Prognosis Prediction in COVID-19 Patients through Deep Feature Space Reasoning
Abstract
1. Introduction
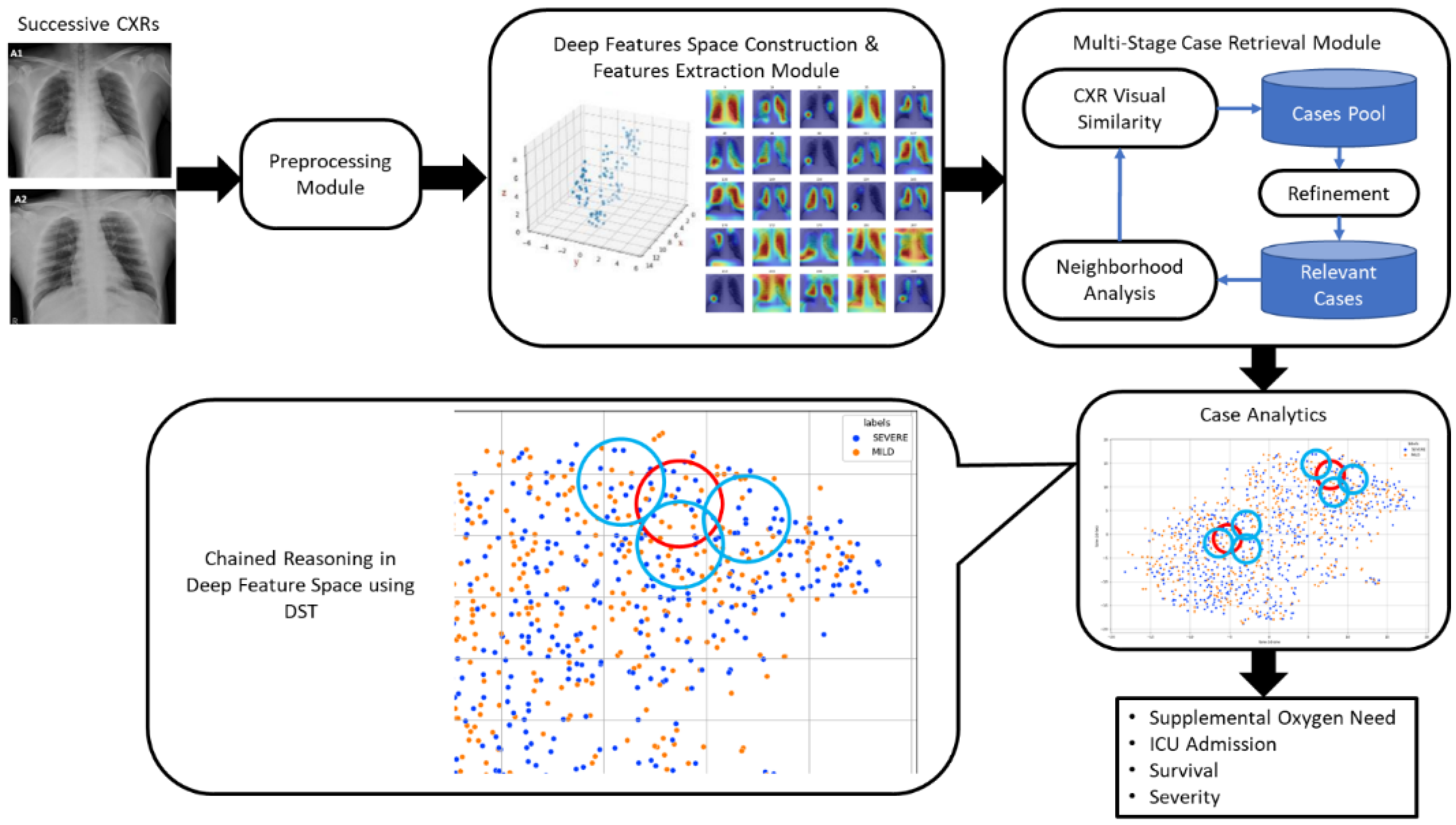
- A COVID-specific deep features space construction and features extraction method is proposed.
- A hybrid representation method is proposed where each patient is represented in the deep feature space using visual features encoding and the age and comorbidities are associated with every patient as ordinary variables.
- A multi-stage case retrieval method is developed to locate relevant cases of COVID-19 patients based on CXR and clinical records.
- A deep feature space reasoning method based on Dempster–Shafer theory is developed, which combines evidence to determine disease progression as well as predict prognosis using relevant past cases involving clinical variables and CXRs.
2. Related Work
3. Materials and Methods
3.1. Study Population
3.2. Analysis of Chest Radiographs and Associated Records
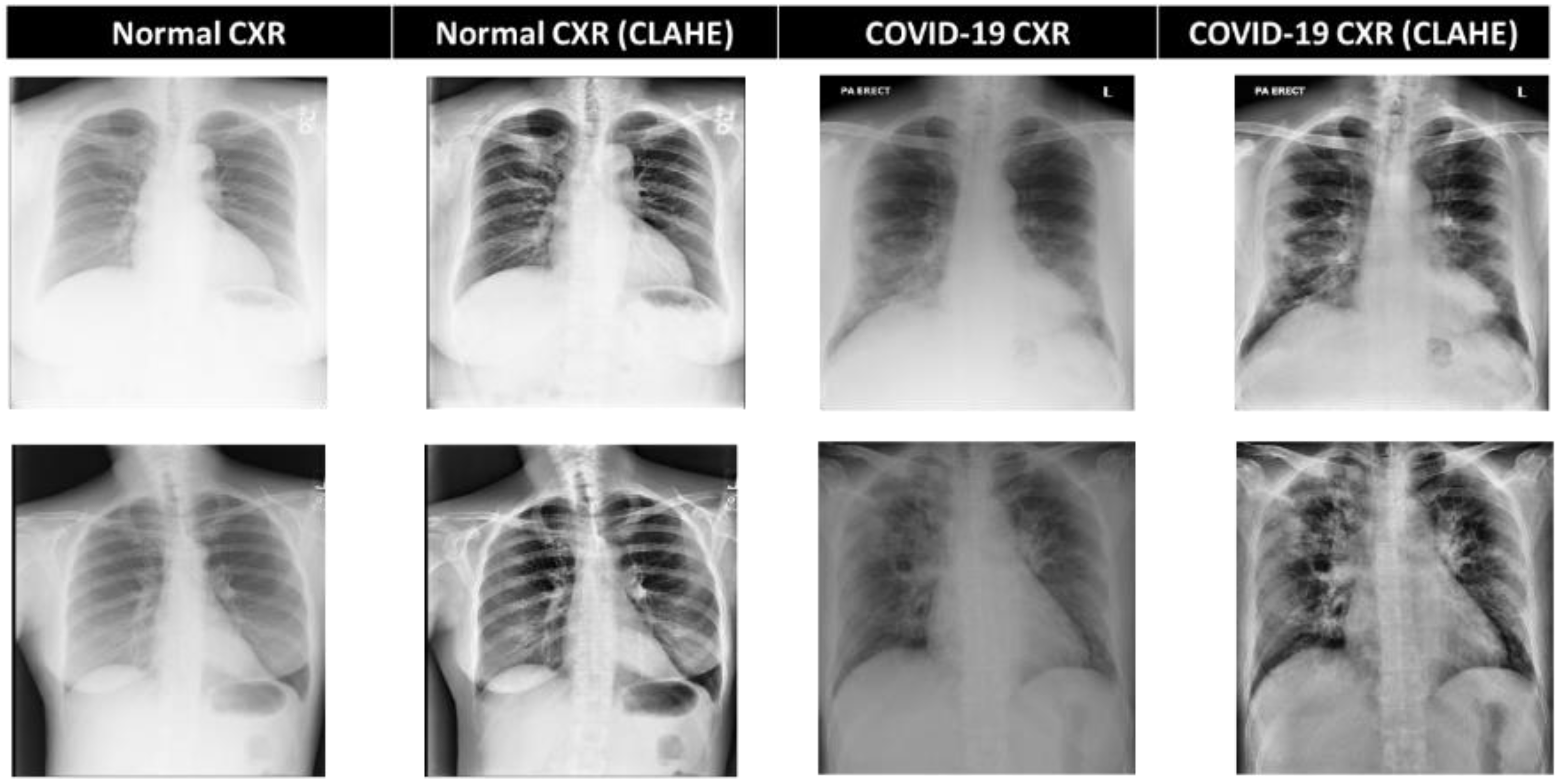
3.3. CXR Preprocessing
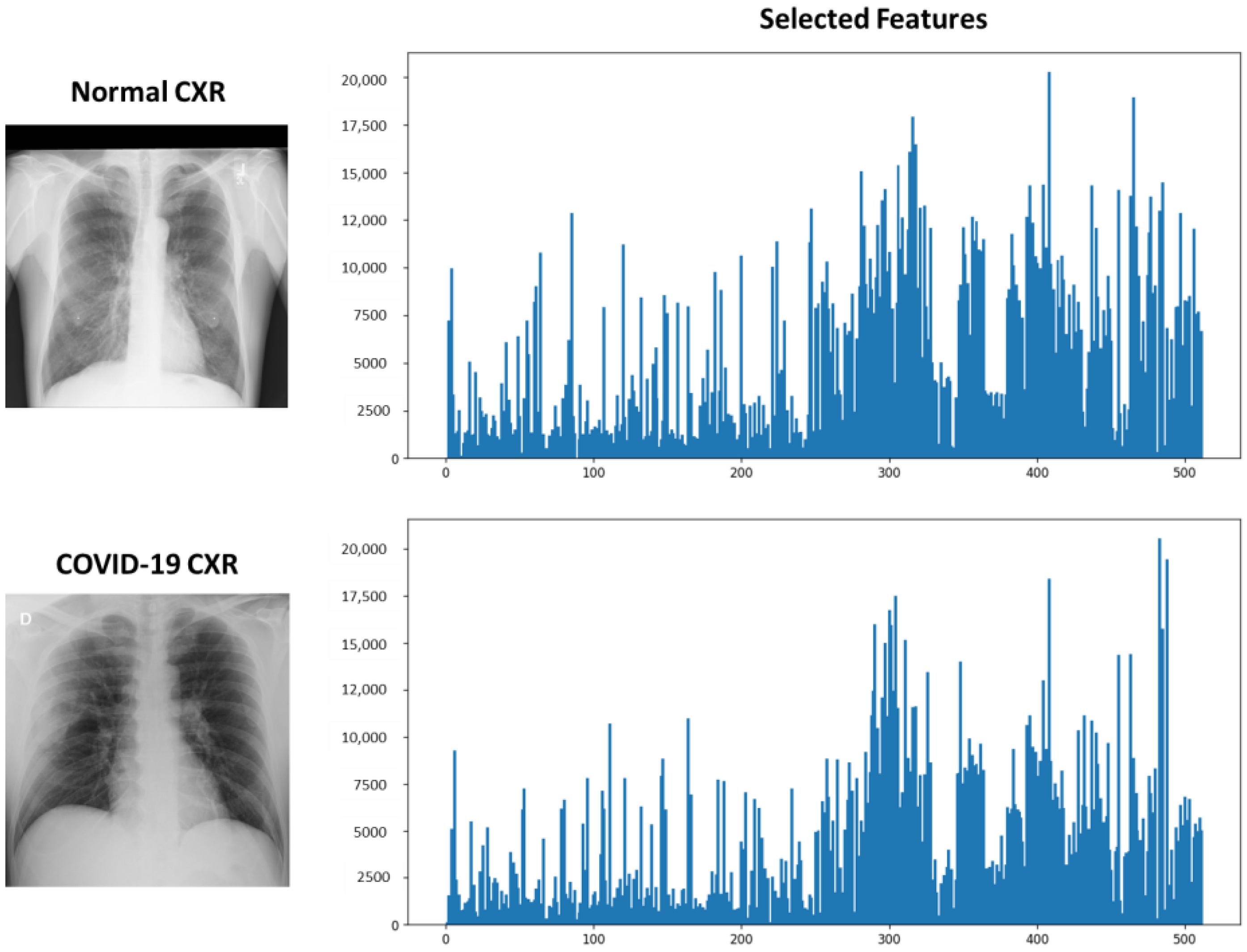
3.4. COVID-Specific Deep Feature Space Construction and Features Extraction
| Algorithm 1: COVID-Specific Deep Feature Space Construction | |
| 1: | Input: Image Set (T) consisting of |
| 2: |
|
| 3: |
|
| 4: | Output: |
| 5: |
|
| 6: | Preparation: |
| 7: |
|
| 8: |
|
| 9: | Steps: |
| 10: |
|
| 11: |
|
| 12: | |
| 13: |
|
| 14: |
|
| 15: |
|
| 16: |
|
| 17: |
|
| 18: |
|
| Algorithm 2: Features Extraction and Representation | |
| 1: | Input: CXR |
| 2: | Output: Feature Vector Fx |
| 3: | Steps: |
| 4: |
|
| 5: |
|
| 6: |
|
| 7: |
|
| 8: | |
| 9: | |
| 10: |
|
| 11: |
|
3.5. Case Retrieval Using CXR Similarity
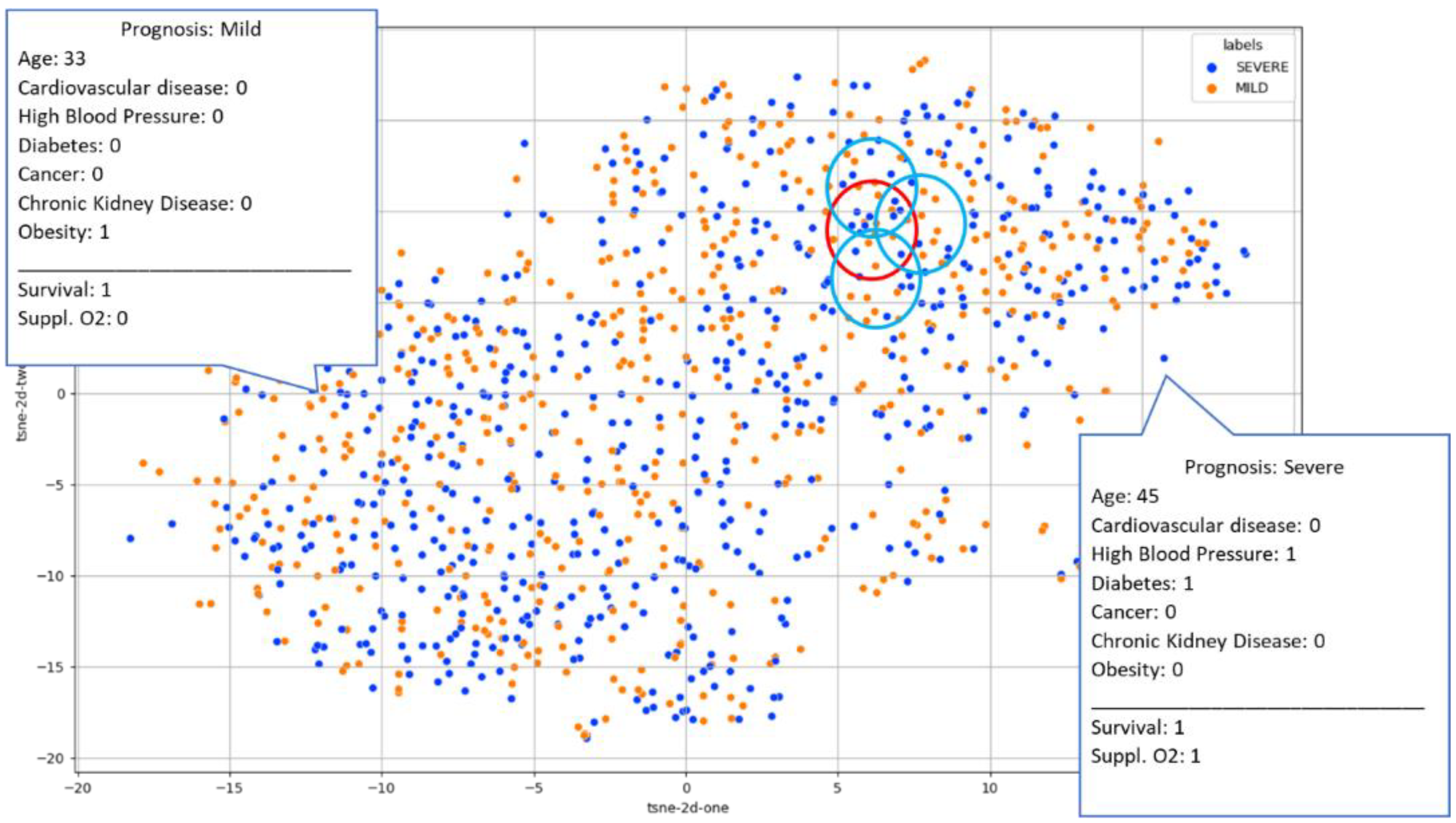
