A deep learning algorithm for detection of oral cavity squamous cell carcinoma from photographic images: A retrospective study. Eclinicalmedicine 2020, 27, 100558. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Sunny, S.; Li, S.; Gurushanth, K.; Mendonca, P.; Mukhia, N.; Liang, R. Bayesian deep learning for relia-ble oral cancer image classification. Biomed. Opt. Express 2021, 12, 6422–6430. [Google Scholar] [CrossRef]
- Dey, D.; Chatterjee, B.; Dalai, S.; Munshi, S.; Chakravorti, S. A deep learning framework using convolution neural network for classification of impulse fault patterns in transformers with increased accuracy. IEEE Trans. Dielectr. Electr. Insul. 2017, 24, 3894–3897. [Google Scholar] [CrossRef]
- Dou, Q.; Chen, H.; Yu, L.; Zhao, L.; Qin, J.; Wang, D.; Mok, V.C.; Shi, L.; Heng, P.-A. Automatic Detection of Cerebral Microbleeds from MR Images via 3D Convolutional Neural Networks. IEEE Trans. Med Imaging 2016, 35, 1182–1195. [Google Scholar] [CrossRef]
- Camalan, S.; Mahmood, H.; Binol, H.; Araújo, A.L.D.; Santos-Silva, A.R.; Vargas, P.A.; Gurcan, M.N. Convolu-tional neural network-based clinical predictors of oral dysplasia: Class activation map analysis of deep learning results. Cancers 2021, 13, 1291. [Google Scholar] [CrossRef]
- Yu, M.; Yan, H.; Xia, J.; Zhu, L.; Zhang, T.; Zhu, Z.; Lou, X.; Sun, G.; Dong, M. Deep convolutional neural networks for tongue squamous cell carcinoma classification using Raman spectroscopy. Photodiagnosis Photodyn. Ther. 2019, 26, 430–435. [Google Scholar] [CrossRef]
- Chatterjee, S.; Nawn, D.; Mandal, M.; Chatterjee, J.; Mitra, S.; Pal, M.; Paul, R.R. Augmentation of Statistical Features in Cytopathology Towards Computer Aided Diagnosis of Oral PrecancerlCancer, Chennai, India, 22–24 March 2018; pp. 206–212. [CrossRef]
- Yamaguchi, S.; Lee, C.; Karaer, O.; Ban, S.; Mine, A.; Imazato, S. Predicting the Debonding of CAD/CAM Composite Resin Crowns with AI. J. Dent. Res. 2019, 98, 1234–1238. [Google Scholar] [CrossRef]
- Tanriver, G.; Tekkesin, M.S.; Ergen, O. Automated Detection and Classification of Oral Lesions Using Deep Learning to Detect Oral Potentially Malignant Disorders. Cancers 2021, 13, 2766. [Google Scholar] [CrossRef]
- Jeyaraj, P.R.; Panigrahi, B.K.; Nadar, E.R.S. Classifier Feature Fusion Using Deep Learning Model for Non-Invasive Detection of Oral Cancer from Hyperspectral Image. IETE J. Res. 2020, 68, 1–12. [Google Scholar] [CrossRef]
- Indhumathiand, J.; Dhanalakshmi, P. Oral Squamous Cell Carcinoma Classification using Deep Boltzmann Machine and GLCM Features Semantic Scholar. Oral Squamous Cell Carcinoma Classification Using Deep Boltzmann Machine and GLCM Features. 1 January 2013. Available online: https://www.semanticscholar.org/paper/Oral-Squamous-Cell-Carcinoma-Classification-using-Indhumathiand-Dhanalakshmi/ba3a75083410af0eb9954a54f5b0f46c11e40cef (accessed on 5 August 2022).