3.6. Case Analytics via Deep Feature Space Reasoning
| Algorithm 3: Deep Feature Space Reasoning for Prediction | |
| 1: | Input: |
| 2: |
|
| 3: | Output: |
| 4: |
|
| 5: | Steps: |
| 6: |
|
| 7: |
|
| 8: |
|
| 9: |
|
| 10: |
|
| 11: |
|
| 12: |
|
| 13: | |
| 14: | where is the normalized distance between X and Y. |
| 15: |
|
| 16: | where M is the number of comorbidities considered. |
| 17: |
|
| 18: | |
| 19: |
|
| 20: |
|
| 21: | , , , |
| 22: |
|
| 23: |
|
| 24: |
|
| 25: |
|
| 26: |
|
| 27: |
|
3.7. Progression and Prognosis Prediction
4. Experimental Results and Analysis
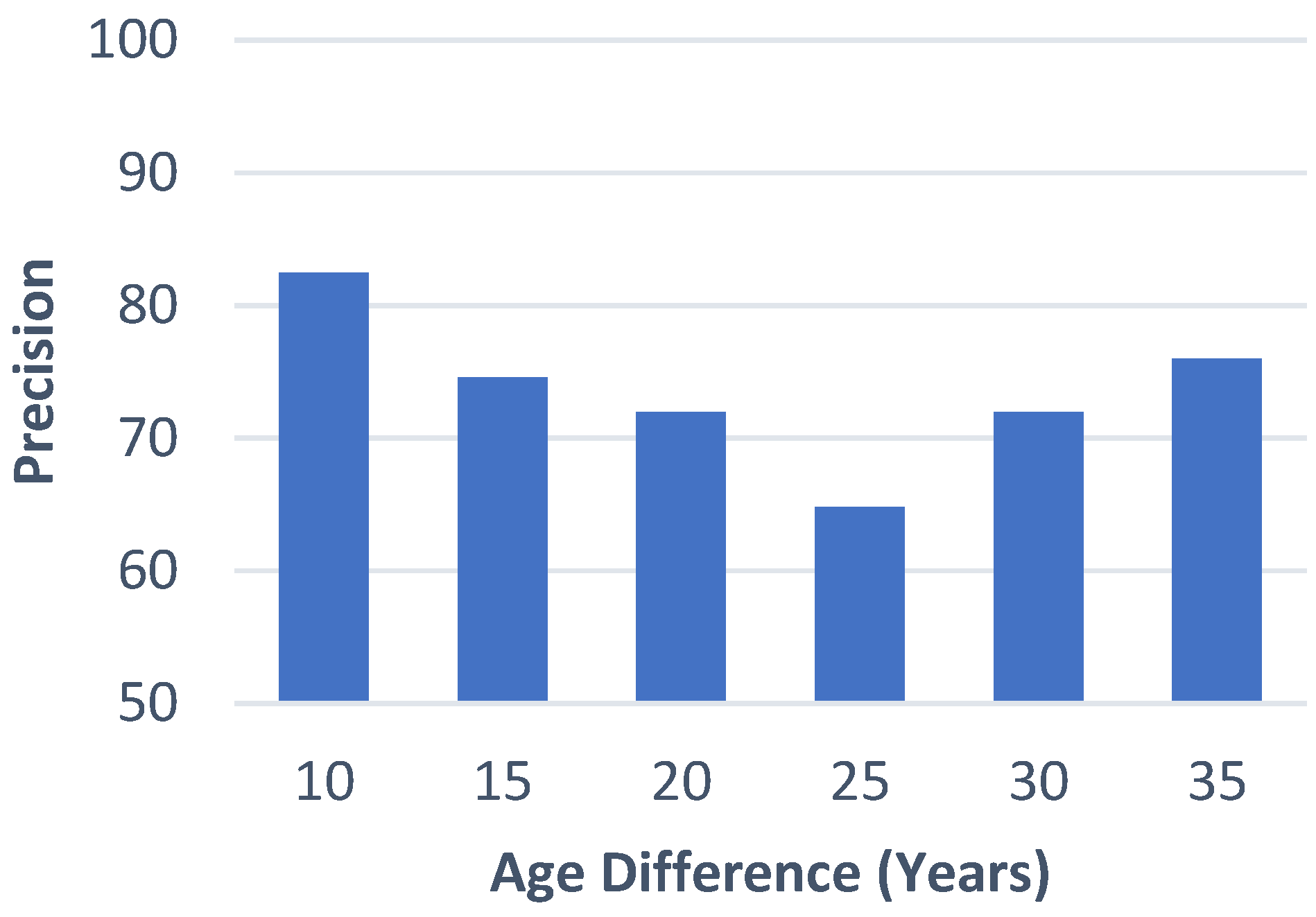
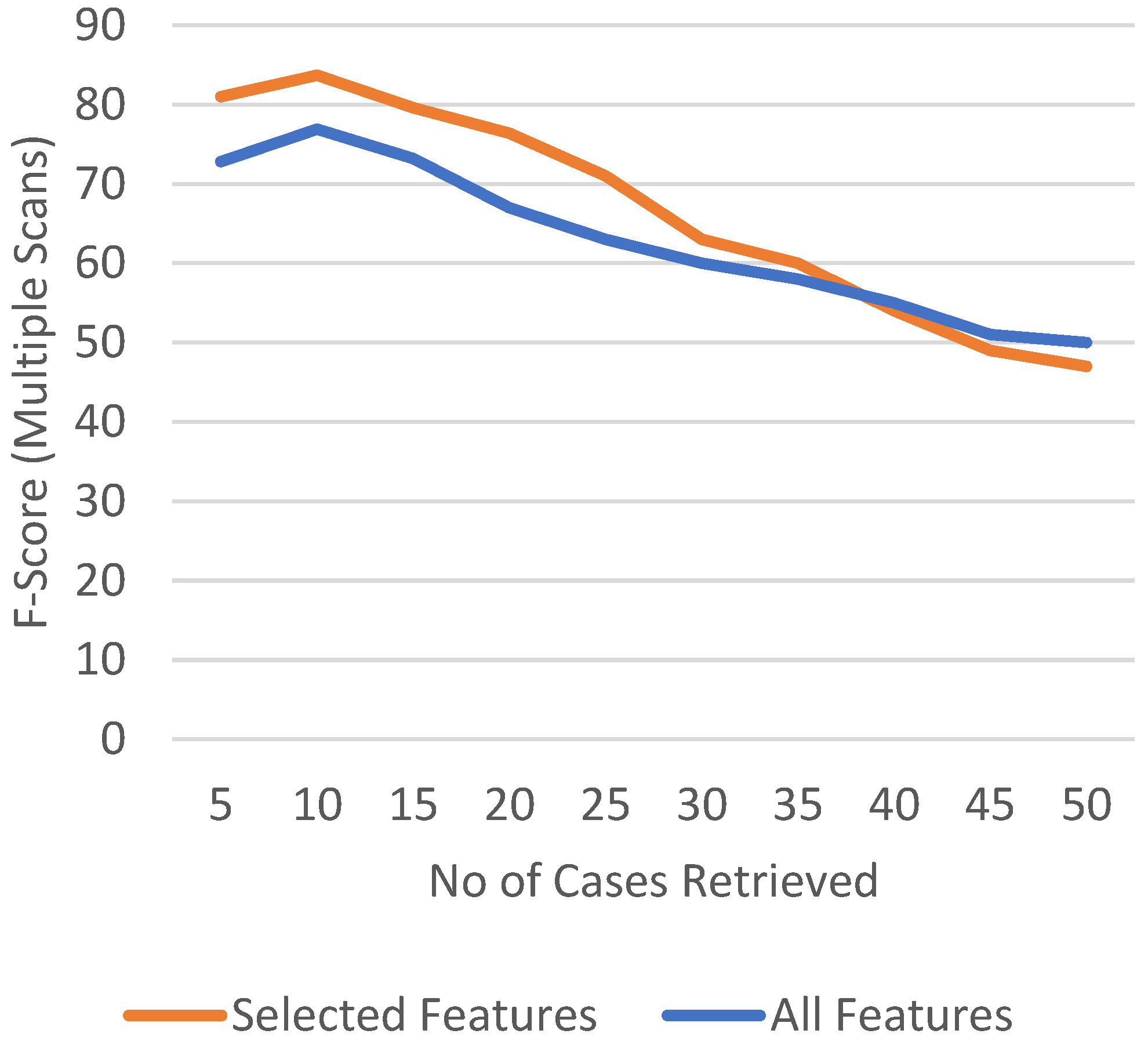
4.1. Case Retrieval Performance
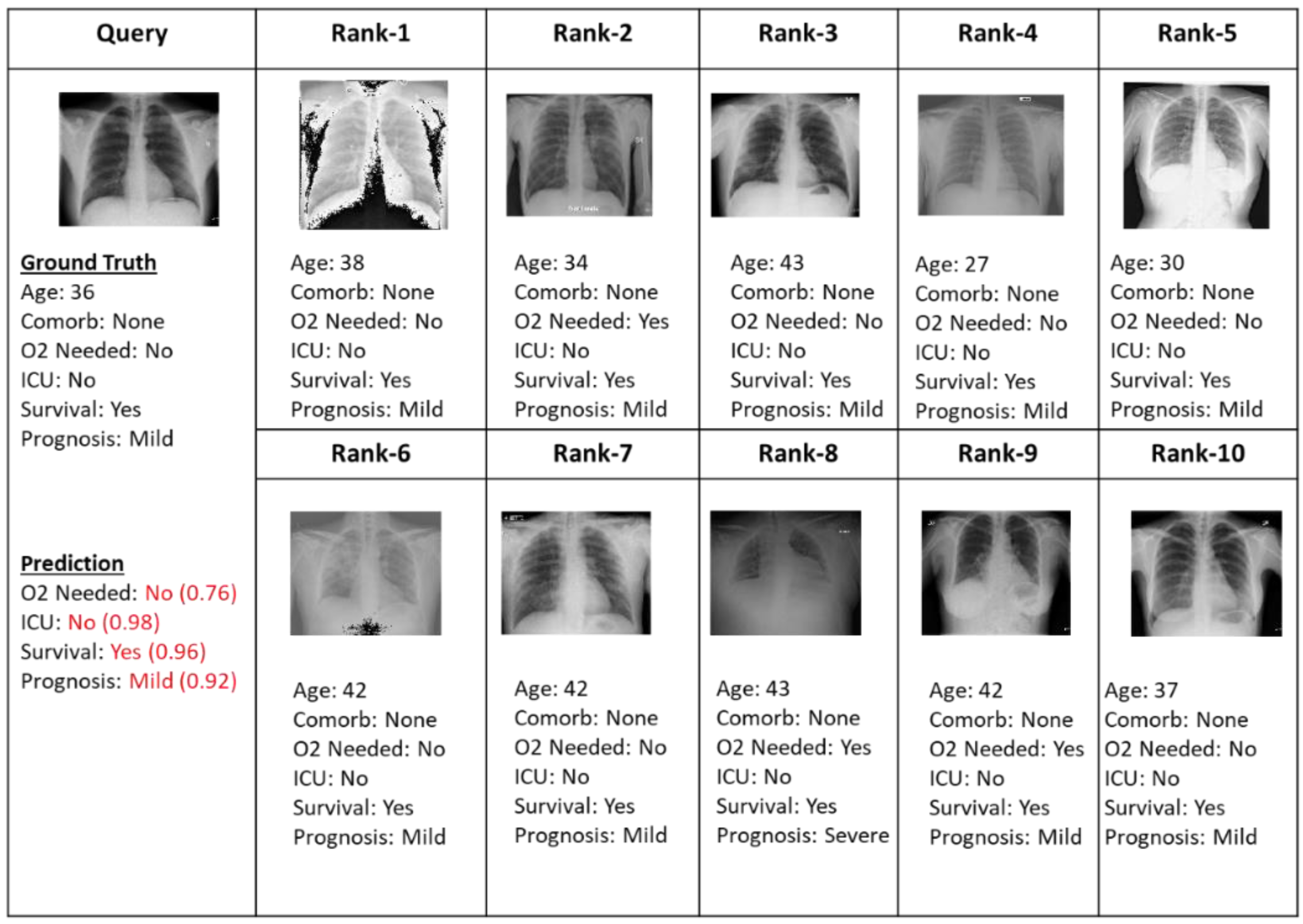
4.2. Retrieved Cases (Images with Clinical Records)
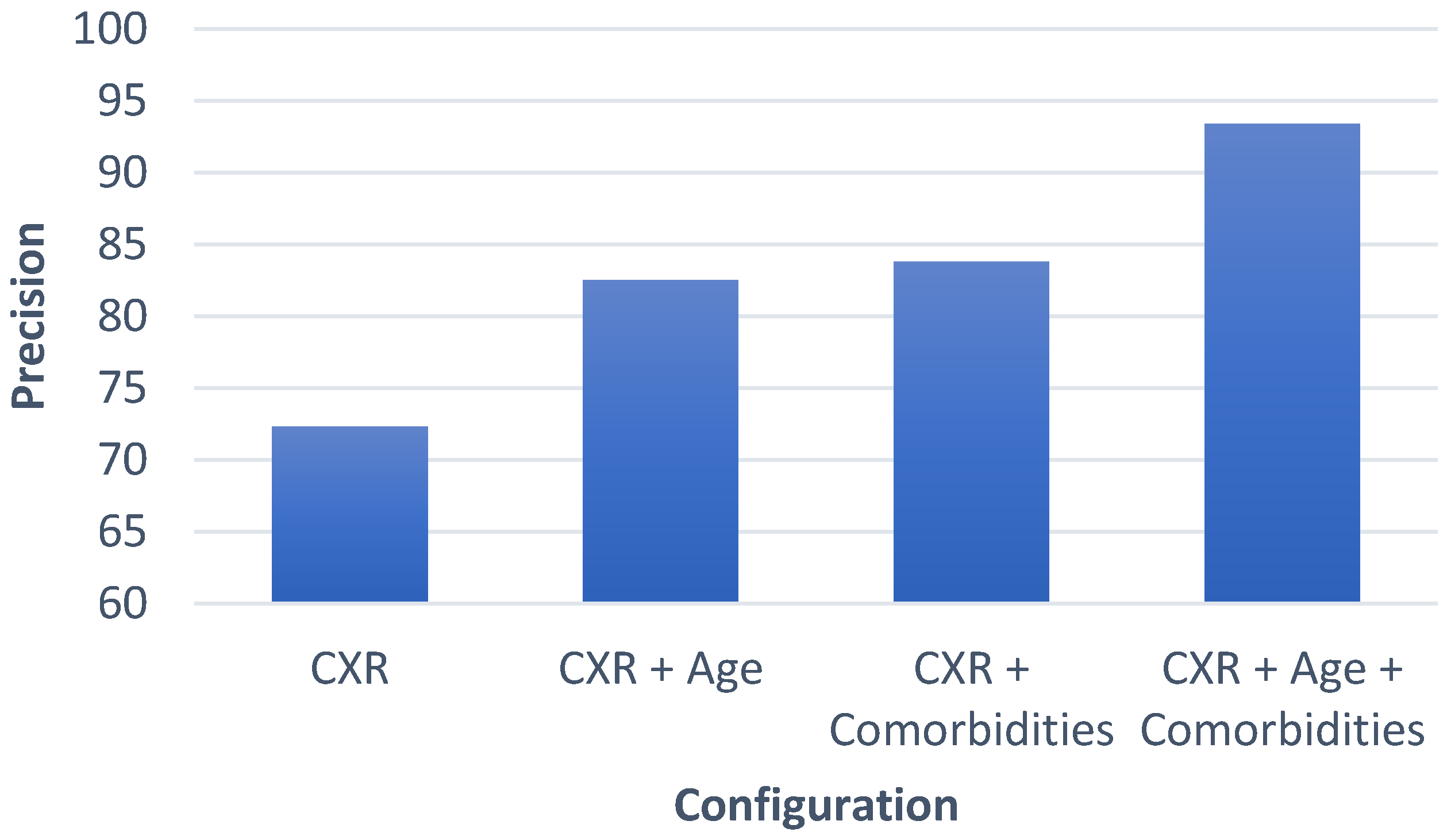
4.3. Progression/Prognosis Prediction
4.4. Comparison with Similar Methods
4.5. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmad, J.; Sajjad, M.; Mehmood, I.; Baik, S.W. SiNC: Saliency-injected neural codes for representation and efficient retrieval of medical radiographs. PLoS ONE 2017, 12, e0181707. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, J.; Muhammad, K.; Lee, M.Y.; Baik, S.W. Endoscopic Image Classification and Retrieval using Clustered Convolutional Features. J. Med. Syst. 2017, 41, 196. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Vasu, B.; Hoogs, A. X-MIR: EXplainable Medical Image Retrieval. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 440–450. [Google Scholar]
- Fang, J.; Fu, H.; Liu, J. Deep triplet hashing network for case-based medical image retrieval. Med. Image Anal. 2021, 69, 101981. [Google Scholar] [CrossRef] [PubMed]
- Ismael, A.M.; Şengür, A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021, 164, 114054. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, J.; Saudagar, A.K.J.; Malik, K.M.; Ahmad, W.; Khan, M.B.; Hasanat, M.H.A.; AlTameem, A.; AlKhathami, M.; Sajjad, M. Disease Progression Detection via Deep Sequence Learning of Successive Radiographic Scans. Int. J. Environ. Res. Public Health 2022, 19, 480. [Google Scholar] [CrossRef] [PubMed]
- Shan, F.; Gao, Y.; Wang, J.; Shi, W.; Shi, N.; Han, M.; Xue, Z.; Shen, D.; Shi, Y. Abnormal lung quantification in chest CT images of COVID-19 patients with deep learning and its application to severity prediction. Med. Phys. 2021, 48, 1633–1645. [Google Scholar] [CrossRef]
- Gong, K.; Wu, D.; Arru, C.D.; Homayounieh, F.; Neumark, N.; Guan, J.; Buch, V.; Kim, K.; Bizzo, B.C.; Ren, H. A multi-center study of COVID-19 patient prognosis using deep learning-based CT image analysis and electronic health records. Eur. J. Radiol. 2021, 139, 109583. [Google Scholar] [CrossRef]
- Zebin, T.; Rezvy, S. COVID-19 detection and disease progression visualization: Deep learning on chest X-rays for classification and coarse localization. Appl. Intell. 2021, 51, 1010–1021. [Google Scholar] [CrossRef]
- Sun, C.; Hong, S.; Song, M.; Li, H.; Wang, Z. Predicting COVID-19 disease progression and patient outcomes based on temporal deep learning. BMC Med. Inform. Decis. Mak. 2021, 21, 45. [Google Scholar] [CrossRef]
- Feng, Z.; Yu, Q.; Yao, S.; Luo, L.; Zhou, W.; Mao, X.; Li, J.; Duan, J.; Yan, Z.; Yang, M. Early prediction of disease progression in COVID-19 pneumonia patients with chest CT and clinical characteristics. Nat. Commun. 2020, 11, 4968. [Google Scholar] [CrossRef]