- Martino, F.; Bloisi, D.D.; Pennisi, A.; Fawakherji, M.; Ilardi, G.; Russo, D.; Merolla, F. Deep learning-based pix-el-wise lesion segmentation on oral squamous cell carcinoma images. Appl. Sci. 2020, 10, 8285. [Google Scholar] [CrossRef]
- Deif, M.A.; Attar, H.; Amer, A.; Elhaty, I.A.; Khosravi, M.R.; Solyman, A.A.A. Diagnosis of Oral Squamous Cell Carcinoma Using Deep Neural Networks and Binary Particle Swarm Optimization on Histopathological Images: An AIoMT Approach. Comput. Intell. Neurosci. 2022, 2022, 6364102. [Google Scholar] [CrossRef] [PubMed]
- Folmsbee, J.; Liu, X.; Brandwein-Weber, M.; Doyle, S. Active deep learning: Improved training efficiency of convolutional neural networks for tissue classification in oral cavity cancer. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–8 April 2018; pp. 770–773. [Google Scholar] [CrossRef]
- Li, Y.; Bai, S.; Carroll, W.; Dayan, D.; Dort, J.C.; Heller, K.; Jour, G.; Lau, H.; Penner, C.; Prystowsky, M.; et al. Validation of the Risk Model: High-Risk Classification and Tumor Pattern of Invasion Predict Outcome for Patients with Low-Stage Oral Cavity Squamous Cell Carcinoma. Head Neck Pathol. 2012, 7, 211–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Haron, N.; Zain, R.B.; Ramanathan, A.; Abraham, M.T.; Liew, C.S.; Ng, K.G.; Cheng, L.C.; Husin, R.B.; Chong, S.M.Y.; Thangavalu, L.A.; et al. M-Health for Early Detection of Oral Cancer in Low- and Middle-Income Countries. Telemed. e-Health 2020, 26, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.B.; Ejbali, R.; Zaied, M. Detection and classification of dental caries in x-ray images using deep neural networks. In Proceedings of the International Conference on Software Engineering Advances (ICSEA), Rome, Italy, 21–25 August 2016; p. 236. [Google Scholar]
- Kirubabai, M.P.; Arumugam, G. Deep Learning Classification Method to Detect and Diagnose the Cancer Regions in Oral MRI Images. Med. Leg. Update 2021, 21, 462–468. [Google Scholar]
- Kim, D.W.; Lee, S.; Kwon, S.; Nam, W.; Cha, I.H.; Kim, H.J. Deep learning-based survival prediction of oral can-cer patients. Sci. Rep. 2019, 9, 1–10. [Google Scholar]
- Shaban, M.; Khurram, S.A.; Fraz, M.M.; Alsubaie, N.; Masood, I.; Mushtaq, S.; Rajpoot, N.M. A novel digital score for abundance of tumour infiltrating lymphocytes predicts disease free survival in oral squamous cell carcino-ma. Sci. Rep. 2019, 9, 13341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uthoff, R.D.; Song, B.; Sunny, S.; Patrick, S.; Suresh, A.; Kolur, T.; Keerthi, G.; Spires, O.; Anbarani, A.; Wilder-Smith, P.; et al. Point-of-care, smartphone-based, dual-modality, dual-view, oral cancer screening device with neural network classification for low-resource communities. PLoS ONE 2018, 13, e0207493. [Google Scholar] [CrossRef]
- Xu, S.; Liu, Y.; Hu, W.; Zhang, C.; Liu, C.; Zong, Y.; Chen, S.; Lu, Y.; Yang, L.; Ng, E.Y.K.; et al. An Early Diagnosis of Oral Cancer based on Three-Dimensional Convolutional Neural Networks. IEEE Access 2019, 7, 158603–158611. [Google Scholar] [CrossRef]
- Panigrahi, S.; Das, J.; Swarnkar, T. Capsule network based analysis of histopathological images of oral squamous cell carcinoma. J. King Saud Univ.—Comput. Inf. Sci. 2020, 34, 4546–4553. [Google Scholar] [CrossRef]
- Ariji, Y.; Fukuda, M.; Kise, Y.; Nozawa, M.; Yanashita, Y.; Fujita, H.; Ariji, E. Contrast-enhanced computed to-mography image assessment of cervical lymph node metastasis in patients with oral cancer by using a deep learning system of artificial intelligence. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 127, 458–463. [Google Scholar] [CrossRef]
- Miki, Y.; Muramatsu, C.; Hayashi, T.; Zhou, X.; Hara, T.; Katsumata, A.; Fujita, H. Classification of teeth in cone-beam CT using deep convolutional neural network. Comput. Biol. Med. 2017, 80, 24–29. [Google Scholar] [CrossRef]
- Heinrichs, B.; Eickhoff, S.B. Your evidence? Machine learning algorithms for medical diagnosis and prediction. Hum. Brain Mapp. 2019, 41, 1435–1444. [Google Scholar] [CrossRef] [Green Version]
- Altmann, A.; Toloşi, L.; Sander, O.; Lengauer, T. Permutation importance: A corrected feature importance measure. Bioinformatics 2010, 26, 1340–1347. [Google Scholar] [CrossRef] [Green Version]
- Shah, N.D.; Steyerberg, E.W.; Kent, D.M. Big data and predictive analytics: Recalibrating expectations. JAMA 2018, 320, 27–28. [Google Scholar] [CrossRef] [PubMed]