- Singh, S.; Karimi, S.; Ho-Shon, K.; Hamey, L. Show, tell and summarise: Learning to generate and summarise radiology findings from medical images. Neural Comput. Appl. 2021, 33, 7441–7465. [Google Scholar] [CrossRef]
- Rädsch, T.; Eckhardt, S.; Leiser, F.; Pandl, K.D.; Thiebes, S.; Sunyaev, A. What Your Radiologist Might be Missing: Using Machine Learning to Identify Mislabeled Instances of X-ray Images. In Proceedings of Proceedings of the 54th Hawaii International Conference on System Sciences (HICSS), Honolulu, HI, USA, 5–8 January 2021. [Google Scholar]
- Nave, O.; Shemesh, U.; HarTuv, I. Applying Laplace Adomian decomposition method (LADM) for solving a model of COVID-19. Comput. Methods Biomech. Biomed. Eng. 2021, 24, 1618–1628. [Google Scholar] [CrossRef] [PubMed]
- Maghdid, H.S.; Asaad, A.T.; Ghafoor, K.Z.; Sadiq, A.S.; Mirjalili, S.; Khan, M.K. Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. In Proceedings of the Multimodal Image Exploitation and Learning 2021, Virtual. 12–17 April 2021; p. 117340E. [Google Scholar]
- Bukhari, S.U.K.; Bukhari, S.S.K.; Syed, A.; Shah, S.S.H. The diagnostic evaluation of Convolutional Neural Network (CNN) for the assessment of chest X-ray of patients infected with COVID-19. MedRxiv 2020. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, S.; Siegelman, J.; Alderson, P.O.; Folio, L.S.; Folio, L.R.; Antani, S.K. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access 2020, 8, 115041–115050. [Google Scholar] [CrossRef]
- Sedik, A.; Iliyasu, A.M.; El-Rahiem, A.; Abdel Samea, M.E.; Abdel-Raheem, A.; Hammad, M.; Peng, J.; El-Samie, A.; Fathi, E.; El-Latif, A. Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections. Viruses 2020, 12, 769. [Google Scholar] [CrossRef]