- Alabi, R.O.; Youssef, O.; Pirinen, M.; Elmusrati, M.; Mäkitie, A.A.; Leivo, I.; Almangush, A. Machine learning in oral squamous cell carcinoma: Current status, clinical concerns and prospects for future—A systematic review. Artif. Intell. Med. 2021, 115, 102060. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Datta, S.; Chaudhuri, B.B. Handling data irregularities in classification: Foundations, trends, and future challenges. Pattern Recognit. 2018, 81, 674–693. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Habas, P.A.; Zurada, J.M.; Lo, J.Y.; Baker, J.A.; Tourassi, G.D. Training neural network clas-sifiers for medical decision making: The effects of imbalanced datasets on classification performance. Neural Netw. 2008, 21, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Xiao, C.; Choi, E.; Sun, J. Opportunities and challenges in developing deep learning models using electronic health records data: A systematic review. J. Am. Med Inform. Assoc. 2018, 25, 1419–1428. [Google Scholar] [CrossRef] [Green Version]
- Lasko, T.A.; Denny, J.C.; Levy, M.A. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PLoS ONE 2013, 8, e66341. [Google Scholar] [CrossRef]
- Panigrahi, S.; Swarnkar, T. Machine learning techniques used for the histopathological image analysis of oral cancer—A review. Open Bioinform. J. 2020, 13, 106–118. [Google Scholar] [CrossRef]
- Cho, J.; Lee, K.; Shin, E.; Choy, G.; Do, S. How much data is needed to train a medical image deep learning system to achieve necessary high accuracy? arXiv 2015, arXiv:1511.06348. [Google Scholar]
- Kleppe, A.; Skrede, O.-J.; De Raedt, S.; Liestøl, K.; Kerr, D.J.; Danielsen, H.E. Designing deep learning studies in cancer diagnostics. Nat. Rev. Cancer 2021, 21, 199–211. [Google Scholar] [CrossRef]
- Wang, K.-J.; Makond, B.; Chen, K.-H. A hybrid classifier combining SMOTE with PSO to estimate 5-year survivability of breast cancer patients. Appl. Soft Comput. 2014, 20, 15–24. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent reporting of a multivariable prediction mod-el for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. J. Br. Surg. 2015, 102, 148–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shamim, M.Z.M.; Syed, S.; Shiblee, M.; Usman, M.; Ali, S.J.; Hussein, H.S.; Farrag, M. Automated Detection of Oral Pre-Cancerous Tongue Lesions Using Deep Learning for Early Diagnosis of Oral Cavity Cancer. Comput. J. 2020, 65, 91–104. [Google Scholar] [CrossRef]
- Al-Rawi, N.; Sultan, A.; Rajai, B.; Shuaeeb, H.; Alnajjar, M.; Alketbi, M.; Mashrah, M.A. The Effectiveness of Artificial Intelligence in Detection of Oral Cancer. Int. Dent. J. 2022, 72, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, S.A.; Nagaraj, R.; Mitra, S. Classification of dental diseases using CNN and transfer learning. In Proceedings of the 2017 5th International Symposium on Computational and Business Intelligence (ISCBI), Dubai, United Arab Emirates, 11–14 August 2017; IEEE: Piscataway, NJ, UJSA, 2017; pp. 70–74. [Google Scholar]
- Li, X.; Wang, Y.; Cai, Y. Automatic annotation algorithm of medical radiological images using convolutional neural network. Pattern Recognit. Lett. 2021, 152, 158–165. [Google Scholar] [CrossRef]
- Mehta, S.; Shelling, A.; Muthukaruppan, A.; Lasham, A.; Blenkiron, C.; Laking, G.; Print, C. Predictive and prognostic molecular markers for cancer medicine. Ther. Adv. Med. Oncol. 2010, 2, 125–148. [Google Scholar] [CrossRef] [Green Version]











| Acronym | Definition |
|---|---|
| AC | Ameloblastic Carcinoma |
| AF | Ameloblastic Fibroma |
| AI | Artificial Intelligence |
| ANN | Artificial Neural Network |
| BCC | Basal-Cell-Based Lip Cancer |
| BDT | Boosted Decision Tree |
| BP | Back-Propagation |
| BSC | Basaloid Squamous Carcinoma |
| CAD | Computer-Aided Detection |
| CCOC | Clear Cell Odontogenic Carcinoma |
| CNN | Convolutional Neural Network |
| CT | Computed Tomography |
| DBM | Deep Boltzmann Machine |
| DT | Decision Tree |
| DL | Deep Learning |
| DNN | Deep Neural Network |
| FOM | Floor of Mouth |
| GCOC | Ghost cell odontogenic carcinoma |
| KNN | K-Nearest Neighbor |
| LOC | Lab-On-a-Chip |
| LSTM | Long Short-Term Memory |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| OC | Oral Cancer |
| OCS | Odontogenic Carcinosarcoma |
| OCT | Optical Coherence Tomography |
| OPMD | Oral Potentially Malignant Disorders |
| OSCC | Oral Squamous Cell Carcinoma |
| PIOC | Primary Intraosseous Carcinoma |
| PIOSCC | Primary intraosseous Squamous Cell Carcinoma |
| SCC | Squamous Cell Carcinoma |