- Luz, E.; Silva, P.; Silva, R.; Silva, L.; Guimarães, J.; Miozzo, G.; Moreira, G.; Menotti, D. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Res. Biomed. Eng. 2022, 38, 149–162. [Google Scholar] [CrossRef]
- Gupta, A.; Gupta, S.; Katarya, R. InstaCovNet-19: A deep learning classification model for the detection of COVID-19 patients using Chest X-ray. Appl. Soft Comput. 2021, 99, 106859. [Google Scholar] [CrossRef]
- Guadiana-Alvarez, J.L.; Hussain, F.; Morales-Menendez, R.; Rojas-Flores, E.; García-Zendejas, A.; Escobar, C.A.; Ramírez-Mendoza, R.A.; Wang, J. Prognosis patients with COVID-19 using deep learning. BMC Med. Inform. Decis. Mak. 2022, 22, 78. [Google Scholar] [CrossRef]
- Signoroni, A.; Savardi, M.; Benini, S.; Adami, N.; Leonardi, R.; Gibellini, P.; Vaccher, F.; Ravanelli, M.; Borghesi, A.; Maroldi, R. End-to-end learning for semiquantitative rating of covid-19 severity on chest x-rays. arXiv 2020, arXiv:2006.04603. [Google Scholar]
- Cohen, J.P.; Dao, L.; Roth, K.; Morrison, P.; Bengio, Y.; Abbasi, A.F.; Shen, B.; Mahsa, H.K.; Ghassemi, M.; Li, H. Predicting covid-19 pneumonia severity on chest x-ray with deep learning. Cureus 2020, 12, e9448. [Google Scholar] [CrossRef] [PubMed]
- Iandola, F.; Moskewicz, M.; Karayev, S.; Girshick, R.; Darrell, T.; Keutzer, K. Densenet: Implementing efficient convnet descriptor pyramids. arXiv 2014, arXiv:1404.1869. [Google Scholar]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the 33rd AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; pp. 590–597. [Google Scholar]