| SCCOC | Squamous Cell Carcinoma of the Oral Cavity |
| SOC | Sclerosing Odontogenic Carcinoma |
| SVM | Super Vector Machine |
| VGG | Visual Geometry Group |
| WHO | World Health Organization |
| Reference | Year | One-Phrase Summary | ML | DL | OC | FD |
|---|---|---|---|---|---|---|
| Our paper | - | This review offers a thorough assessment of DL and ML models for diagnosing OC | H | H | H | H |
| [3] | 2020 | In order to lower the frequency of OC in India, this study emphasized the significance of early identification, adequate treatment, and prevention. | N | N | H | H |
| [26] | 2021 | This review concluded that it is essential to differentiate between malignant and benign cells while diagnosing OSCC. Additionally, it included a general summary of the elements of a delayed OC assessment. | L | M | L | N |
| [27] | 2020 | The results of SCCOC therapy in a newly published major series were presented in this study. It has been established that improved early detection techniques and understanding, as well as increased education concerning the risk factors connected to lifestyle choices, are essential for both the primary and secondary prevention of OC. | N | N | L | L |
| [28] | 2012 | The authors noted that men and people aged over 65 years had greater rates of lip cancer. The results demonstrated that SCC patients displayed typical clinical and epidemiological features to those identified in prior investigations. | N | N | H | N |
| [29] | 2020 | This study was the first comprehensive evaluation that aimed to assess the clinicopathological characteristics of PIOSCC and potential etiological factors related to its prognosis. | N | N | H | H |
| [30] | 2019 | According to the results of this study, an OPMD may serve as a risk factor for the development of OC. Although there are active clinical trials and recommendations to remove high-risk lesions, there are currently no effective chemopreventive strategies available. | N | N | M | H |
| [31] | 2021 | The findings of this study implied that cutting-edge AI methods can contribute in an unobtrusive way to the early detection of OC. | H | H | M | M |
| [32] | 2003 | According to this research, ameloblastoma cases should be thoroughly examined to discover small histological changes that could indicate aggressive behavior by comparing the tumors’ histologic pattern to their biological behavior. | N | N | L | L |
| [33] | 2022 | This study’s conclusions demonstrated the need for all patients with non-healing lip lesions to have a full physical examination that includes an intraoral examination and a review of their medical history. When lip cancer is accurately diagnosed and staged, it can be treated quickly and with the best surgical procedure possible for the greatest results. | N | N | L | N |
| [17] | 2022 | This research demonstrated the value of ML applications for the prognosis and treatment of potentially malignant (pre-cancerous) oral lesions. | H | M | L | L |
| [18] | 2022 | ML and DL classification techniques for OC detection were studied in this study. Several studies revealed that the ML model works admirably in diagnostic and prognostic investigations of oral cancer. To be used in routine clinical practice, these models need to be enhanced to increase their interpretability and they require external evaluation for generalizability utilizing deep hybrid learning approaches. | H | H | L | L |
| [19] | 2021 | Most OC outcomes can be predicted using ML algorithms with good accuracy. Furthermore, this study concluded that because these outcomes are uncommon, it is necessary to use class imbalance strategies to handle the skewness of the data. | H | H | L | L |
| Ref. | ML Approaches Used | Data Set | Computation Tools | Features Extracted/Features Selected | Feature Extraction Approach | Key Contribution | Limitations | Performance Evaluation Metrics |
|---|---|---|---|---|---|---|---|---|
| [144] | LR, DT, SVM, and K-NN | 467 OSCC patients | MATLAB R2020a | Prognostic features | PCA and bivariate analysis | It will allow the clinicians to predict the progression of the disease. | Genetic profiling, biomarker analysis, and sophisticated histopathology imaging were missing from this paper. | Acc = 0.705, specificity = 0.841, sensitivity = 0.41 |
| [163] | SVM, GMM | The 1194 cells were taken from 341 healthy and 429 OSF with dysplasia photos. | Snake tool for image segmentation | Hyperchromasia, and nuclear texture, 23 characteristics were derived from segmented biopsy pictures. | Active contour method of gradient vector flow (GVF). | A median filtering technique is suggested for image pre-processing to get rid of the noise. | Expert’s topic expertise and the right image processing were absent. | Acc = 99.66% |
| [164] | CNN, Gabor filter, Random forests | High-grade = 15 Low-grade = 25 and Healthy = 2 subjects. | Computer aided automatic tools | Texture-based features | Gabor feature extraction | The identification of keratin pearls and the segmentation of subepithelial and epithelial layers can be used for oral precancerous screening and OSCC grading, respectively. | Very little research was carried out on cytopathological and histological pictures to identify the keratin pearl structure. | Acc = 99.88% |
| [165] | ANN | 211 cases with OSCC were identified between 1990 and 2000. | Statistical tools | Age and gender of the patient during the time of diagnosis were considered when data were analyzed. | Peri-tumoral inflammatory infiltrate with local recurrence | This study’s goal was to ascertain whether patients with OSCC may have their 5-year survival rate and incidence rate of LR affected by the presence and grade of PTI | It was not possible to determine involvement in other age-related cancers. | Specificity = 90.59%, sensitivity = 67.74%, Acc = 78.56% |
| [166] | LR, linear SVM | A total of 34 patients were enlisted for tissue biopsies of suspected oral epithelial lesions. | - | Spectral, time-resolved, and autofluorescence features. | Linear discriminant analysis | They created a CAD system that used ML to automatically distinguish between malignant and healthy oral tissue using data from in vivo widefield maFLIM endoscopy. | In this study, numerous spectra per individual were used as separate datasets, resulting in training and testing sets that were not genuinely independent. | F1 score = 0.85, specificity = 74%, sensitivity = 94% |
| [131] | SVM, RF, LR, and K-NN | High-definition cytology photos | Telectology platform | Mitotic figures, hyperchromatic nucleus, multiple nuclei, etc. | Field of view extraction method | This study thus prove the value of tele cytology for accurate, remote diagnosis and the application of autonomous ANN-based assessment to increase its efficiency. | According to the limitations of traditional cytology, OPML can only be recognized with a poor sensitivity of approximately 18%. | It demonstrated an accuracy result of 84 to 86% in the identification of oral lesions |
| [167] | LR, RF, SVM, NB | 145 patients suffering from early stage OTSCC. | GridSearchCV, StratifiedKFold, and sklearn Python tools. | Simple clinical and pathologic characteristics linked to patients’ prognoses were the factors used for this investigation. | - | They proved that the best approach is not to create an application that blends ML algorithms with an EHR system. | Lack of large training sets and samples. | The best results were achieved by the random forest model (specificity = 75%: sensitivity = 85%; AUC = 0.786. |
| [168] | KNN | Using a cytology-on-a-chip method, 999 patients had OSCC and PMOLs. | Data visualization tools, cytopathology tools | 144 cellular/nuclear features were gathered from single-cell analyses. | PCA | The results of the present study demonstrated the benefit of a POC-amenable cytology platform that can detect and monitor oral lesions throughout the full spectrum of OED diagnoses. | The present study was limited by the fact that past investigations of cytology adjuncts and POCOCT, in general, focused primarily on PMOL examination in secondary conditions or clinical settings, where malignant and dysplastic lesions could be significantly more prevalent compared to the primary clinical setting. | Acc = 99.3 % |
| Ref. | DL Approaches Used | Data Set | Computation Tools | Features Extracted/Features Selected | Feature Extraction Approach | Key Contribution | Limitations | Performance Evaluation Metrics |
|---|---|---|---|---|---|---|---|---|
| [202] | CNN | 160 oral cancer images | Tensorflow | Texture features | Local binary pattern | This paper developed a methodology using the DL algorithm to classify oral images into either normal or abnormal images. | The DL models used in this paper were quite outdated. | Acc = 0.99, specificity = 0.99, sensitivity = 0.98 |