- Fridadar, M.; Amer, R.; Gozes, O.; Nassar, J.; Greenspan, H. COVID-19 in CXR: From detection and severity scoring to patient disease monitoring. IEEE J. Biomed. Health Inform. 2021, 25, 1892–1903. [Google Scholar] [CrossRef] [PubMed]
- Blain, M.; Kassin, M.T.; Varble, N.; Wang, X.; Xu, Z.; Xu, D.; Carrafiello, G.; Vespro, V.; Stellato, E.; Ierardi, A.M. Determination of disease severity in COVID-19 patients using deep learning in chest X-ray images. Diagn. Interv. Radiol. 2021, 27, 20. [Google Scholar] [CrossRef] [PubMed]
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.; Dahly, D.L.; Damen, J.A.; Debray, T.P. Prediction models for diagnosis and prognosis of covid-19: Systematic review and critical appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef]
- Gentilotti, E.; Savoldi, A.; Compri, M.; Górska, A.; De Nardo, P.; Visentin, A.; Be, G.; Razzaboni, E.; Soriolo, N.; Meneghin, D. Assessment of COVID-19 progression on day 5 from symptoms onset. BMC Infect. Dis. 2021, 21, 883. [Google Scholar] [CrossRef]
- Karthik, K.; Kamath, S.S. A deep neural network model for content-based medical image retrieval with multi-view classification. Vis. Comput. 2021, 37, 1837–1850. [Google Scholar] [CrossRef]
- Haq, N.F.; Moradi, M.; Wang, Z.J. A deep community based approach for large scale content based X-ray image retrieval. Med. Image Anal. 2021, 68, 101847. [Google Scholar] [CrossRef]
- Ahmad, J.; Muhammad, K.; Baik, S.W. Medical Image Retrieval with Compact Binary Codes Generated in Frequency Domain Using Highly Reactive Convolutional Features. J. Med. Syst. 2017, 42, 24. [Google Scholar] [CrossRef]
- Choe, J.; Hwang, H.J.; Seo, J.B.; Lee, S.M.; Yun, J.; Kim, M.-J.; Jeong, J.; Lee, Y.; Jin, K.; Park, R. Content-based Image Retrieval by Using Deep Learning for Interstitial Lung Disease Diagnosis with Chest CT. Radiology 2022, 302, 187–197. [Google Scholar] [CrossRef]