| [203] | DeepSurv, Cox proportional hazard model (CPH), random survival forest (RSF) | 255 images | Python packages such as Lasagne, tensorboard_logger, etc. | 9 features: T stage, N stage, HG, PNI, ENE, LVP, OR, BM, and RM. | - | This model can be effective in predicting with higher accuracy and can guide clinicians both in choosing treatment options and avoiding unnecessary treatments | Statistical methods such as regression trees and classification might be intuitive for clinicians, but they suffer from poor performance and high variance. | DeepSurv showed the finest performance with Acc = 0.810 |
| [204] | TILAb. ResNet50, DenseNet, Inception-v3, Xception | 70 cases, containing 10 control cases and 60 OSCC patients. | Tensor-flow, OpenSlide, Sickit-Learn, Matplotlib, NumP, and Pandas. | Histological and pathological features | Patch-based feature extraction approach | The proposed framework for automated quantification of TILs, computation of their abundance score, and its prognostic analysis of patient survival using OSCC histology images is the first of its kind. | Difficulties in managing OSCC patients include early recurrence, frequent lymph node metastases, and extra nodal extension. | AUC = 0.98 |
| [205] | VGG-16 CNN | 170 image pairs | Android studio | Pre-cancerous and cancerous lesions | - | Created low-cost but powerful smartphones are promising developments for the creation of low-cost, portable, simple-to-use autofluorescence imaging devices for oral cancer detection. | It would not work for professionals who are in remote areas. | Acc = 0.94 |
| [206] | 3DCNN, 2DCNN | 7000 CT images of early oral cancers | Caffe, CT | Topology features such as pixel and audio | The 3DCNN automatically extracts features from the dataset | The results proved that 3DCNN can better identify benign and malignant lesions of early oral cancers | Due to space limitations, this paper only discussed a single sequence of images, without combining different imaging modalities. | 3DCNN AUC = 0.801 |
| [207] | CNN, Capsnet | 82 malignant and 68 benign slide images were obtained from the GDC portal. | Tensorflow | Visual features | Artisanal feature extraction method | The capsule network is suitable for identifying histopathological images in early stage oral cancer. | CNN is not resilient to significant input data modifications | Sensitivity = 0.9778 Specificity = 0.9692 ACC = 97.35% |
| [208] | CNN | 45 OSCC patients had CT scans of 127 cervical lymph nodes that were verified to be positive and 314 cervical lymph nodes that were discovered to be negative. | DIGITS library was used to implement the AlexNet architecture on the Caffe framework. | Image features | - | This study evaluated the efficacy of DL image categorization for the detection of lymph node metastases | The image segmentation was carried out manually; hence the model did not work in real time. | Acc = 0.782 Sensitivity = 0.754 Specificity = 0.81 |
| [209] | CNN (AlexNet) | Cone-beam CT (CBCT) 3D dental imaging | Veraviewepocs 3D, Alphard VEGA | The test ROIs were classified into seven tooth types by the trained network | It was carried out through convolution and pooling layers. | The proposed method is advantageous in obtaining high classification accuracy without the need for precise tooth segmentation. | The major limitations of this study were the small amount of evaluation data and the independent evaluation of slice images. | Acc = 0.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dixit, S.; Kumar, A.; Srinivasan, K. A Current Review of Machine Learning and Deep Learning Models in Oral Cancer Diagnosis: Recent Technologies, Open Challenges, and Future Research Directions. Diagnostics 2023, 13, 1353. https://doi.org/10.3390/diagnostics13071353
Dixit S, Kumar A, Srinivasan K. A Current Review of Machine Learning and Deep Learning Models in Oral Cancer Diagnosis: Recent Technologies, Open Challenges, and Future Research Directions. Diagnostics. 2023; 13(7):1353. https://doi.org/10.3390/diagnostics13071353
Chicago/Turabian StyleDixit, Shriniket, Anant Kumar, and Kathiravan Srinivasan. 2023. "A Current Review of Machine Learning and Deep Learning Models in Oral Cancer Diagnosis: Recent Technologies, Open Challenges, and Future Research Directions" Diagnostics 13, no. 7: 1353. https://doi.org/10.3390/diagnostics13071353
APA StyleDixit, S., Kumar, A., & Srinivasan, K. (2023). A Current Review of Machine Learning and Deep Learning Models in Oral Cancer Diagnosis: Recent Technologies, Open Challenges, and Future Research Directions. Diagnostics, 13(7), 1353. https://doi.org/10.3390/diagnostics13071353