- De Falco, I.; De Pietro, G.; Sannino, G. Classification of COVID-19 chest X-ray images by means of an interpretable evolutionary rule-based approach. Neural Comput. Appl. 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Siegel, E.; Shen, D. Deep Learning and Medical Image Analysis for COVID-19 Diagnosis and Prediction. Annu. Rev. Biomed. Eng. 2022, 24, 179–201. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.K.; Choi, J.W.; Jiao, Z.; Wang, D.; Wu, J.; Yi, T.Y.; Halsey, K.C.; Eweje, F.; Tran, T.M.L.; Liu, C. An automated COVID-19 triage pipeline using artificial intelligence based on chest radiographs and clinical data. NPJ Digit. Med. 2022, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Rana, A.; Singh, H.; Mavuduru, R.; Pattanaik, S.; Rana, P.S. Quantifying prognosis severity of COVID-19 patients from deep learning based analysis of CT chest images. Multimed. Tools Appl. 2022, 81, 18129–18153. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Huang, C.; Bao, S.; Fan, T.; Sun, Z.; Wang, Y.; Jiang, H.; Wang, S. Study on the prognosis predictive model of COVID-19 patients based on CT radiomics. Sci. Rep. 2021, 11, 11591. [Google Scholar] [CrossRef]
- Soda, P.; D’Amico, N.C.; Tessadori, J.; Valbusa, G.; Guarrasi, V.; Bortolotto, C.; Akbar, M.U.; Sicilia, R.; Cordelli, E.; Fazzini, D. AIforCOVID: Predicting the clinical outcomes in patients with COVID-19 applying AI to chest-X-rays. An Italian multicentre study. Med. Image Anal. 2021, 74, 102216. [Google Scholar] [CrossRef]
- COVID Chest XRay Dataset. Available online: https://github.com/ieee8023/covid-chestxray-dataset (accessed on 12 July 2022).
- Ieracitano, C.; Mammone, N.; Versaci, M.; Varone, G.; Ali, A.-R.; Armentano, A.; Calabrese, G.; Ferrarelli, A.; Turano, L.; Tebala, C. A fuzzy-enhanced deep learning approach for early detection of COVID-19 pneumonia from portable chest X-ray images. Neurocomputing 2022, 481, 202–215. [Google Scholar] [CrossRef]
- LJPvd, M.; Hinton, G. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 2008, 9, 9. [Google Scholar]
- Ejaz, H.; Alsrhani, A.; Zafar, A.; Javed, H.; Junaid, K.; Abdalla, A.E.; Abosalif, K.O.; Ahmed, Z.; Younas, S. COVID-19 and comorbidities: Deleterious impact on infected patients. J. Infect. Public Health 2020, 13, 1833–1839. [Google Scholar] [CrossRef]
- Adab, P.; Haroon, S.; O’Hara, M.E.; Jordan, R.E. Comorbidities and covid-19. BMJ 2022, 377, o1431. [Google Scholar] [CrossRef]
- Shafer, G. Dempster-shafer theory. Encycl. Artif. Intell. 1992, 1, 330–331. [Google Scholar]
- Jiao, Z.; Choi, J.W.; Halsey, K.; Tran, T.M.L.; Hsieh, B.; Wang, D.; Eweje, F.; Wang, R.; Chang, K.; Wu, J. Prognostication of patients with COVID-19 using artificial intelligence based on chest X-rays and clinical data: A retrospective study. Lancet Digit. Health 2021, 3, e286–e294. [Google Scholar] [CrossRef] [PubMed]
- Schalekamp, S.; Huisman, M.; van Dijk, R.A.; Boomsma, M.F.; Freire Jorge, P.J.; de Boer, W.S.; Herder, G.J.M.; Bonarius, M.; Groot, O.A.; Jong, E. Model-based prediction of critical illness in hospitalized patients with COVID-19. Radiology 2021, 298, E46–E54. [Google Scholar] [CrossRef] [PubMed]



| SNo | Comorbidities | Description |
|---|---|---|
| 1 | Cardiovascular disease | COVID-19 can cause stress on the heart and blood vessels, leading to a higher likelihood of cardiovascular events such as heart attack or stroke. |
| 2 | High Blood Pressure | High blood pressure can also worsen COVID-19 outcomes as it increases the risk of severe illness. |
| 3 | Diabetes | Diabetes can affect the body’s ability to fight off the virus and manage symptoms, while obesity can increase the risk of hospitalization and respiratory failure. |
| 4 | Cancer | Cancer patients, especially those undergoing treatment, have a weakened immune system which can make them more susceptible to severe COVID-19. |
| 5 | Chronic Kidney Disease | Chronic Kidney Disease increases the risk of hospitalization, mechanical ventilation, and death in COVID-19 patients as it affects the body’s ability to clear waste and fluid. |
| 6 | Obesity | Obesity can put additional strain on the respiratory system, making it harder for the body to fight off the virus and manage symptoms. This can increase the risk of respiratory failure and the need for mechanical ventilation. |
| Model Parameters | Description | Model Parameters | Description |
|---|---|---|---|
| T | Labeled image set | τ | Threshold value |
| FS | Feature subspace | VE | Visual similarity |
| X | Input CXR | DE | Normalized distance between X and Y |
| Y | Target CXR to be compared for relevance | CE | Evidence corresponding to comborbidities |
| Fx | Deep features of X | Ag | Evidence for Age |
| NAI | Neuronal activation index | Sx | Probability of survival of patient x |
| Ix | ICU admission | Px | Prognosis (mild vs. severe) |
| Ox | Need for supplemental oxygen |
| Prognosis (Single Scan) | Precision | Recall | F-Measure |
|---|---|---|---|
| Supplemental Oxygen | 0.85 | 0.796 | 0.822 |
| ICU Admission | 0.84 | 0.78 | 0.809 |
| Survival | 0.86 | 0.725 | 0.787 |
| Severity | 0.79 | 0.77 | 0.780 |
| Overall | 0.835 | 0.768 | 0.799 |
| Prognosis (Multiple Scans) | Precision | Recall | F-Measure |
|---|---|---|---|
| Supplemental Oxygen | 0.86 | 0.802 | 0.830 |
| ICU Admission | 0.846 | 0.815 | 0.830 |
| Survival | 0.882 | 0.79 | 0.833 |
| Severity | 0.936 | 0.784 | 0.853 |
| Overall | 0.881 | 0.798 | 0.837 |
| Severity Prediction Methods | Precision | Recall | F-Measure |
|---|---|---|---|
| Jiao et al. [47] (CXR + Clinical) | 0.853 | 0.738 | 0.830 |
| Schalekamp et al. [48] (CXR + Comorbidities) | - | - | 0.826 |
| Gong et al. [8] (CT + Age + Comorbidities) | 0.702 | 0.905 | 0.788 |
| Feng et al. [11] (CT + Clinical) | - | - | 0.820 |
| Proposed Method (Single Scan) | 0.835 | 0.768 | 0.799 |
| Proposed Method (Multiple Scans [2+]) | 0.881 | 0.798 | 0.837 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, J.; Saudagar, A.K.J.; Malik, K.M.; Khan, M.B.; AlTameem, A.; Alkhathami, M.; Hasanat, M.H.A. Prognosis Prediction in COVID-19 Patients through Deep Feature Space Reasoning. Diagnostics 2023, 13, 1387. https://doi.org/10.3390/diagnostics13081387
Ahmad J, Saudagar AKJ, Malik KM, Khan MB, AlTameem A, Alkhathami M, Hasanat MHA. Prognosis Prediction in COVID-19 Patients through Deep Feature Space Reasoning. Diagnostics. 2023; 13(8):1387. https://doi.org/10.3390/diagnostics13081387
Chicago/Turabian StyleAhmad, Jamil, Abdul Khader Jilani Saudagar, Khalid Mahmood Malik, Muhammad Badruddin Khan, Abdullah AlTameem, Mohammed Alkhathami, and Mozaherul Hoque Abul Hasanat. 2023. "Prognosis Prediction in COVID-19 Patients through Deep Feature Space Reasoning" Diagnostics 13, no. 8: 1387. https://doi.org/10.3390/diagnostics13081387
APA StyleAhmad, J., Saudagar, A. K. J., Malik, K. M., Khan, M. B., AlTameem, A., Alkhathami, M., & Hasanat, M. H. A. (2023). Prognosis Prediction in COVID-19 Patients through Deep Feature Space Reasoning. Diagnostics, 13(8), 1387. https://doi.org/10.3390/diagnostics13081387








